catabolic core of metabolism
1/26
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
27 Terms
uniporter
glucose transporter family membrers glut 1-14
GLUT 1
ubiquitous expression
basal glucose uptake
GLUT 2
liver ,pancreas, brain, kidney, small intestine
high capacity, low affinity glucose transporter
glucose sensing role in islets
glut 3
brain, placenta
basal glucose uptake
glut 4
muscle , fat , heart, hippocampal neurons
insulin responsive glucose transpoter
symporters
sodium glucose co transporters
SLGT1
small intestine
absorption of glucose from small intestine
SLGT 2
kidney
glucose resorption from glomerular ultrafiltration
glycolysis
glucose is phosphorylated upon entry into the cell by hexokinase to G6P and this stops glucose diffusing back across the transporter
G6P is isomerised to F-6-P and phosphorylated to form F-1,6-BP
this invests 2 molecules of ATP
one 6c molecule is lysed into 2 3C
the coenzyme NAD+ is reduced to NADH
4 molecules of ATP produced- net 2
two mplecule sof pyruvate produced
glucose uptake
facilitated diffusion across non insulin dependent glucose transporters
insulin responsive tissues express GLUT 4 -hormonal control of glucose uptake
hexokinase
allosterically inhibited by glucose 6 phosphate
if pathway is slow and G6P conc rise, not wasting atp to phosphorylate glucose cannot be used yet
different tissue specific hexokianse have diff affinities for glucose enabling glycolysis to occur prefentially in extrahepatic tissue
phosphofructokinase
what does it do and what is it inhibited and activated by
catalyses first step of glycolysis- the rate limiting step
allosterically inhibited by atp, citrate and protons
activated by AMP
pyruvate kinase
alloserically activated by F-1,6BP
allosterically inhibied by ATP and alanine
intermediates used by anabolic pathways to generate…
nucelotides
lipids
amino acids
other organic molecules
pyruvate dehydrogenase complex
enzyme linking glycolysis to krebs cycle
inhibited by high conc of atp, acetyl coA and NADH
citrate synthase
atp is an allosteric negative regulator
regulation important as oxaloacetate is utilised in gluconeogenesis in the liver
isocitrate dehydrogenase
ADP is a positive allosteric regulator
ATP and NADH are negative allosteric regulators
converts isocitrate to alpha ketoglutarate
alpha ketoglurate dehydrogenase complex- what is it inhibited by
inhibited by high conc of its own products, NADH and succinyl coA and additional inhibtion by high atp levels
ETC creating a proton gradient
COMPLEX 1- NADH from the krebs cycle donates 2 electrons to complex i.
4 hydrogen ions are pumped out matrix into intermembrane space
COMPLEX II- FADH2 from the Krebs cycle is used in the conversion of succinate to fumarate by succinate dehydrogenase.
no direct pump of hydrogen ions
COMPLEX III- coenzyme q transfers electrons to complex iii. complex iii shuttles two electrons from coenzyme q onto cytochrome c.
4 hydrogen ions are pumped out of the matric into the intermembrane space
COMPLEX IV- accepts electrons from cytochrome c. (haem group in cytochrome c carries out electron transport)
electrons then shuttled through complex iv and onto oxygen producing water (oxygen = final electron acceptor
COMPLEX V ATP SYNTHASE- i-iv used to establish proton gradient.
proton motive force drives ATP synthase
catalyses addition of phosphate groups onto ADP to ATP
ATP synthase structure
multi protein complex embedded in the inner mitochondiral membrane that extends into the mitochondrial matrix
central core of the complex turns due to rotational forces provided by the movement of hydrogen ions through channels in the membrane embedded portion
beta subunits contain active site and catalyse ADP—→ ATP
there are 3 beta subunits and so each full rotation of the central core results in synthesis of 3 ATP
the krebs cycle products for every turn of the cycle
2xCO2
3x NADH
1x FADH2
1x GTP
what are intermediates used by anabolic pathways to generate
nucleotides
lipids
amino acids
other organic molecules
what is the actual ATP yield
30
NADH and other substrates from cytosol need to enter mitochondria- shuttle mechanisms result in a loss of mitchondria (-2ATP)
Protons utilised in the transport of molecules across the inner membrane
proton leak (uncontrolled backflow of protons through the inner mitochondrial membrane bypassing ATP synthase) can occur so yields about 2.5ATP per NADH and 1.5 ATP per FADH2
proton gradient drives a pump not a fixed reaction (slippage- complex transport fewer protons than what is thermodynamically expected)
so lower than calc
brown apidose tissue
contain a large number of mitochondria
enables endogenous heat production to maintain body temp independent of env temps
important in newborns who cannot yet produce heat through shivering
highly vascularised
hibernators have a large amount of BAT due to need to rewarm from hypothermic torpor through hibernation
brown vs beige vs white
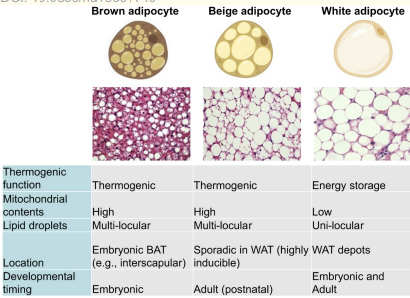
uncoupling the electron transport chain from ATP production
allow protons to leak back into the mitochondrial matrix without passing through ATP synthase by creating alternative pathways
this dissipates the proton gradient that normally powers ATP synthase releasing heat energy instead
eg thermogenin (UCP1) is found in brown apidose tissue and is activated by norepinephrin and free fatty acids