L17: Cytoplasmic control of gene expression (mRNA transport, turnover + rate of translation)
1/17
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
18 Terms
What is involved in the transport of mRNA?
Processed mRNA is packaged and transported out of the nucleus with only fully processed mRNA, capped, intron-free, polyadenylated, is transported for translation, regulated by:
Identification of “export ready” mRNA
Transport of mRNA to cytoplasm is highly selective
Role of nuclear pore complex (~30 different pore proteins)
What is meant by “export-ready” mRNA?
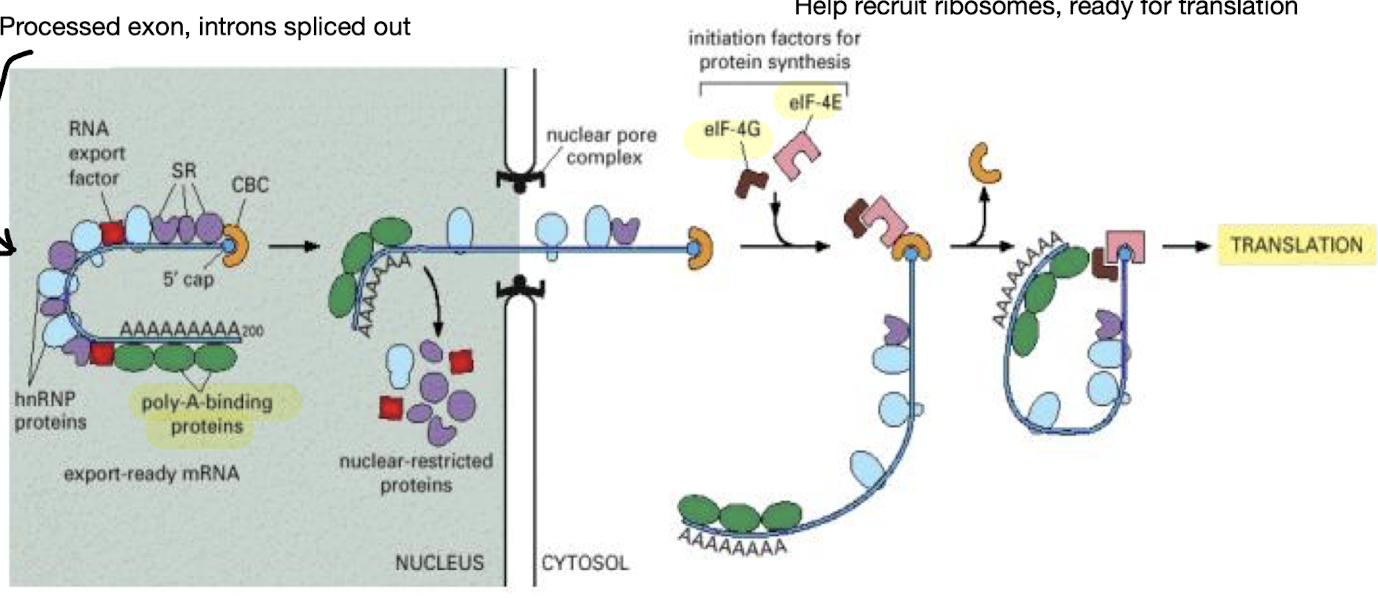
Mature mRNA with cap and polyA carrying residual SR proteins
Export is active transport (GTP hydrolysis) through nuclear pore complex
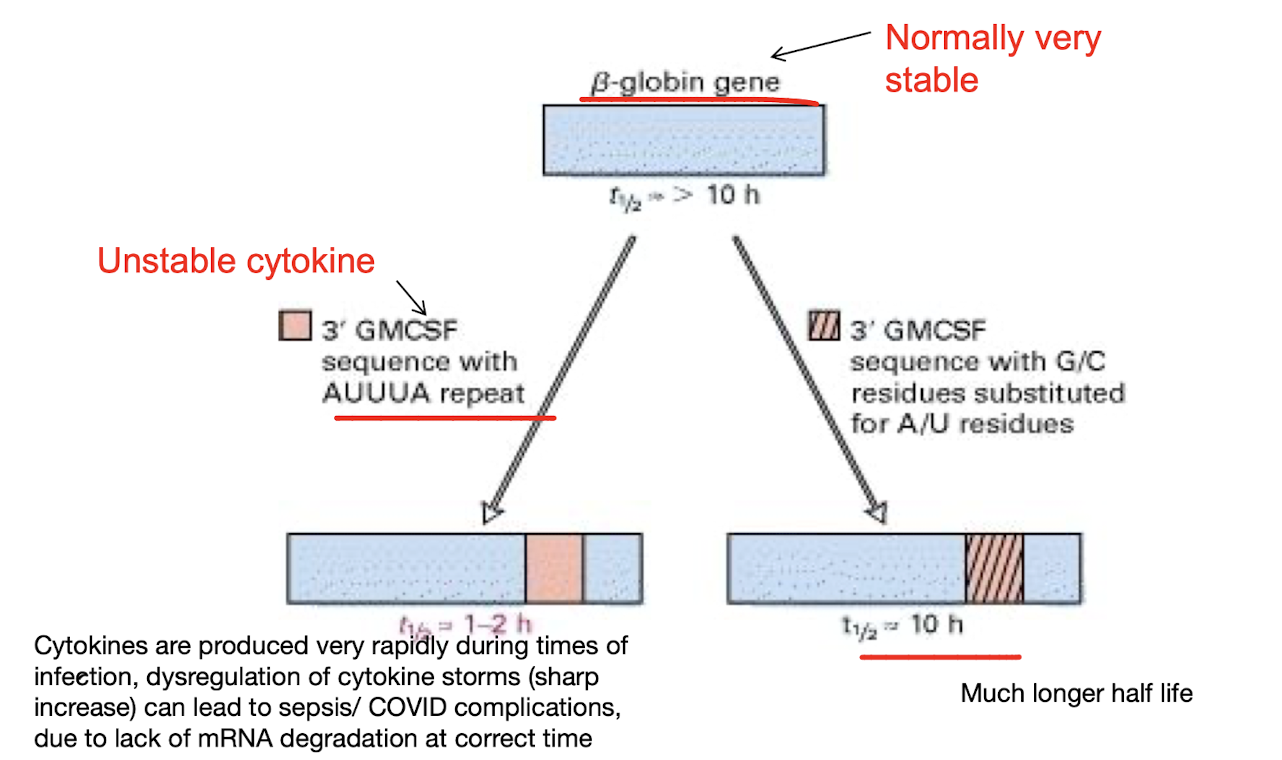
What 2 things control mRNA stability which regulates gene expression?
- Specific sequences in the mRNA that may affect stability e.g. a sequence of AUUUA repeats at 3’ end reduces the mRNA stability
- Rate of mRNA degradation i.e. mRNAs with short or no poly(A) tails are rapidly degraded by exonucleases (digests at either end i.e. chop off A at polyA tail) or by endonucleases (digest within certain site)
What effects the amount of mRNA?
Rate of synthesis
Rate of degradation e.g. mRNAs encoding cyclins for the cell cycle are extremetly stable and rise for 40 minutes until no longer needed where they are then rapidly degraded by specific pathways.
What is the destabilising effect of AUUUA?
It promotes mRNA degradation by targeting it for rapid turnover, significantly reducing its stability
They are Adenine-Uracil rich elements (ARE)
ARE-binding protein (ABP) helps recruit mRNA degradation machinery
Why is mRNA degradation important?
1. Regulates gene expression by rapidly changing the amount of an mRNA available for translation
2. Functions in quality control to ensure mRNA pool is “translatable”
Rate of mRNA degradation controlled by 2 factors
1 . Presence of CAP and polyA tail:
mRNAs with short or no poly(A) tails are rapidly degraded
- e.g. HISTONES - NO POLY (A) tail VERY SHORT T½
2. Degradation rate of mRNAs changes in response to extracellular signals e.g. transferrin receptor (TfR) degradation in presence of
different [iron] – controls Iron metabolism
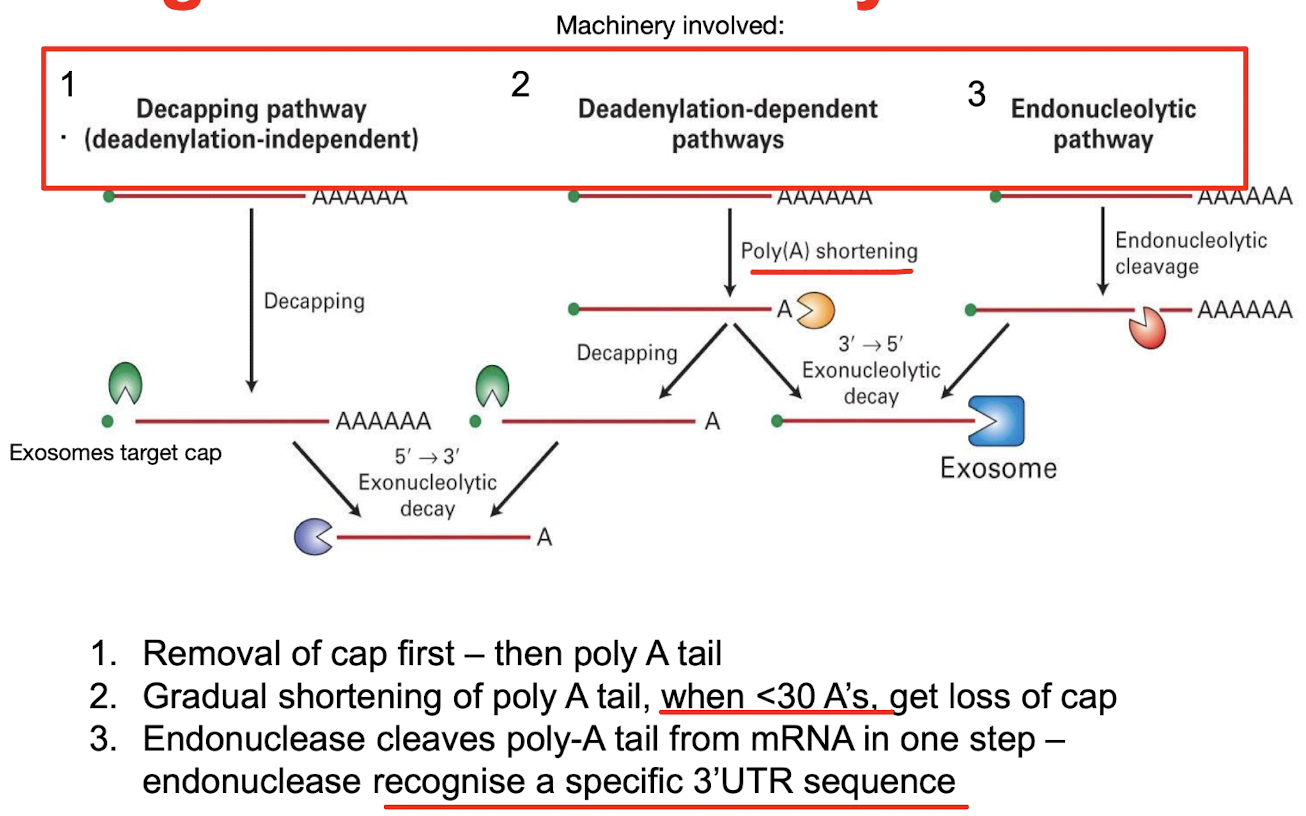
What machinery is involved in the degradation of eukaryotic mRNA?
Decapping pathway - removal of cap then poly A tail by exonucleases
Deadenylation-dependent pathway - gradual poly A shortening until <30 mRNAs
Endonucleatic pathway - cleaves the poly A tail in one step by recognition of a specific 3’ UTR sequence
How is Iron taken up into the eukaryotic cell?
Very insoluble, circulated Fe requires Transferrin (HOLO-TF)
Gets taken up into the cell by the TfR (transferrin receptor) which controls its availability
Stored in cell as the intracellular protein Ferritin (out of action form which binds Fe ions, preventing accumulation) , ferritin mRNA translation controls its availability
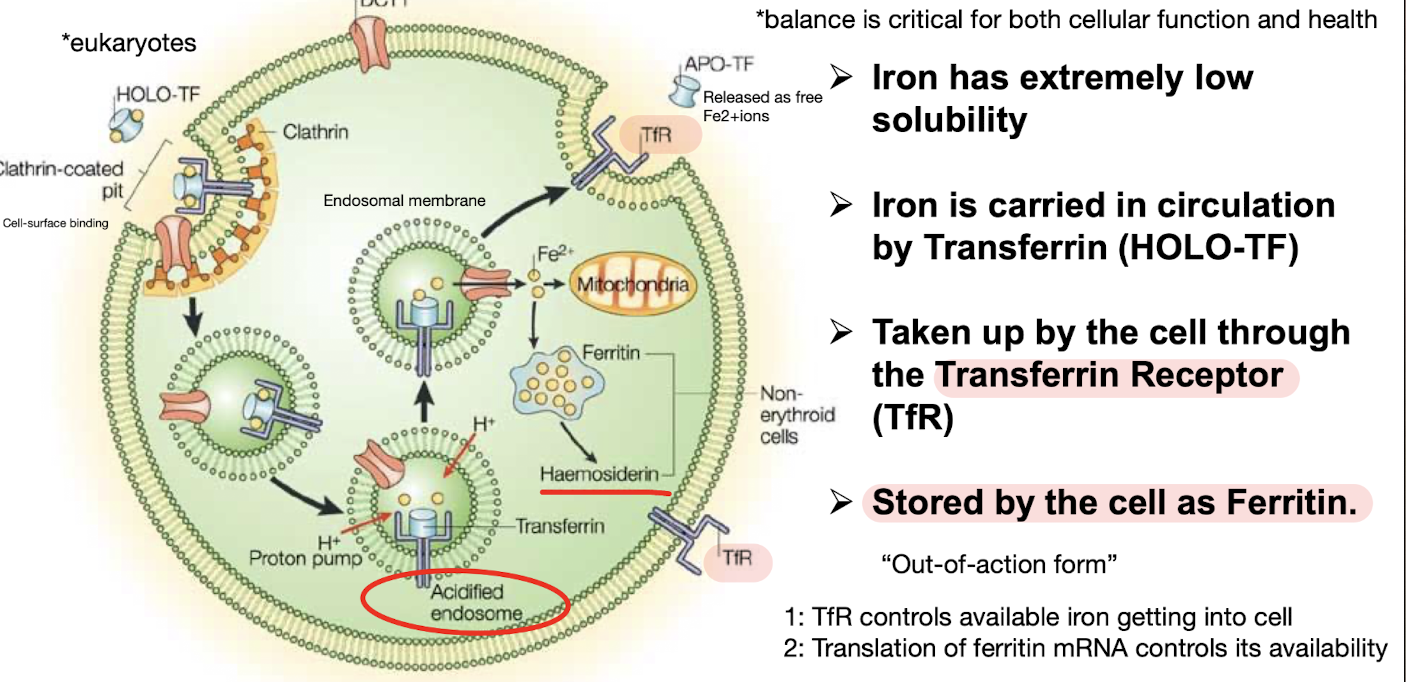
Why have cells evolved tightly regulated gene expression mechanisms for iron homeostasis?
Iron is required for essential cellular mechanisms e.g. oxygen transport (as heme in Hb), DNA biosynthesis, ATP generation.
Dysregulated iron metabolism contributes to iron overload diseases and cancer
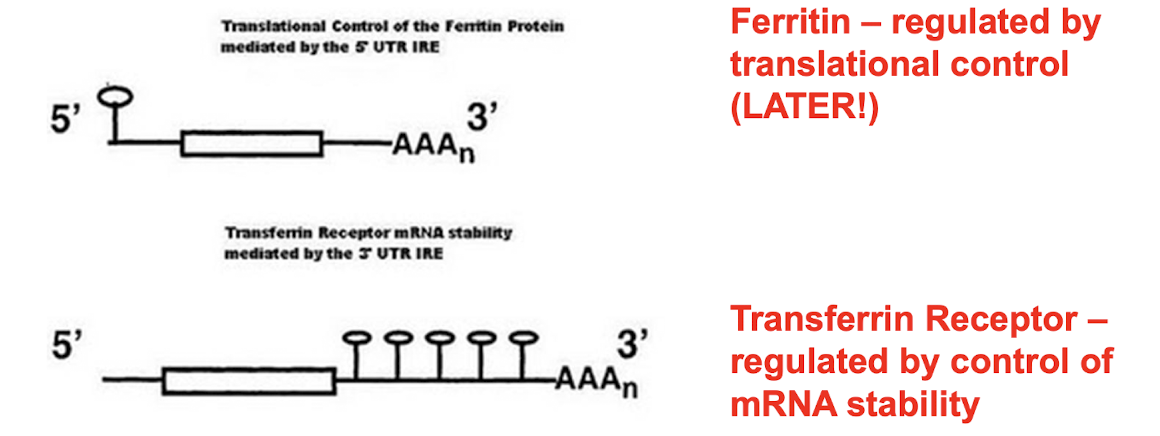
What does Iron homeostasis require?
Tight regulation of Ferritin and TfR using IRE’s (Iron response elements)
Ferritin - regulated by translational control
TfR - regulated by control of mRNA stability
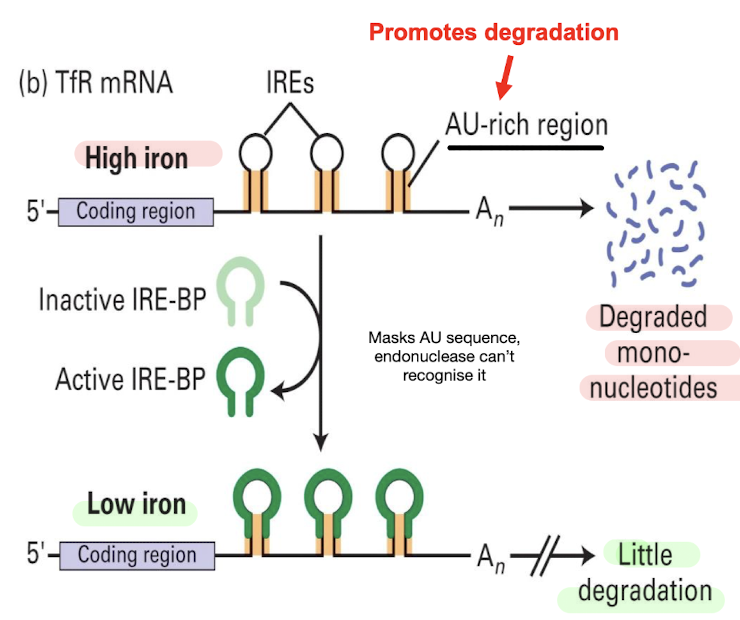
How do iron levels control stability of TfR mRNA?
High Fe levels decrease the stability of TfR-encoding mRNA due to endonuclease binding to the AU region of IRE → decrease Fe intake into cell
Low Fe levels involves mRNA 3’ UTR IREs and IRE-binding protein (IRE-BP):
Confirmational change activates IRE-BP which binds to IRE blocking recognition of AU region by endonucleases, preventing degradation → increased TfR levels + Fe uptake
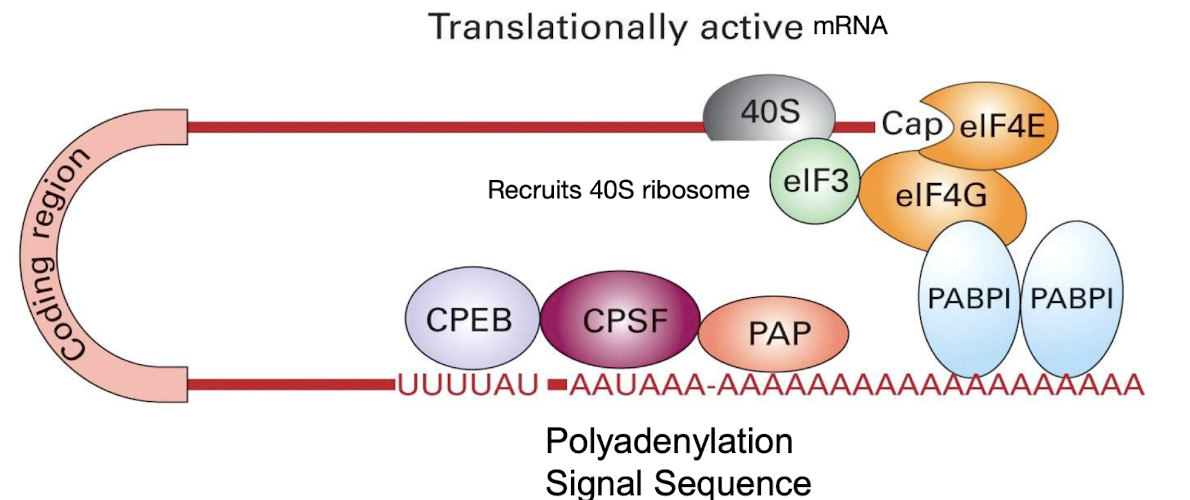
How is translation regulated?
•Ribosomes must attach to mRNA to initiate translation
•Attach by recognising CAP – role of elFs
•Role of Poly A tail – circularises mRNA
•Repressed by Protein Binding to 3’-UTR e.g. Maskin
•Repressed by Protein Binding the 5’-UTR e.g. aconitase binding to IRE
What is involved in translation initiation? - review
Requires a 5’ CAP, polyA tail and a 40S Ribosome
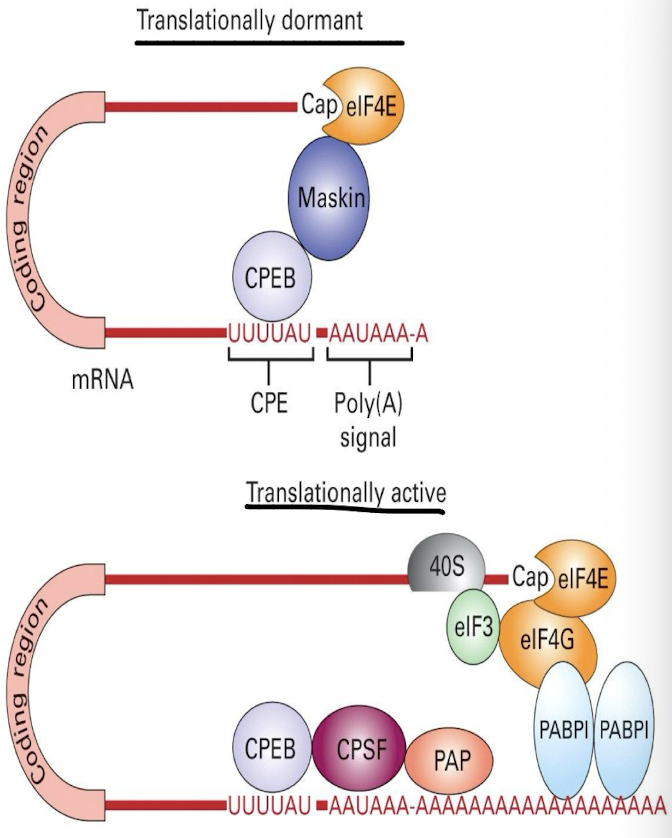
After PolyA extension by PAP, multiple copies of Poly A Binding Proteins (PABPs) bound to the PolyA tail also interact with eIF4G
eIF4G functions with other eIFs to bind to the 5’ CAP & the 40S ribosome subunit to aid in initiation of translation
What is an example of protein binding to 3’ UTR that leads to repression of translation?
Binding of CPEB (cytoplasmic Polyadenylation Element binding protein) to Maskin
Maskin binds to eIF4E at 5’ cap preventing eIF4G from binding → blocking transcription + translation
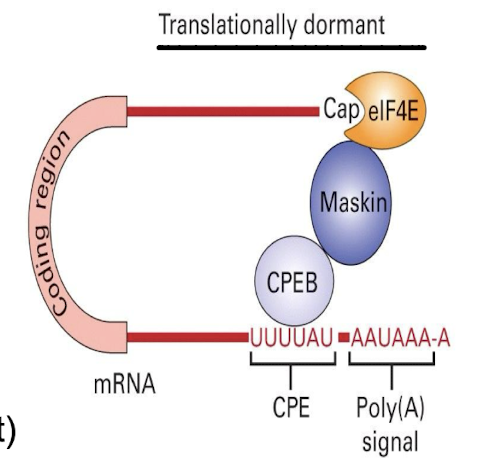
How is translational inhibition reversed?
By phosphorylation of CPEB → binds to CPSF (allows for recruition of polyadenylation PABP1) which promotes the release of Maskin, by allowing eIF4G to bind to eIF4E, facilitating translation initiation.
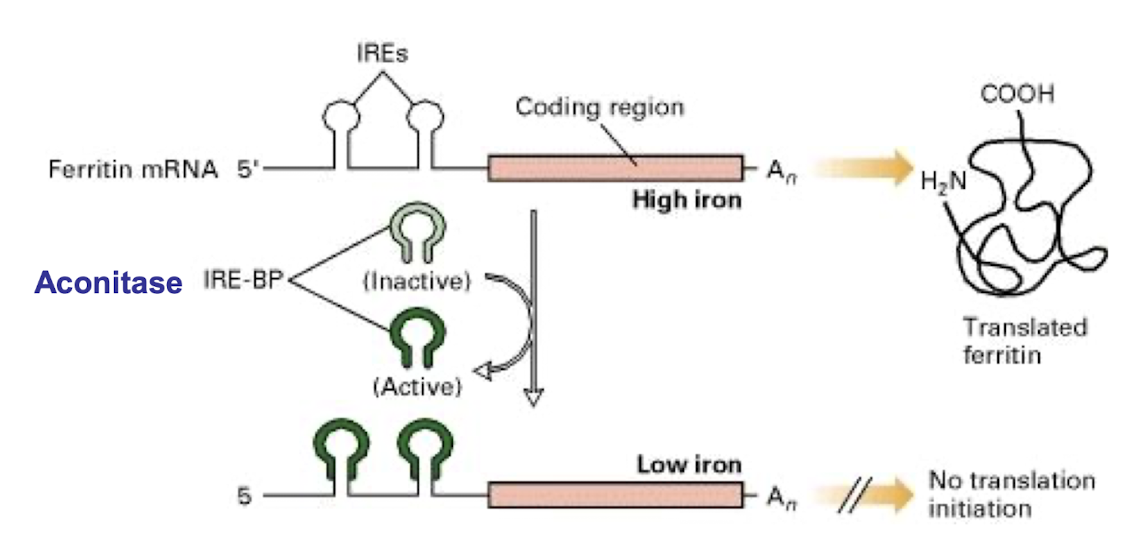
How is the regulation of Ferritin translation carried out?
Regulated by mRNA 5’ UTR IRE-BP in response to iron levels:
Low intracellular Fe: Repression of ferritin mRNA translation by aconitase RP, free Fe available as cofactor for Fe-requiring enzymes
Excess intracellular Fe: Derepression of ferritin mRNA translation, free Fe bound by newly-made ferritin for intracellular storage, preventing excess Fe and toxicity
What is the role of aconitase for controlling Ferritin?
At Low Iron concentration –
Ferritin REPRESSION
➢ Repressor protein (aconitase) binds IRE
➢ Prevents the 40S subunit from binding.
At high Iron concentrations -
Ferritin DE-REPRESSION
➢ Aconitase, binds Fe-S cluster.
➢conformational change prevents its binding to IRE
➢ loop unfolds and translation takes place