Biochemistry - Amino Acids, Peptides, and Proteins
1/67
Earn XP
Description and Tags
chapter 1
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
68 Terms
amino acid molecular structure
- amino group (-NH2)
- carboxyl group (-COOH)
- hydrogen atom
- R group (side chain) [specific to each amino acid = determines amino acid properties & functions]
alpha vs gamma carbon
alpha carbon: amino & carboxyl bonded to same carbon
gamma carbon: amino & carboxyl bonded 3 carbons away from each other
alpha carbon
- chiral (stereogenic) center; 4 different groups attached
- exception: glycine (hydrogen as R-group) is achiral
chiral amino acids
- L-amino acids [amino group on left] = S configuration
- exception: cysteine is D-amino acid [amino group on right] = R configuration
(L-amino acids in eukaryotic proteins)
Nonpolar, nonaromatic side chains (7 amino acids)
glycine
alanine
valine
leucine
isoleucine
methionine
proline
Glycine (Gly = G)
- R-group: single hydrogen atom
- smallest amino acid, achiral
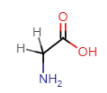
Alanine (Ala = A)
- alkyl R-group: carbon atom
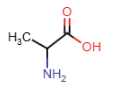
Valine (Val = V)
- alkyl R-group: 3 carbon atoms
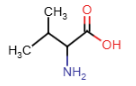
Leucine (Leu = L)
- alkyl R-group: 4 carbon atoms
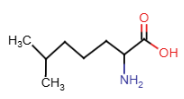
Isoleucine (Ile = I)
- alkyl R-group: 4 carbon atoms
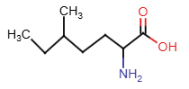
Methionine (Met = M)
- R-group: 3 carbon atoms and sulfur atom
- one of two amino acids with sulfur in R-group
- methyl group attached to sulfer
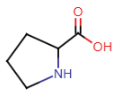
Proline (Pro = P)
- R-group: 3 carbon atoms and amino group
- cyclic amino acid; amino group part of R-group
- 5-membered ring
- constraints on flexibility due to ring
Aromatic side chains (3 amino acids)
tryptophan
phenylalanine
tyrosine
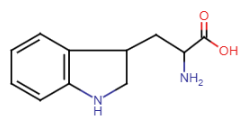
Tryptophan (Trp = W)
- largest of aromatic side chains
- R-group: double ring system with nitrogen atom
Phenylalanine (Phe = F)
- smallest of aromatic side chains
- R-group: benzene ring and CH2 group
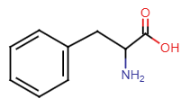
Tyrosine (Tyr = Y)
- added OH to phenylalanine
- R-group: benzene ring, CH2 group, OH group
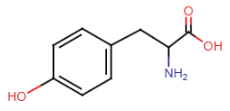
Polar side chains (5 amino acids)
serine
threonine
asparagine
glutamine
cysteine
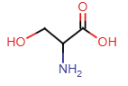
Serine (Ser = S)
- OH group in side chain = highly polar, participate in hydrogen bonding
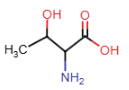
Threonine (Thr = T)
- OH group in side chain = highly polar, participate in hydrogen bonding
Asparagine (Asn = N)
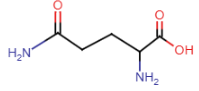
- amide side chain
- amide nitrogens don’t gain'/lose protons w/ changes in pH, don’t become charged
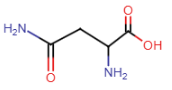
Glutamine (Gln = Q)
- amide side chain
- amide nitrogens don’t gain'/lose protons w/ changes in pH, don’t become charged
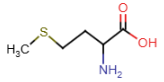
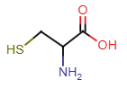
Cysteine (Cys = C)
- contains thiol in side chain
- thiol: SH
- SH bond is weaker than OH, sulfur more electronegative than oxygen = thiol group prone to oxidation
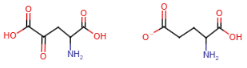
Negatively charged (acidic) side chains (2 amino acids)
aspartic acid (aspartate)
glutamic acid (glutamate)
Aspartic acid (aspartate) (Asp = D)
- COOH group in side chain
- aspartate is the deprotonated form (COO-)
~related to asparagine
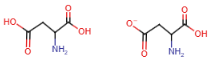
Glutamic acid (glutamate) (Glu = E)
- COOH group in side chain
- glutamate is the deprotonated form (COO-)
~related to glutamine
Positively charged (basic) side chains (3 amino acids)
lysine
arginine
histidine
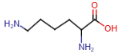
Lysine (Lys = K)
- terminal primary amino group
- side chain with positively charged nitrogen atoms
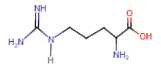
Arginine (Arg = R)
- 3 nitrogen atoms in side chain
- positive charge delocalized over all 3 atoms
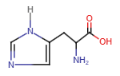
Histidine (His = H)
- aromatic ring (imidazole) with 2 nitrogen atoms
- side chain with positively charged nitrogen atoms
How can histidine aquire positive charge?
- side chain pKa is ~6
= at physiological pH, 1 nitrogen atom is protonated & other isn’t
= under more acidic conditions, 2nd nitrogen is protonated
= side chain positive charge
Hydrophobic amino acids
- amino acids with long alkyl side chains (alanine, isoleucine, leucine, valine, phenylalanine)
- found interior of proteins
Hydrophilic amino acids
- amino acids with charged side chains (pos: histidine, arginine, lysine) (neg: glutamate, aspartate) (glutamine and asparagine amides)
- found surface of proteins
Amino acids neither philic nor phobic
glycine cysteine proline tryptophan tyrosine serine threonine
amphoteric species amino acids
- acidic carboxylic acid group & basic amino group
- can either accept or donate proton depending on environment pH
Ionizable groups
- gain protons under acidic conditions (low pH)
- lose protons under basic conditions (high pH)
pKa & pH
- pKa = pH where half molecules in species are deprotonated
- [protonated ionizable group] = [deprotonated ionizable group]
- [HA] = [A-]
- majority protonated: pH<pKa
- majority deprotonated: pH>pKa
2 pKa values on amino acids
3 pKa values if side chain is ionizable
- pKa1: carboxyl group ~2
- pKa2: amino group ~9-10
positively charged under acidic conditions
- pH 1 < pKa of amino & carboxylic acid groups
- amino group fully protonated = positively charged
- carboxylic acid group fully protonated = neutral
Zwitterions (dipolar ions) at intermediate pH
- electrically neutral molecule; positive and negative charges neutralize one another
- carboxyl group in conjugate base form & deprotonated = COO-
- amino group in conjugate acid form & protonated = NH3+,
negatively charged under basic conditions
- at highly basic pH
- carboxylate group remains deprotonated (COO-)
- amino group deprotonates (NH2)
titration of Glycine, without side chains
- fully protonated glycine with positive charge
- solution titrated with NaOH
- carboxyl group deprotonates first (more acidic than amino group)
- 0.5 equivalents of base added; protonated glycine conc = zwitterion conc [pH = pKa1]
- 1.0 equivalent of base added; carboxylate group fully deprotonated, only zwitterion form exists, pH increases rapidly [pH = isoelectric point (pI) of glycine]
- 1.5 equivalents of base added; zwitterion form conc = fully deprotonated conc [pH = pKa2]
- 2.0 equivalents of base added; amino acid deprotonated
![<p>- fully protonated glycine with positive charge<br>- solution titrated with NaOH<br>- carboxyl group deprotonates first (more acidic than amino group)<br>- 0.5 equivalents of base added; protonated glycine conc = zwitterion conc [pH = pKa1]<br>- 1.0 equivalent of base added; carboxylate group fully deprotonated, only zwitterion form exists, pH increases rapidly [pH = isoelectric point (pI) of glycine]<br>- 1.5 equivalents of base added; zwitterion form conc = fully deprotonated conc [pH = pKa2]<br>- 2.0 equivalents of base added; amino acid deprotonated</p>](https://knowt-user-attachments.s3.amazonaws.com/13b25f52-d0fc-407b-ad84-931d51473cb6.png)
Isoelectric point (pI)
- pH at which molecule is electrically neutral
- amino acids with acidic side chains: low isoelectric points
- amino acids with basic side chains: high isoelectric points
calculation: pI = (pKaamino group + pKacarboxyl group) / 2
titration of Glutamic acid, with charged side chains
Glutamic acid (2 carboxyl groups + 1 amino group):
- fully deprotonated state; +1 charge
- loses proton from main carboxyl group; electrically neutral
- loses second proton from side chain carboxyl group; -1 charge
calculation: pI = (pKaR-group + pKacarboxyl group) / 2
titration of Lysine, with charged side chains
Lysine (2 amino groups + 1 carboxyl group)
- fully protonated state; +2 charge
- loses proton from carboxyl group; +1
- loses proton from main amino group: electrically neutral
- loses proton from side chain amino group; -1
calculation: pI = (pKaamino group + pKaR-group) / 2
- Peptides
- Dipeptides
- Tripeptides
- Oligopeptides
- Polypeptides
- amino acid subunits (residues)
- two amino acid residues
- three amino acid residues
- small peptides: 20 residues
- longer chains
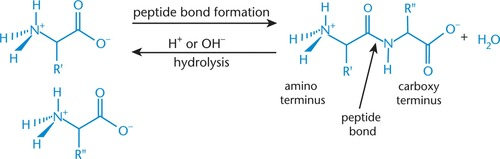
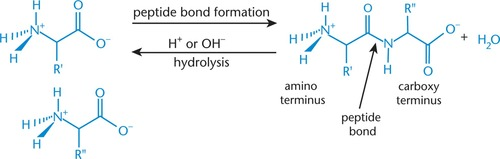
Peptide bond
- joins peptide residues
- specialized form of amide bond between COO- of one amino acid and NH3+ of another amino acid
= functional group C(O)NH-
Peptide bond formation (condensation/dehydration ~acyl sustitution~ reaction)
- nucleophilic amino group (second amino acid) attacks electrophilic carbonyl carbon (first amino acid)
= hydroxyl group of carboxylic acid leaves = peptide (amide) bond
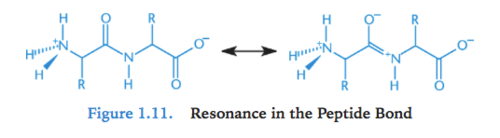
Resonance in peptide bond
- amide group have delocalizable pi electrons in carbonyl & in amino nitrogen lone pair = resonance
- C-N bond in amide has partial double bond character
= protein backbone rotation around C-N bond restricted; makes protein more rigid (sigma bonds are not restricted)
- read from left to right (N-terminus to C-terminus)
N-terminus
- free amino acid end (amino terminus) when peptide bond forms
- drawn on left; read left to right
C-terminus
- free carboxyl end (carboxy terminus) when peptide forms
- drawn on right; read left to right
Peptide/Amide bond hydrolysis (break-down for digestion)
- hydrolytic enzymes (trypsin & chymotrypsin) cleave at certain points in peptide chain
- trypsin: cleaves at carboxyl end of Arg & Lys
- chymotrypsin: cleaves at carboxyl end of Phe, Trp, Tyr
- break amide bond by adding hydrogen atom to amide nitrogen & OH group to carbonyl carbon
Proteins
- polypeptides that range from few to thousands of amino acids
- function as enzymes, hormones, memprane pores/receptors, & elements of cell structure
- made via genetic code
four levels of structure: primary, secondary, tertiary, quaternary
Primary structure
- linear arrangement of amino acids
- stabilized for formation of covalent peptide bonds between adjacent amino acids
- sequencing of DNA that coded protein or from protein itself = structure
Secondary structure
- local structure of neighboring amino acids
- results from hydrogen bonding between nearby amino acids
- alpha-helices & beta-pleated sheets
alpha-helices
- rodlike structure of peptide coiling clockwise
- stabilized by intramolecular hydrogen bonds between carbonyl oxygen and amide hydrogen (4 residues down the chain)
- side chains point away from core
- important component in keratin (skin, hair, fingernails)
beta-pleated sheets
- parallel or antiparallel; form rows/strands = pleated/rippled shape
- stabilized by intramolecular hydrogen bonds between carbonyl oxygen on one chain and amide hydrogen on another chain
- side chains point above/below sheet plane
- fibroin is primary component of silk fibers
Proline (& secondary structure)
- rigid, cyclic structure (=kink in peptide chain)
Rarely found in:
- middle of alpha-helix or beta-pleated sheets
Often found in:
- turns between chains of beta-pleated sheet or as residue at start of alpha-helix
- alpha helices that cross cell membrane
Tertiary structure
- protein’s 3D shape
- determined by hydrophobic/philic interactions between amino acids’ R groups, hydrogen bonding, and acid-base interactions between amino acids with charged R-groups (= salt bridges)
- phobic: interior of proteins
- philic: N-H & C=O bonds get pulled in by phobic and form electrostatic interactions & hydrogen bonds = stabilize protein
- disulfide bonds [2 cysteine molecules oxidize (lose 2 protons & electrons) = cystine]
= create loops in protein chain & determine wavy/curliness of hair (more bonds = more curly)
![<p>- protein’s 3D shape<br>- determined by hydrophobic/philic interactions between amino acids’ R groups, hydrogen bonding, and acid-base interactions between amino acids with charged R-groups (= salt bridges)<br>- phobic: interior of proteins<br>- philic: N-H & C=O bonds get pulled in by phobic and form electrostatic interactions & hydrogen bonds = stabilize protein<br><br>- disulfide bonds [2 cysteine molecules oxidize (lose 2 protons & electrons) = cystine]<br>= create loops in protein chain & determine wavy/curliness of hair (more bonds = more curly)</p>](https://knowt-user-attachments.s3.amazonaws.com/f26b5610-6a0a-4b76-892c-b8e4222434ff.png)
Molten globules
intermediate states in tertiary formation (protein forms in generally less than a second)
denaturation
process of protein losing tertiary structure (& function)
Folding & Solvation layer
- entropy: phobic residues occupy inside, philic residues outside
- solvation layer: forms from neary solvent molecules when solute dissolves in solvent
- enthalpy standpoint: hydrocarbons more favorable in aqueous solution than in organic solvents
- when hydrophobic side chain in aqueous solution:
— water molecules in solvation layer can’t form hydrogen bonds with side chain
— water molecules rearrange to maximize hydrogen bonding
== negative change in entropy (increased order) [unfavorable, nonspontaneous]
- if hydrophobic residues on exterior of protein
— nearby water molecules have more freedom in positioning
— increased entropy in system
— makes solvation system spontaneous
- hydrophobic residues away from water, phydrophilic residues toward water = protein max stability
Quaternary structure
- aggregate of smaller globular peptides (subunits); represents functional form of protein
- only exists for proteins containing more than 1 polypeptide chain (not all proteins have quaternary structure)
- examples: hemoglobin & immunoglobins
—hemoglobin: 4 distinct subunits, each binds to one oxygen molecule
—immunoglobin: 4 subunits
Benefits of quaternary structure
- more stable (reduce area of protein complex)
- reduce DNA amount needed to encode protein complex
- bring catalytic sites closer (one reaction’s intermediates shuttled to another reaction)
- induce cooperativity/allosteric effects (one subunit can undergo conformational/structural changes = enhance or reduce activity of other subunits)
Conjugated proteins
- proteins with covalently attached (prosthetic) molecules
Prosthetic molecules
- organic molecules (vitamins) or metal ions (iron) that determine function of respective proteins & direct protein delivery to certain location (cell membrane, nucleus, lysosome, endoplasmic reticulum)
- proteins with lipis, carbs, and nucleic acid prosthetic groups = lipoprotiens, glycoproteins, and nucleoproteins
Denaturation
- protein unfolding (losing 3D structure)
- irreversible (sometimes reversible)
- unfolded proteins can’t catalyze reactions
- caused heat and solutes
Temperature-based denaturation
- temp increase = kinetic energy increase
- if too high, hydrophobic interactions are overcome and protein unfolds (example: cooking egg whites)
Solute-based denaturation
- urea (solute) denatures proteins by interfering with forces holding protien together
- break disulfide bridges = disrupt teriary & quaternary structures
- reduce cystine to 2 cysteine residues
- overcome hydrogen bonds on other side chain interactions holding alpha-helices and beta-pleated sheets intact
- (SDS / sodium dodecyl sulfate / sodium lauryl sulfate ~detergent) solubilize proteins = disrupt noncovalent bonds & promote denaturation