Lecture 1: Structure, Synthesis & Functions of Nucleic Acids
1/46
Earn XP
Description and Tags
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
47 Terms
Functions of nucleotides (7)
Energy currency in metabolic transactions
Chemical link response of cells to hormones and other extracellular stimuli
Structural components of enzyme cofactors and metabolic intermediates
Constituents of nucleic acids
Some coenzymes contain adenine
Function as second messenger in the cells + work as regulatory molecules (cAMP, cGMP)
Can function as neurotransmitters and ligands
Give examples of nucleotides functioning as neurotransmitters and ligands
ATP binds to P2x receptors in post-synapsis (taste, inflammation, smooth muscle contraction)
ADP binds to P2γ receptors in platelets, promoting clotting (clopidogrel interferes)
Which bonds are hydrolysed to release energy from nucleotides, and how much energy is released?
Ester (α-P) = 14 kJ/mol
Anhydride (β-P, γ-P) = 30.5 kJ/mol
Nucleotide components
nitrogenous base, pentose (sugar with 5 C atoms), phosphate group
A nucleotide without a phosphate group is called…?
A nucleoside
Phosphate group structure
sp3, regular tetrahedron
Nitrogenous bases are derivates of…?
Pyrimidine + purine (heterocyclic rings)
Properties of pyrimidine and purine
aromatic (planar, hydrophobic), basic
Conformation of ribose in nucleic acids
Only β-D-furanose ring form (no equilibrium with aldehydic form)
How many different ribose conformations?
4 different puckered conformations.
4 out of 5 atoms are nearly in a single plane. C-2’ or C-3’ is either on the same (endo) or opposite (exo) side of the plane relative to the C-5’ atom.
The bond between the nitrogenous base and the pentose is called…?
N-glycosidic
The phosphate group and nucleoside are linked by ______________. Therefore, the nucleotide contains a ____________ bond.
An ester bond, phosphodiester
How are successive nucleotides linked in nucleic acids?
Through the formation of phosphodiester bonds between C-3’ and C-5’ of the two nucleotides
Phosphodiester linkages are the ________ of DNA and RNA
covalent backbone
Levels of nucleic acid structure
Primary: The covalent structure and nucleotide sequence
Secondary: Any regular, stable structure taken up by some or all of the nucleotides (e.g. DNA)
Tertiary: The complex folding of large chromosomes or the elaborate folding of large tRNA or rRNA
Chargaff’s rules (4)
For DNA base composition:
[A] = [T] and [G] = [C], therefore [A] + [G] = [T] + [C]
Base composition varies from one species to another
Same composition from different tissues of the same species
No variations with age, nutritional state or changing environment
Watson & Crick model (1953)
Hydrophilic groups exposed to H2O, inner hydrophobic groups and H bonds (fulfils the thermodynamic requirements)
Base complementarity (fulfils [A] + [G] = [T] + [C])
Atoms of complementary nitrogenous bases lie on the same plane (almost perpendicular to the helix axis)
Antiparallel strands
Vertically stacked bases inside the double helix, 3.4 Å apart
Secondary repeat distance of ~ 34 Å (10 bases per helical turn)
DNA structure in aqueous solution vs fibres
Secondary repeat distance altered in aqueous: 10.5 base pairs (36 Å or 3.6 nm) per helical turn
“Space filling” model
Atoms are at the centre of spheres with van der Waals radius
RC = 1.7 Å, RN =1.5 Å
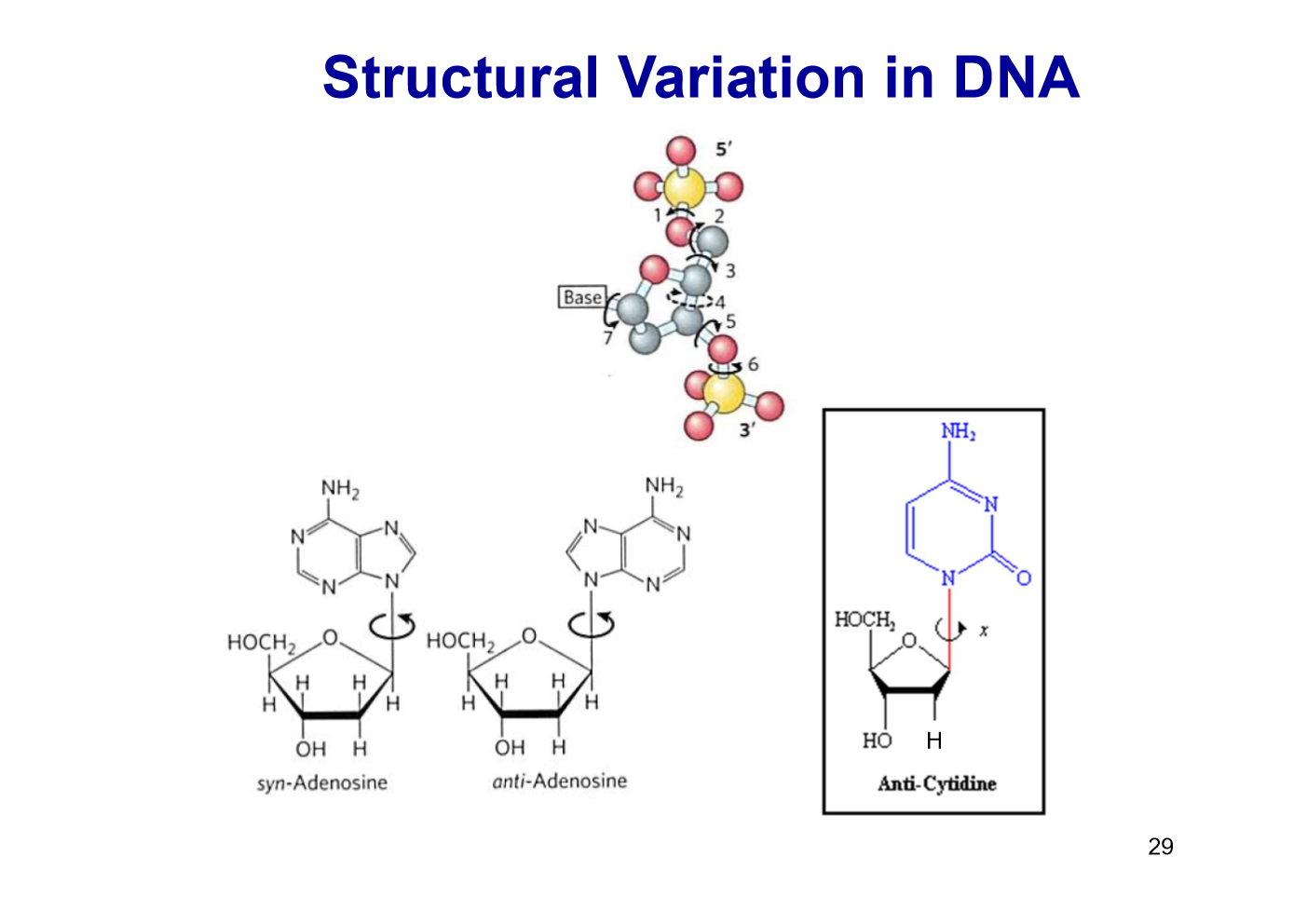
DNA structural variation
Free rotation about C-1’-N-glycosidic bond.
Purines: syn, anti
Pyrimidines: anti (steric interference between sugar and C=O)
Compare the different DNA forms:
A-form | B-form | Z-form | |
|---|---|---|---|
Right-/left-handed? | |||
Helical rise per base pair | |||
Base pairs per turn | |||
Width/diameter | |||
Base tilt normal to helix axis |
A-form | B-form | Z-form | |
|---|---|---|---|
Right-/left-handed? | Right-handed | Right-handed | Left-handed |
Helical rise per base pair | 2.6 Å | 3.4 Å | 3.7 Å |
Base pairs per turn | 11 | 10.5 | 12 |
Width/diameter | 26 Å | 20 Å | 18 Å |
Base tilt normal to helix axis (not perpendicular to helix axis, more tilt) | 20° | 6° | 7° |
Describe Z-form DNA
Structure appears more slender and elongated
Backbone takes on a zigzag appearance
Major groove is barely apparent and minor groove is narrow and deep
May play a role (unknown) in gene expression regulation and in genetic recombination
Conditions for A and B-form DNA
A: solutions devoid of water, DNA crystallisation
B: most stable form under physiological conditions (aqueous solution)
Certain nucleotide sequences fold into __-form DNA more readily than others. Give examples.
Z
Pyrimidines alternating with purines
Purines flip to syn alternating with pyrimidines in anti conformation
C-G alternating with G-C
5-methyl-C and G residues
What kind of nucleotides for hairpins and cruciforms?
hairpin: ssDNA, ssRNA
cruciform: dsDNA
Hoogsteen pairing
Triple helix + quadruplex
Stabilisation occurs between one purine and two pyrimidine bases (in triple helix)
Hoogsteen pairing may originate from…?
virus
if bases are methylated (epigenetic effect)
fragment generated during DNA replication
free nucleotides in nucleus (excision)
DNA denaturation
Reversible disruption of hydrogen bonds between paired bases and of base-stacking interactions causing unwinding of the double helix to form two single strands, completely separated from each other along the entire length or part of the length (partial denaturation) of the molecule
Are covalent bonds broken during denaturation?
No (hydrogen bonds aren’t covalent)
Factors promoting DNA denaturation
high temperature, pH
Melting temperature (tm)
Temperature at which half of the DNA is present as separate single strands
Properties of tm
Each species of DNA has a characteristic tm
Increases with %GC content
dsDNA < DNA:RNA hybrid < dsRNA
(More H-bonds in RNA, due to 2’-OH)
Effect of denaturation of UV light absorption
Hypochromic effect: Close interaction between stacked bases decreases UV light absorption compared to a solution with the same concentration of free nucleotides. Absorption is further decreased when 2 DNA strands are paired.
Hyperchromic effect: Increased UV absorption when a double stranded nucleic acid is denatured.
The transition from dsDNA to denatured ssDNA can be detected by monitoring UV absorption at 260 nm.
Appearance of partially denatured DNA in electron microscopy
Bubbles
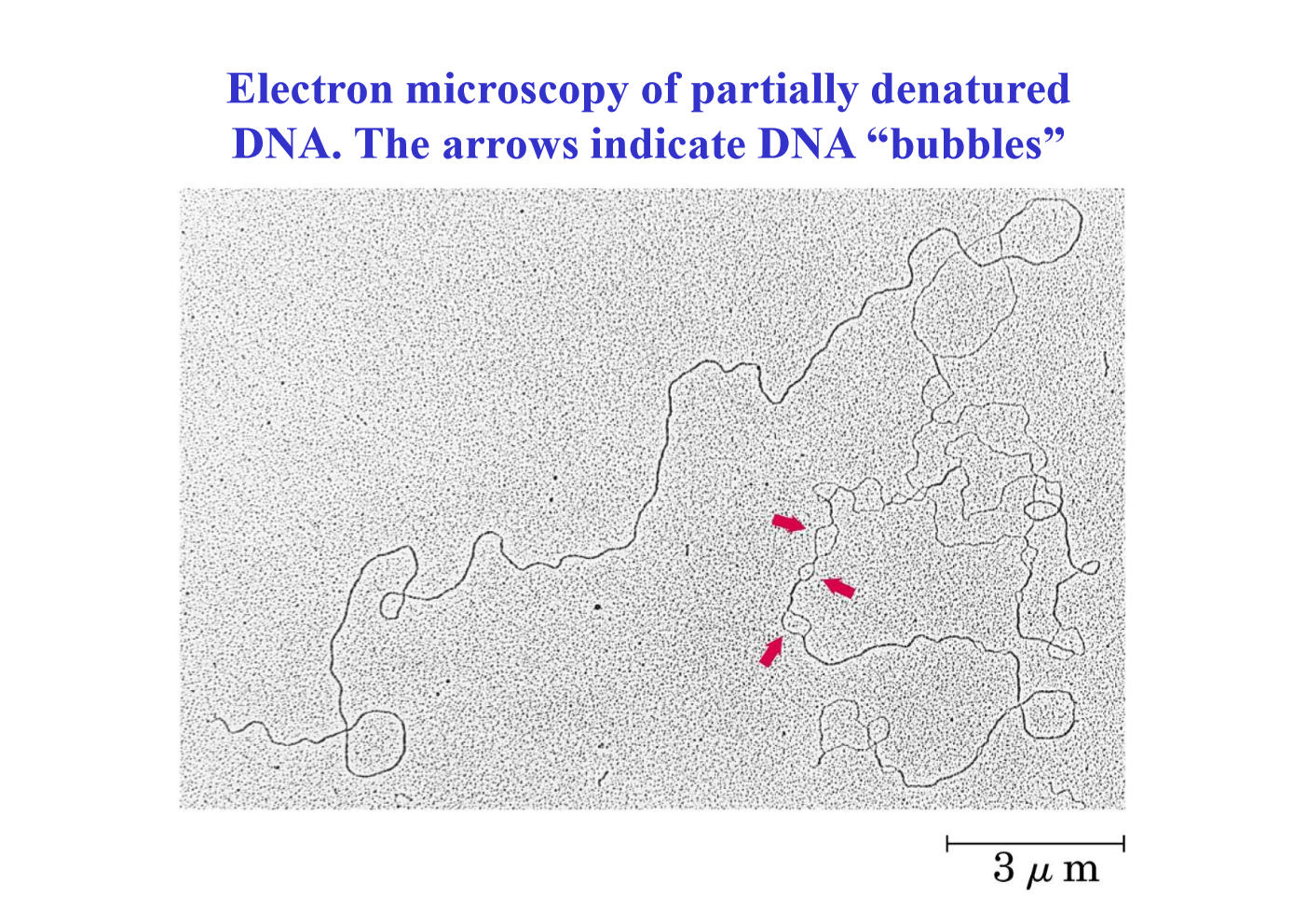
What makes denaturation reversible? What is the process called?
Nitrogen base complementarity
Creates a strong specificity in the interaction between polynucleotides: two complementary DNA strands can “search” for each other in solution and pair, leading to stable double helix
Re-annealing
Which bases are methylated more often?
A and C (more often than G and T)
Methylation is confined to _______?
Specific DNA regions (CpG islands)
All known DNA methyltransferases use __________ as a methyl group donor
S-adenosylmethionine
What % of cytidine residues in DNA are methylated to 5-methylcytidine
5%
DNA methylation plays a role in ______?
Gene expression regulation
PCR (polymerase chain reaction)
Heat to separate strands
Add synthetic DNA oligonucleotide primers; cool
Add thermostable Taq DNA polymerase to catalyse 5’ → 3’ DNA synthesis
Repeat steps 1 - 3
DNA sequencing
Sanger method (dideoxy chain-termination sequencing)
Target DNA denatured
Oligonucleotide primers annealed to template strand (starting point for DNA synthesis)
Mix of dNTPs and ddNTPs + DNA polymerase used to make complementary chains
Chain-terminating ddNTPs result in DNA fragments of different lengths
Fragments separated using gel electrophoresis (shorter fragments migrate faster to other end)
Automated DNA sequencing
Each ddNTP linked to a different fluorescent dye
All 4 ddNTPs added together
Resulting coloured DNA fragments are separated by size in an electrophoretic gel in a capillary tube
All fragments of a given length migrate in a single band
The colour associated with each band is detected with a laser beam
DNA sequence is read by identifying the colour sequences in the bands
Amount of fluorescence in each band is represented as a peak in the computer output
Next-generation pyrosequencing
Requires initial DNA fragmentation; ~ 400-500 nucleotides
4 nucleotides added as pulses one at a time, in repeating sequence
Excess is destroyed by apyrase before next pulse
When the nucleotide binds, pyrophosphate (PP) is released
Sulfurylase uses PP to generate ATP
ATP activates luciferase that reacts with luciferin and releases a detectable signal (peak)
Reversible Terminator Sequencing
Illumina; ~100-200 nucleotides
Primer and template are fixed to solid support
Add blocked, fluorescently (different colours) labelled dNTPs
Fluorescent colour is observed and recorded
Remove labels and blocking groups; wash
Repeat steps 2-4
SMRT Sequencing
Single-Molecule Real Time Sequencing; ~30-40 kb fragments; 10-15% error, mitigated by multiple readings
Zero Mode Wavelength (ZMW) pores: smaller diameter than visible light wavelength (70 nm).
DNA anchored by adapter sequences
Diameter hosts one DNA polymerase
Nucleotide-specific fluorescence emitted and detected as the correct nucleotide is inserted
What are contigs?
SMRT can translate sequences of millions of short DNA fragments into complex and contiguous sequences called contigs, with the help of computerised alignment of overlapping segments