1- Proteins, RP1
1/29
Earn XP
Description and Tags
Biological molecules
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
30 Terms
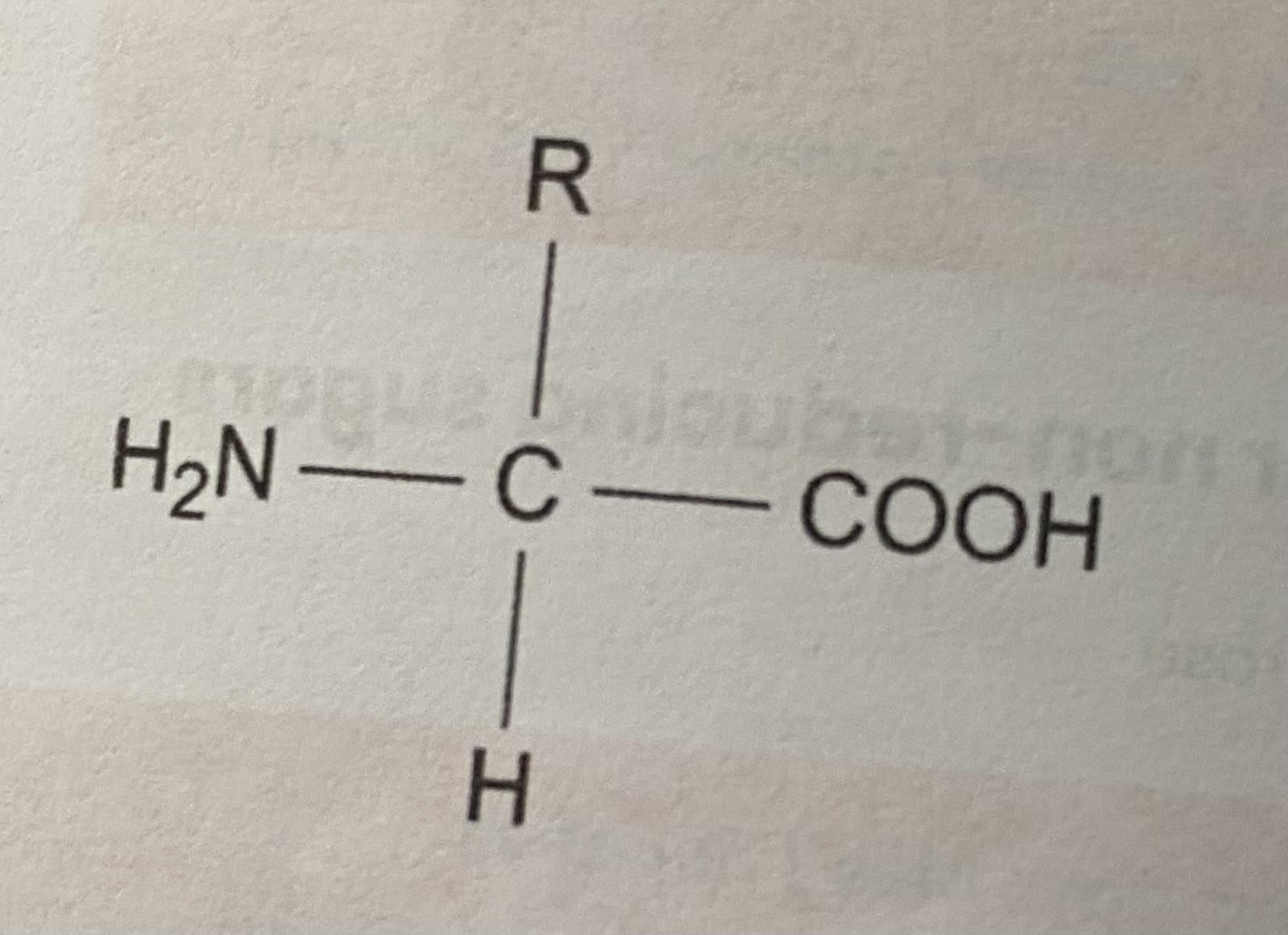
Describe/ draw the general structure of an amino acid
COOH= carboxyl group
R= variable group
H2N= amine group
How many amino acids are common in all organisms? How do they vary?
20 amino acids are common in all organisms
differ only in their side/ variable group
Describe how amino acids join together
condensation reaction
removing a water molecule
between carboxyl group of one and amine group of another
forming a peptide bond
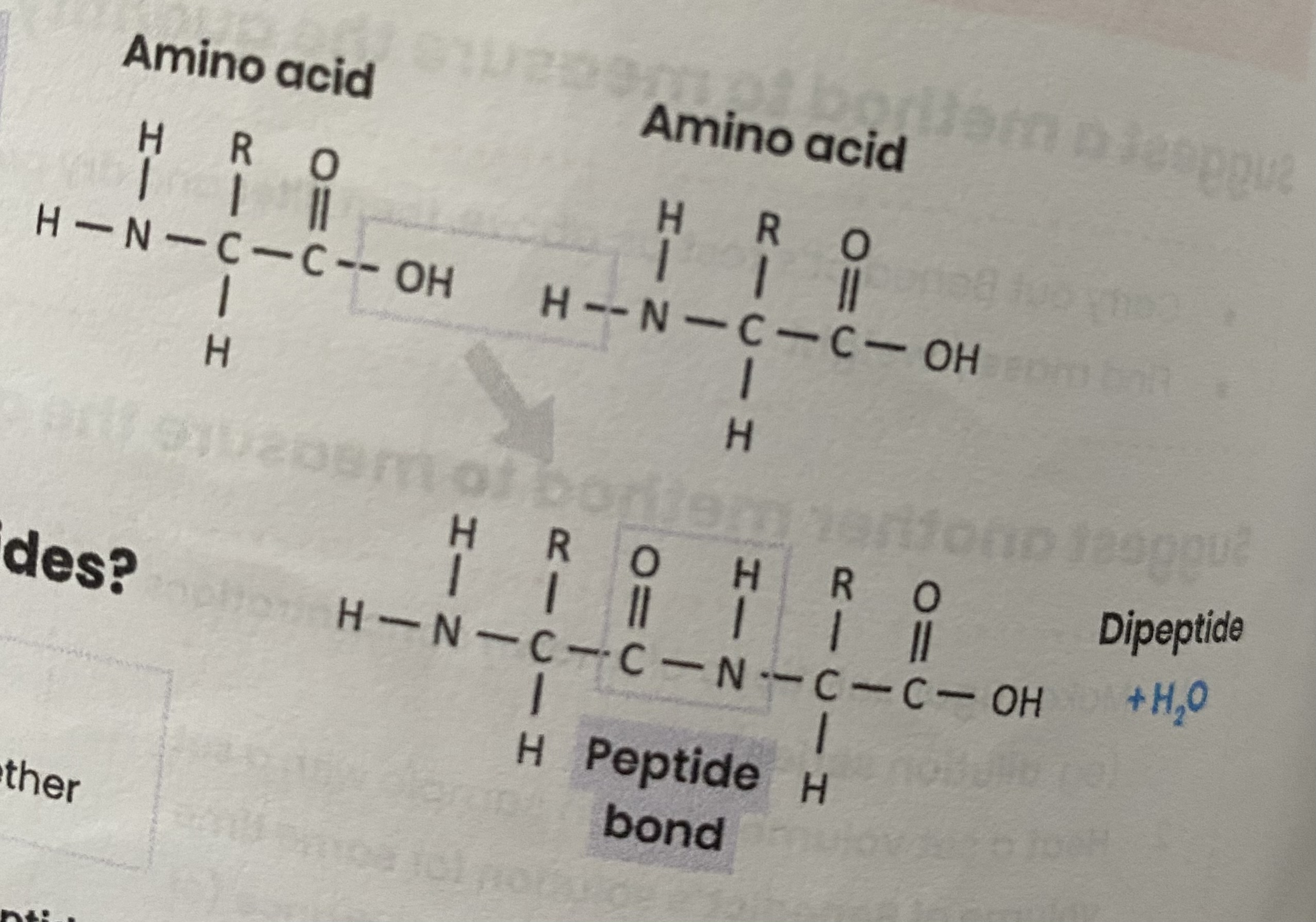
What are dipeptides and polypeptides?
dipeptide= 2 amino acids joined together
polypeptide= many amino acids joined together
> a functional protein may contain one or more polypeptides
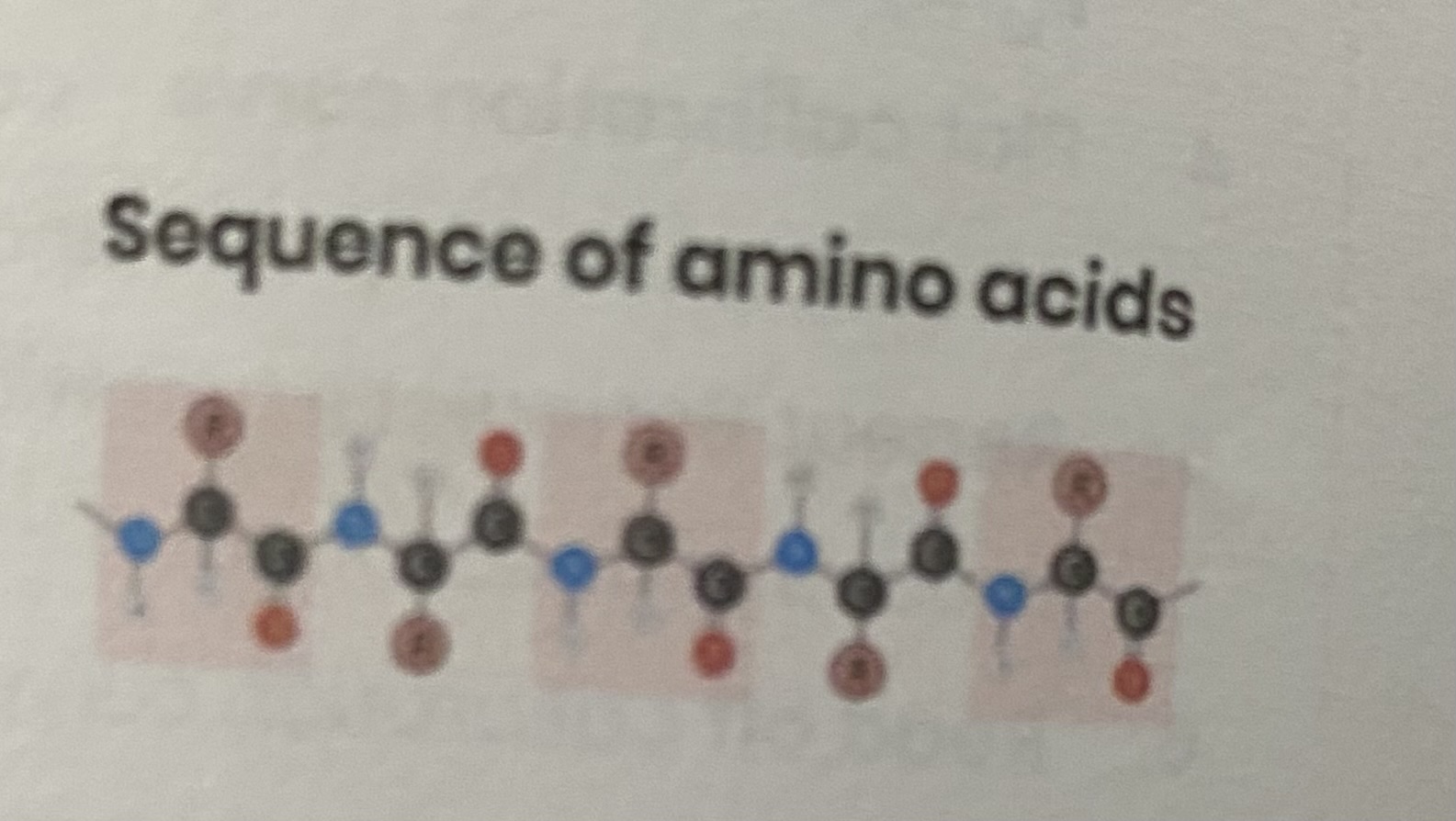
Describe the primary structure of a protein
sequence of amino acids in a polypeptide chain, joined by peptide chain
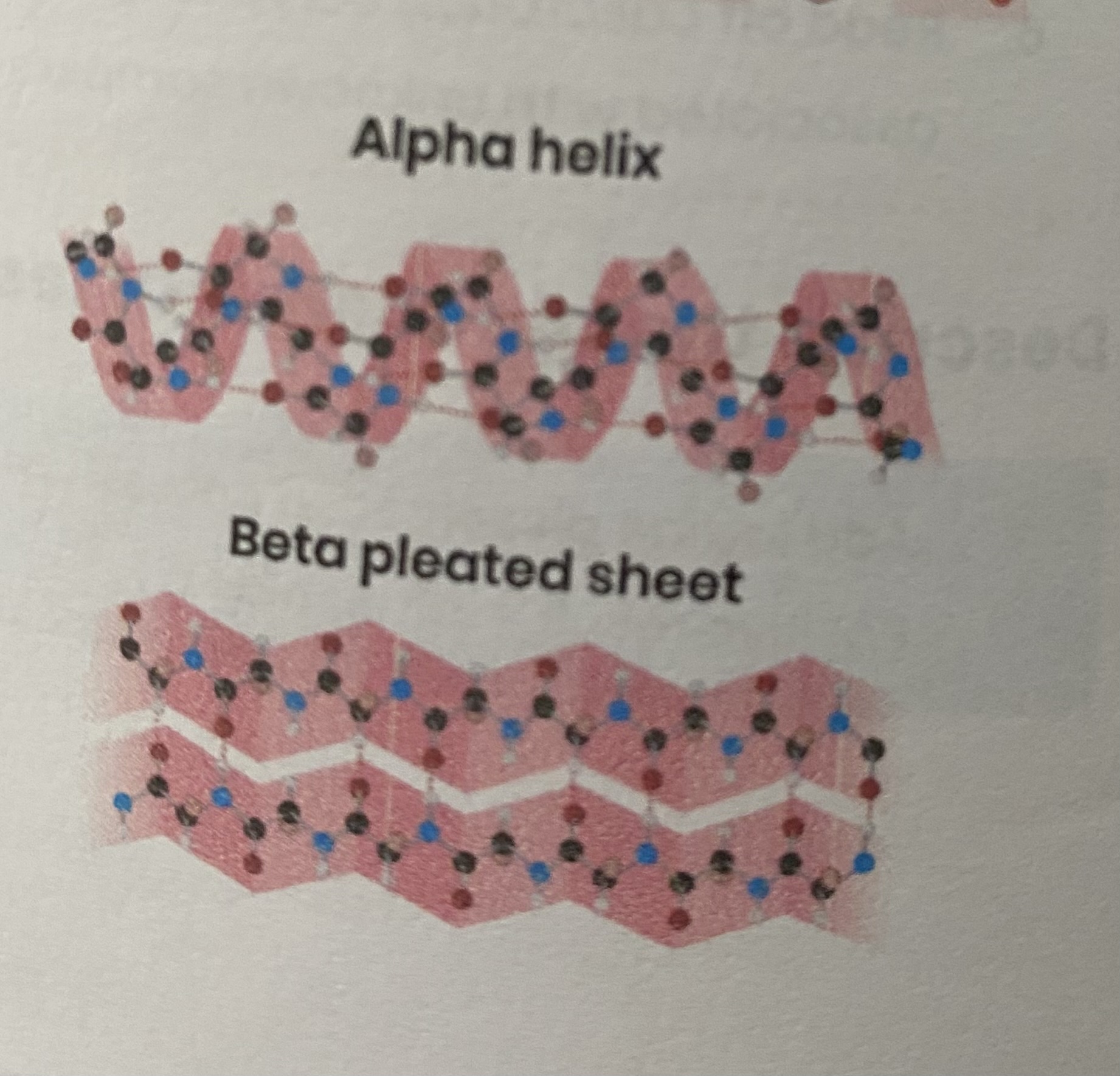
Describe the secondary structure of a protein
folding (repeating patterns) of polypeptide chain e.g. alpha helix/ beta pleated sheet
due to hydrogen bonding between amino acids
between amine group and carboxyl group of different amino acids
Describe the tertiary structure of a protein
3D folding of polypeptide chain
due to interactions between amino acid R groups (dependent on sequence of amino acids)
forming hydrogen, ionic bonds and disulphide bonds
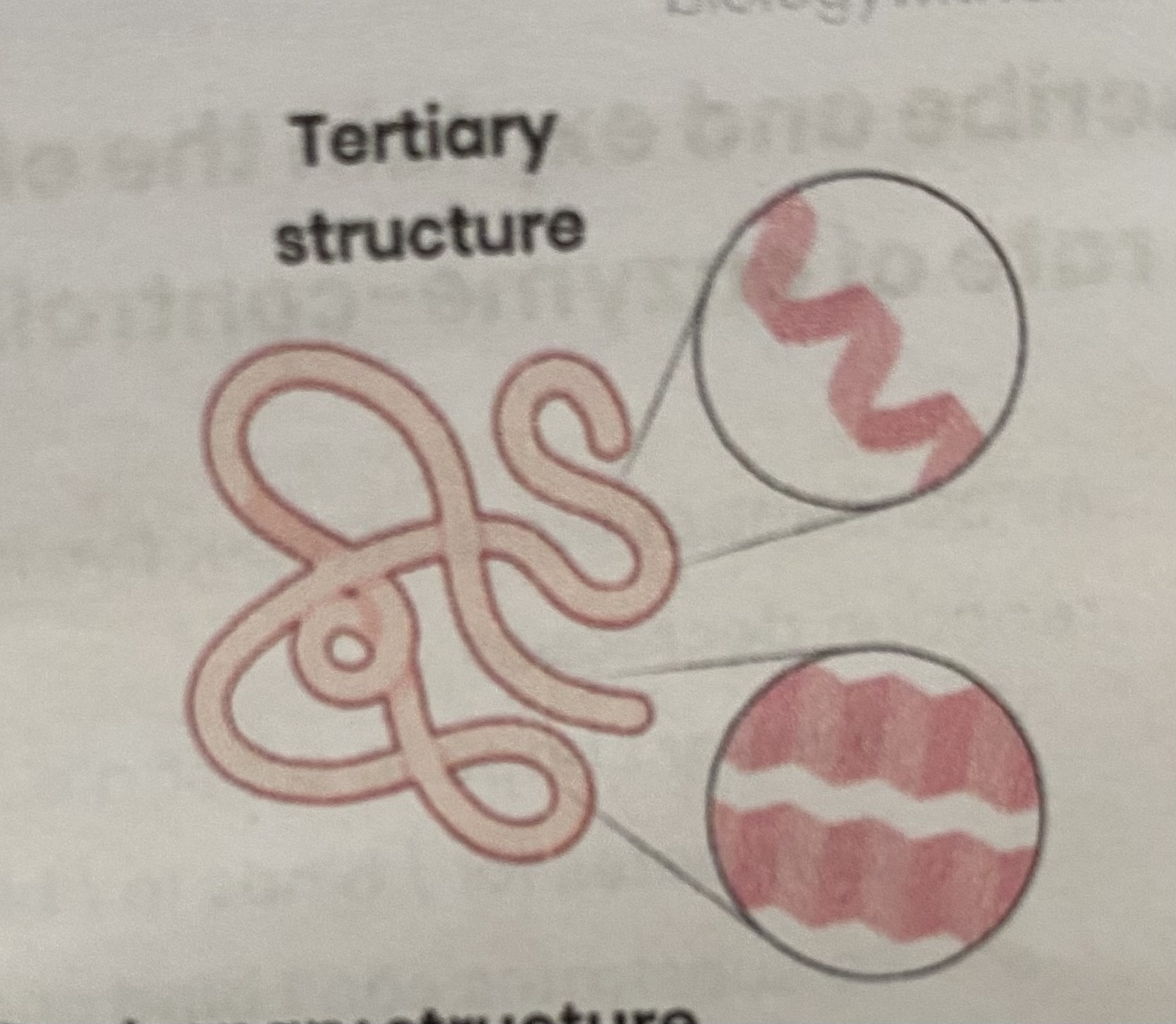
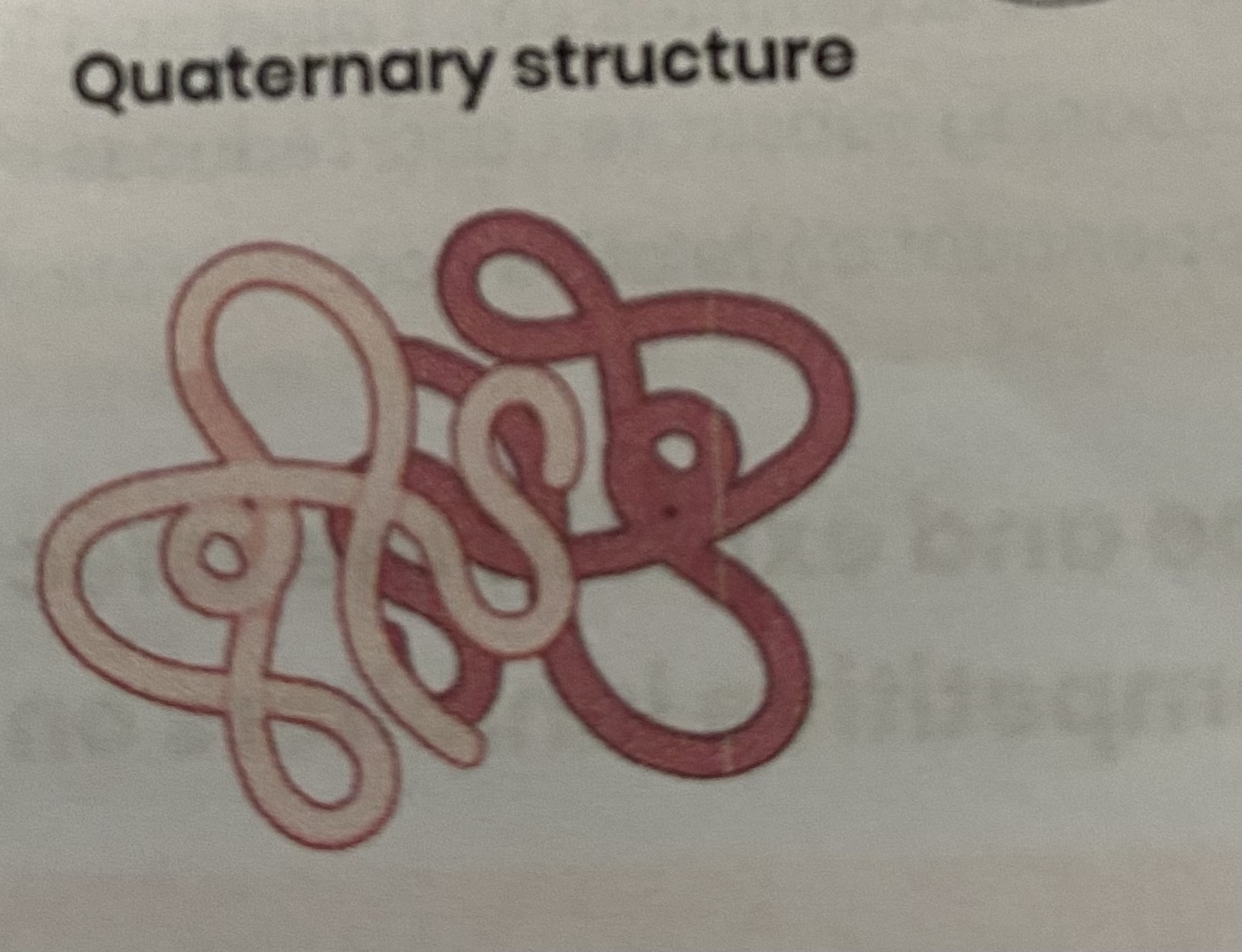
Describe the quaternary structure of a protein
more than one polypeptide chain
formed by interactions between polypeptides (hydrogen, ionic bonds, disulphide bridges)
Describe the test for proteins
add biuret reagent (sodium hydroxide + copper II sulphate)
positive result= purple/lilac colour (negative stays blue)—> indicates presence of peptide bonds
How do enzymes act as biological catalysts?
each enzyme lowers activation energy of reaction it catalyses
to speed up rate of reaction
Enzymes catalyse a wide range of intracellular and extracellular reactions that determine structures and functions from cellular to whole-organism level

Describe the induced-fit model of enzyme action
Substrate binds to (not completely complementary) active site of enzyme
causing active site to change shape (slightly) so it is complementary to substrate
so enzyme- substrate complex forms
causing bonds in substrate to bend/ distort, lowering activation energy
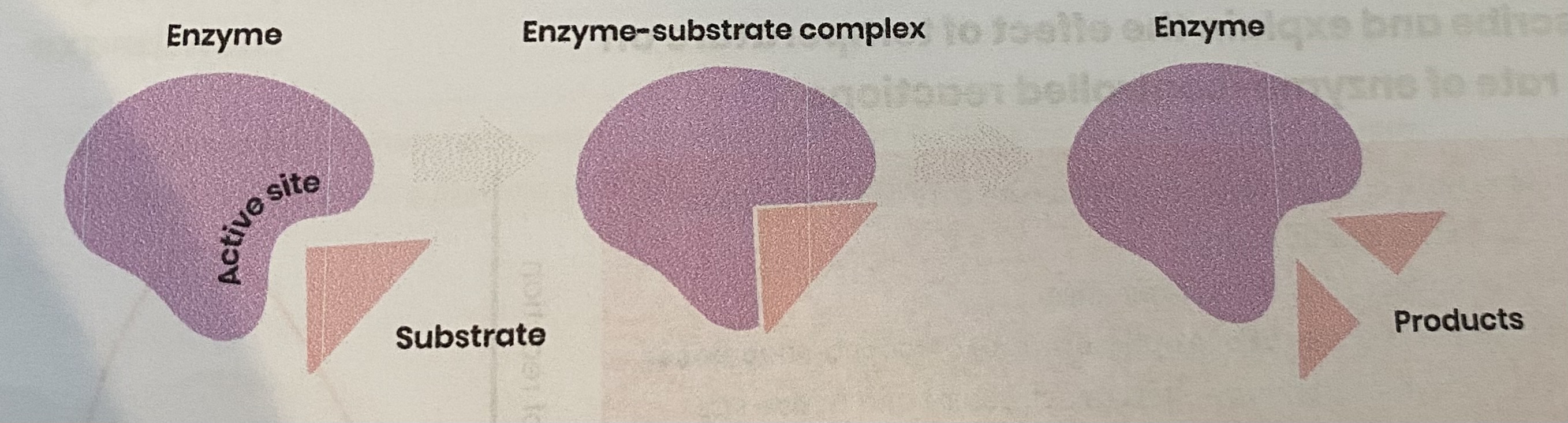
Describe how models of enzyme action have changed over time
initially lock and key model- active site a fixed shape, complementary to one substrate
now induced-fit model
Explain the specificity of enzymes
specific tertiary structure determines shape of active site- dependent on sequence of amino acids (primary structure)
active site is complementary to a specific substrate
only this substrate can bind to active site, inducing fit and forming an E-S complex
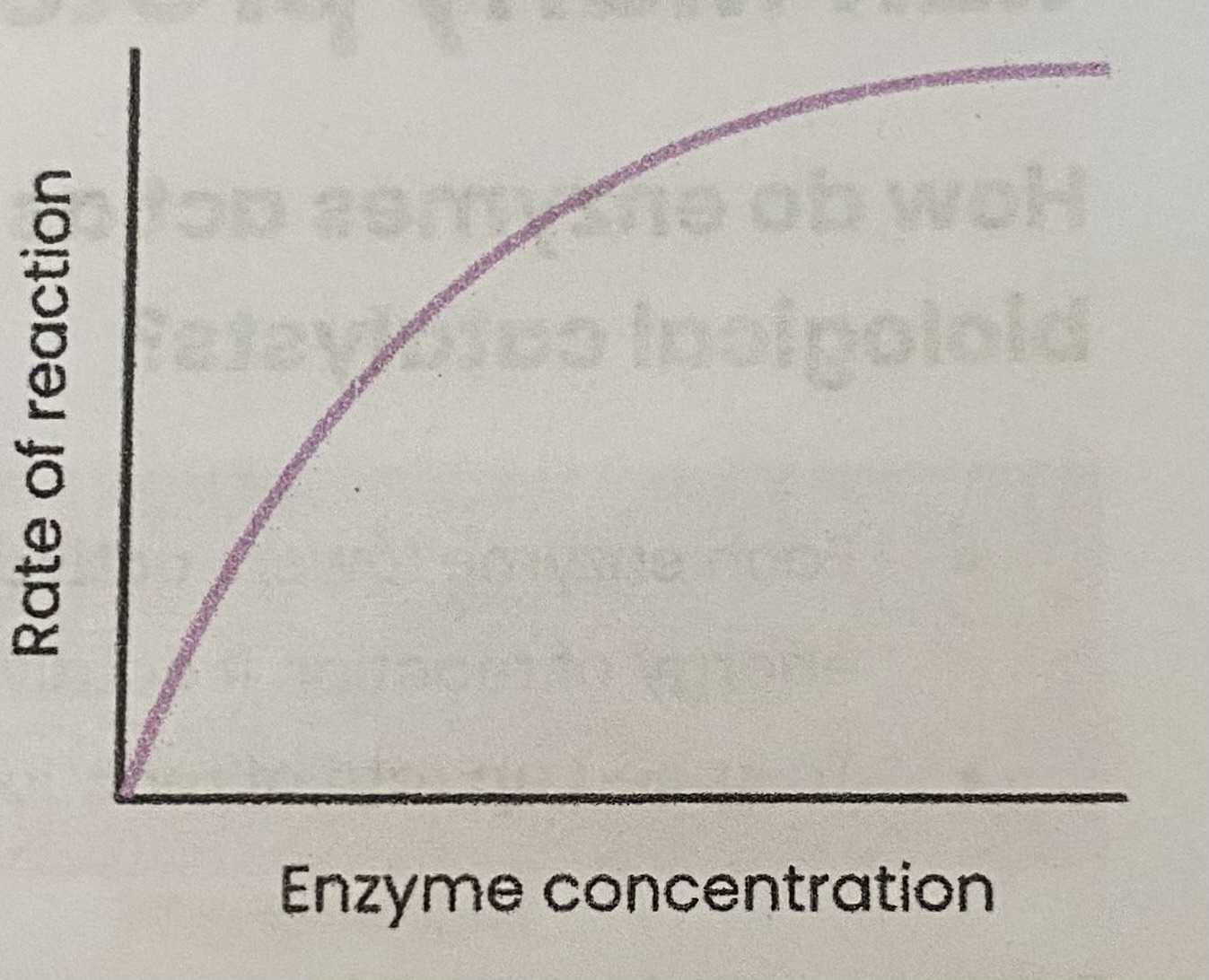
Describe and explain the effect of enzyme concentration on the rate of enzyme-controlled reactions
As enzyme conc increases, rate of reaction increases
> enzyme conc= limiting factor (excess substrate)
> more enzymes so more available active sites
> so more E-S complexes form
At a certain point, rate of reaction stops increasing/ levels off
> substrate conc.= limiting factor (all substrates in use)
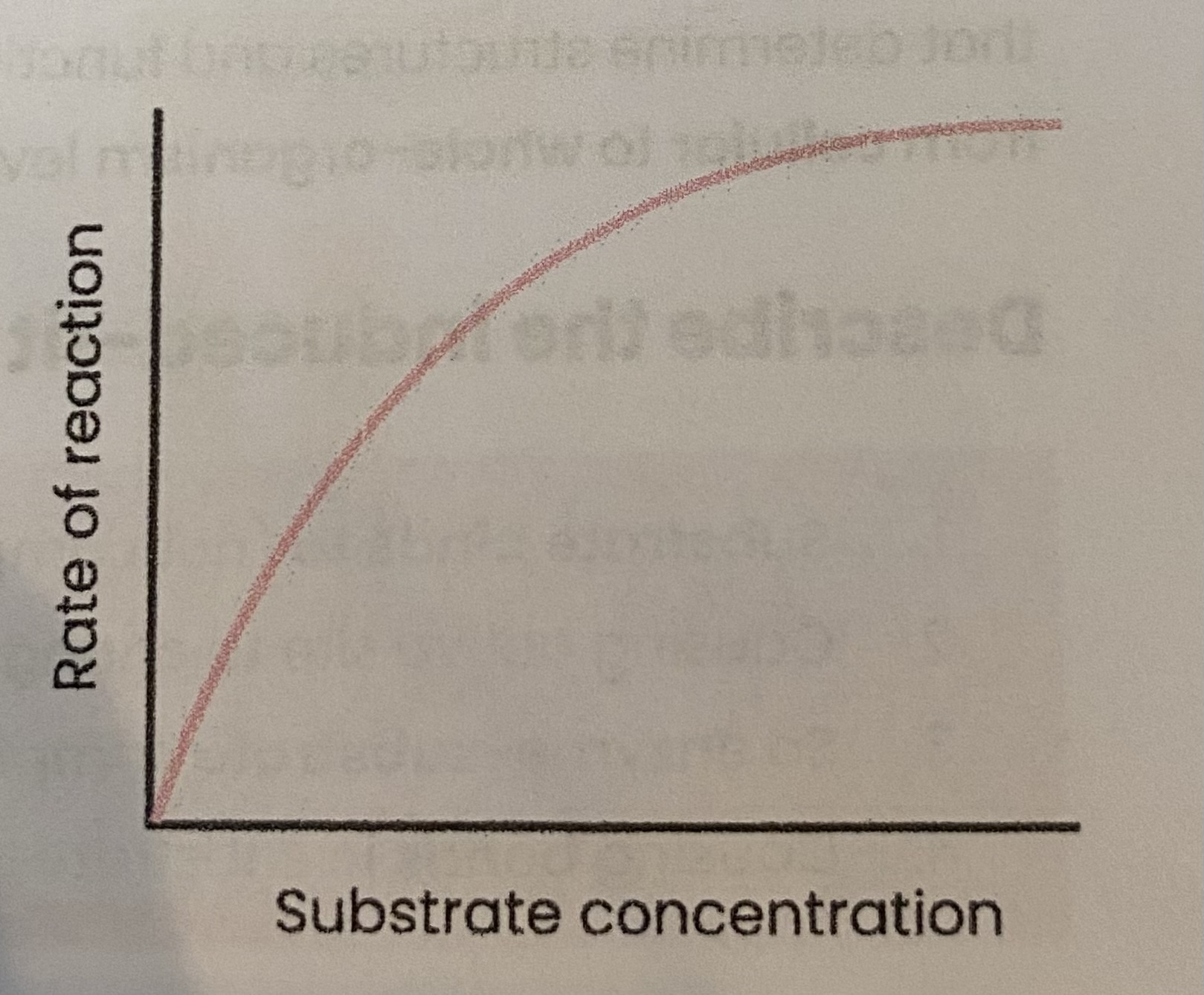
Describe and explain the effect of substrate concentration on the rate of enzyme-controlled reactions
As substrate conc increases, rate of reaction increases
> substrate conc= limiting factor (too few enzymes molecules to occupy all active sites)
> more E-S complexes form
At a certain point, rate of reaction stop increasing/ levels off
> enzyme conc= limiting factor
> as all active sites saturated/ occupied (at a given time)
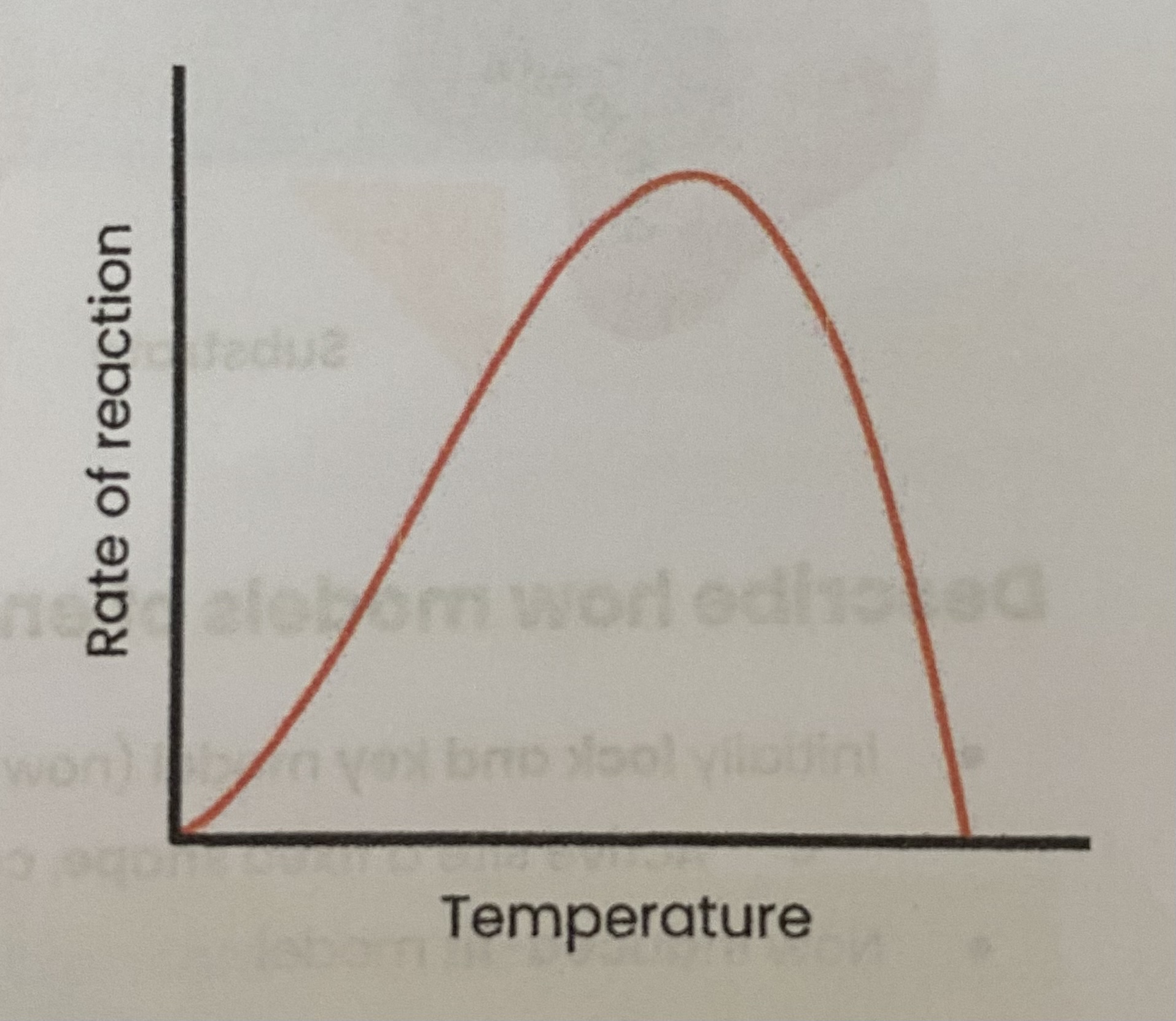
Describe and explain the effect of temperature on the rate of enzyme-controlled reactions
As temp increases, rate of reaction increases
> more kinetic energy
> so more E-S complexes form
As temp increases above optimum, rate of reaction decreases
> enzymes denature- tertiary structure and active sites change shape
> as hydrogen/ ionic bonds break
> so active site no longer complementary
> so fewer E-S complexes form
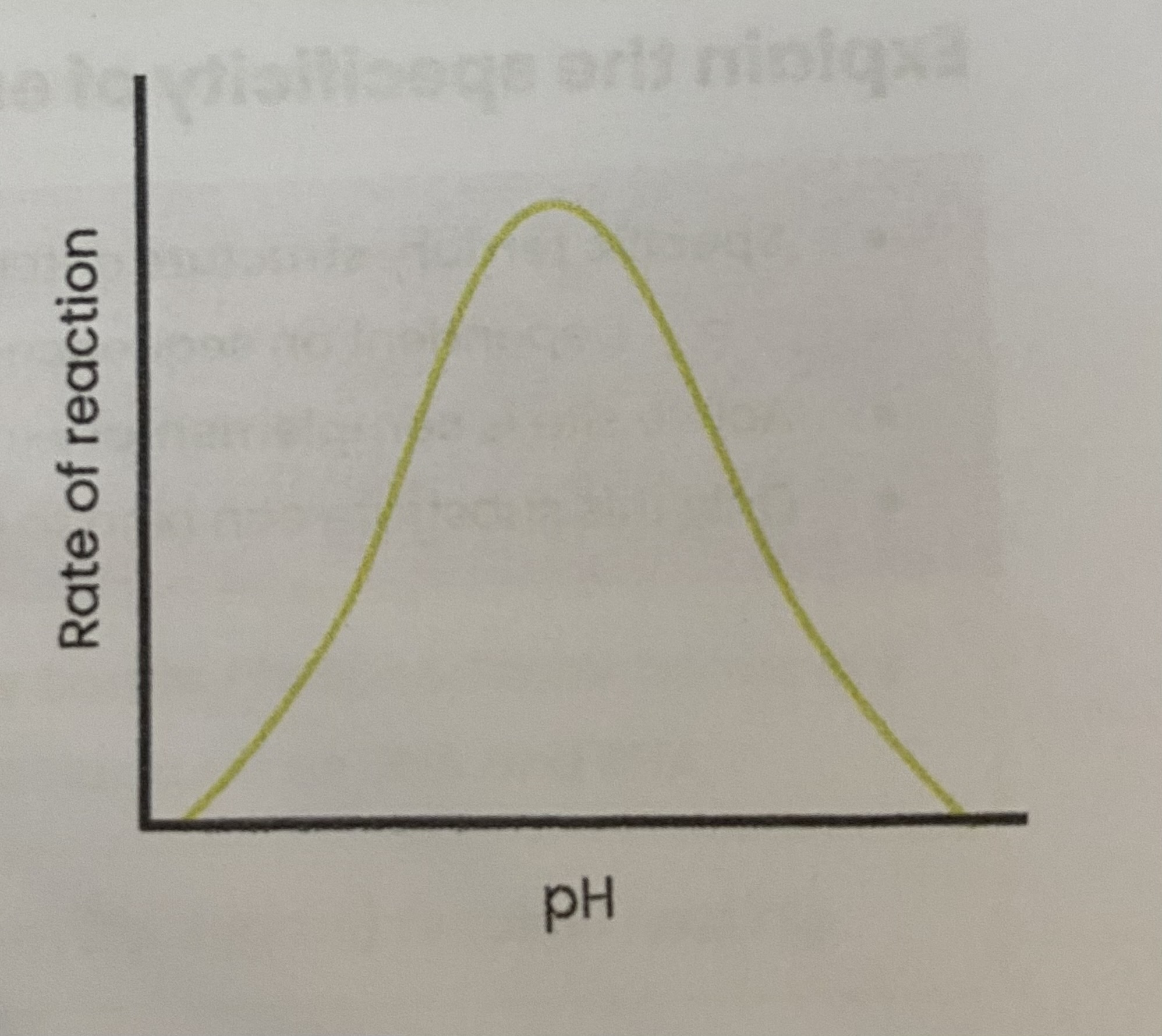
Describe and explain the effect of pH on the rate of enzyme-controlled reactions
As pH increases/ decreases above/ below optimum, rate of reaction decreases
> enzymes denature- tertiary structure and active site change shape
> as hydrogen/ ionic bonds break
> so active site no longer complementary
> so fewer E-S complexes form
Describe and explain the effect of competitive inhibitors on the rate of enzyme-controlled reactions
As conc of competitive inhibitor increases, rate of reaction decreases
> similar shape to substrate
> competes for/ binds to/ blocks active site
> so substrates can’t bind and fewer E-S complexes form
Increasing substrate conc reduces effect of inhibitors (dependent on relative concs of substrate and inhibitor)
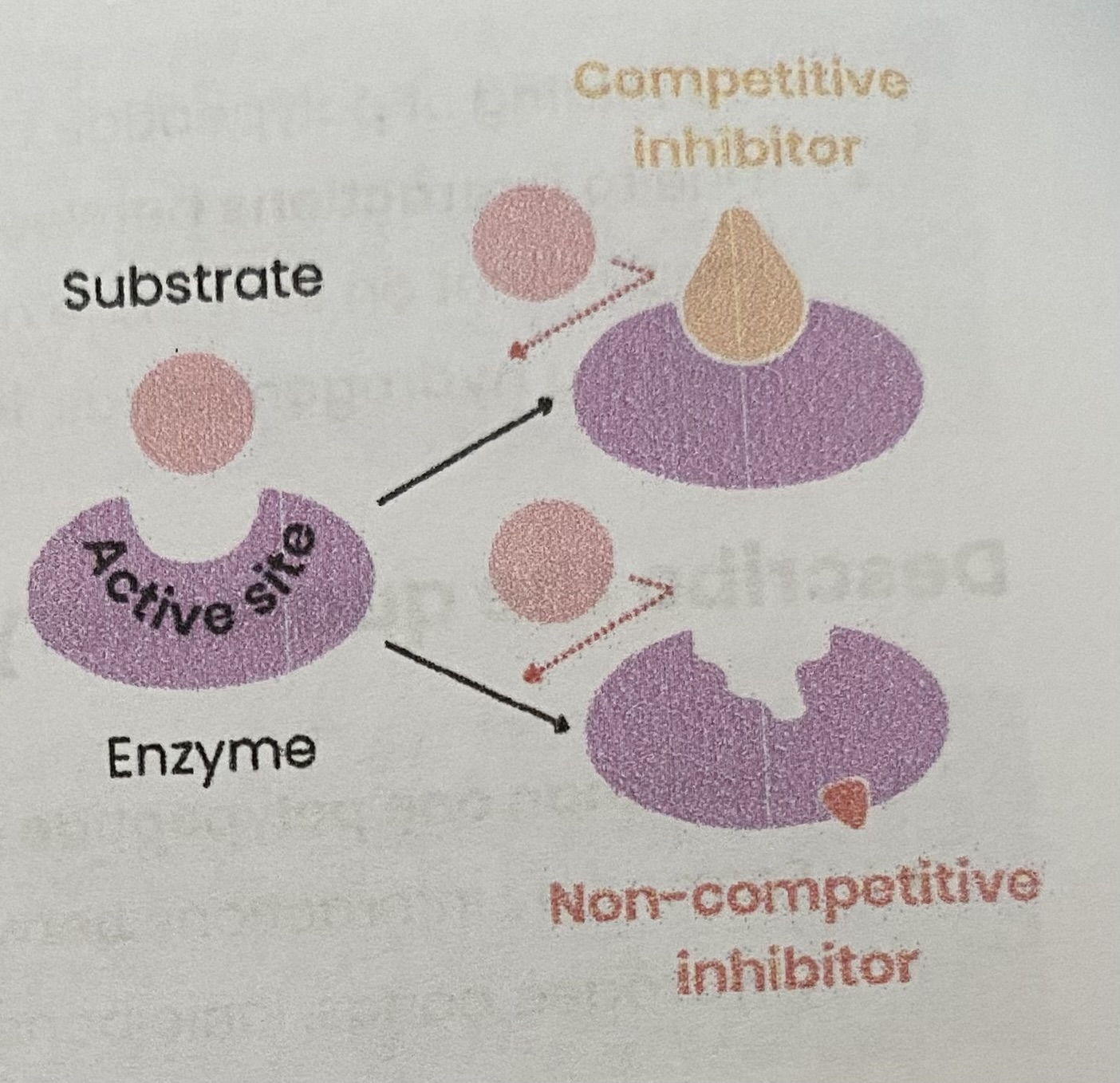
Describe and explain the effect of non-competitive inhibitors on the rate of enzyme-controlled reactions
As conc of non-competitiove inhibitors increases, rate of reaction decreases
> binds to site other than the active site (allosteric site)
> changes enzyme tertiary structure/ active site shape
> so active site no longer complementary to substrate
> so substrates can’t bind so fewer E-S complexes form
Increasing substrate conc has no effect on rate of reaction as change to active site is permanent
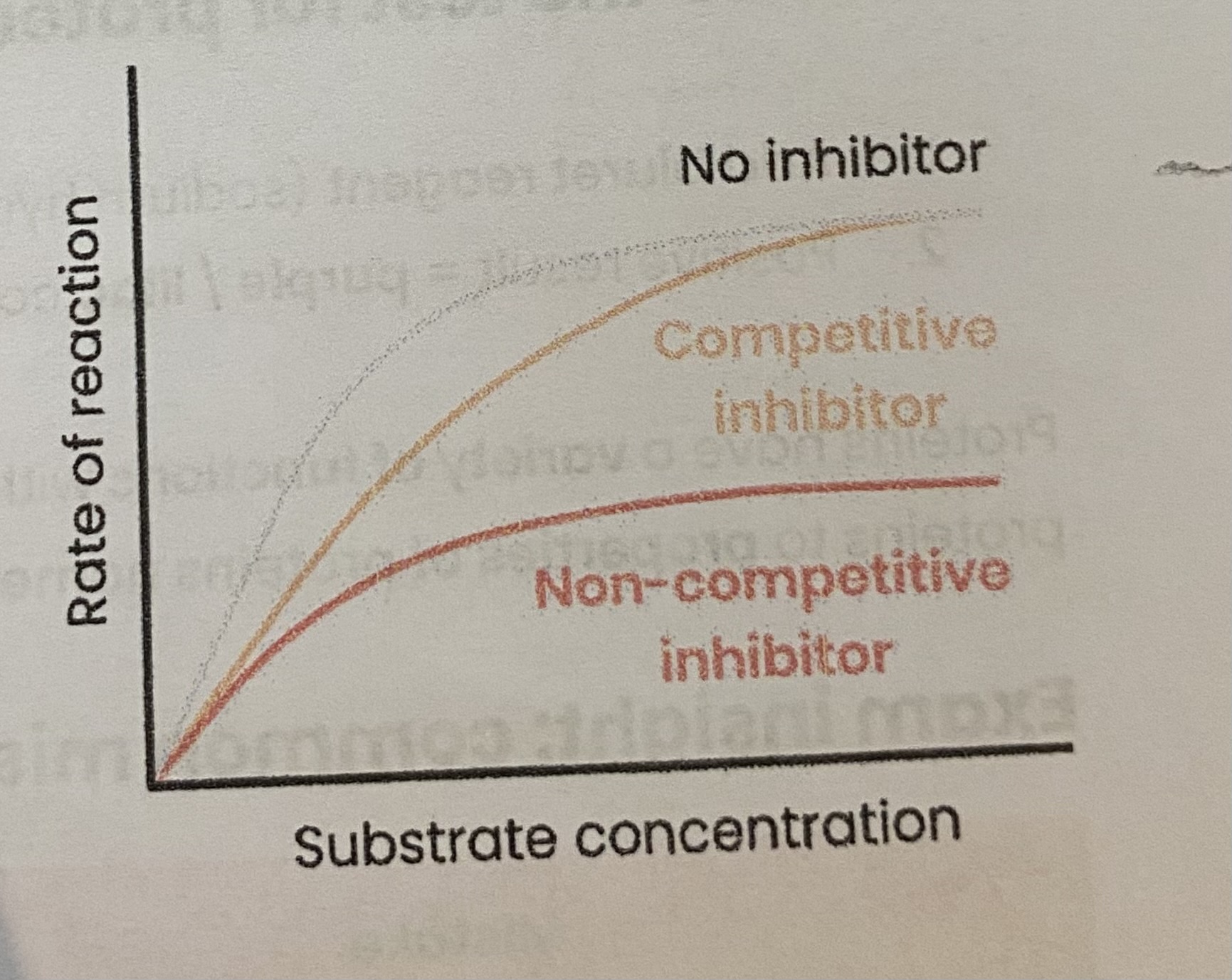
RP1: Effect of a named variable on the rate of an enzyme-controlled reaction
Give examples of variables that could affect the rate of an enzyme-controlled reaction
enzyme conc/ vol
substrate conc/ vol
temperature of solution
pH of solution
inhibitor conc
RP1: Effect of a named variable on the rate of an enzyme-controlled reaction
Describe how temperature can be controlled
use a thermostatically controlled water bath
monitor using a thermometer at regular intervals and add hot/ cold water if temperature fluctuates
RP1: Effect of a named variable on the rate of an enzyme-controlled reaction
Describe how pH can be controlled
use a buffer solution
monitor using a pH meter at regular intervals
RP1: Effect of a named variable on the rate of an enzyme-controlled reaction
Why were the enzyme + substrate solutions left in the water bath for 10 mins before mixing?
so solutions equilibrate/ reach the temperature of the water bath
RP1: Effect of a named variable on the rate of an enzyme-controlled reaction
Describe a control experiment
use denatured enzymes (e.g. by boiling)
everything else same as experiment, e.g. same conc/ vol of substrate (at start) and enzyme, same type/ vol of buffer solution, same temperature
RP1: Effect of a named variable on the rate of an enzyme-controlled reaction
Describe how the rate of an enzyme- controlled reaction can be measured
Measure time taken for reaction to reach a set point, e.g. conc/ vol/ mass/ colour of substrate or product
> rate of reaction= 1/time= S^-1
Measure conc/ vol/ mass/ colour of substrate or product at regular intervals (or using a continuous data logger) throughout reaction
> plot on a graph with time on the x axis and whatever is being measured on the y axis
> draw a langent at t=0 (or any other time for rate at a particular point)
initial rate of reaction= change in y/ change in x = cm³ s^-1
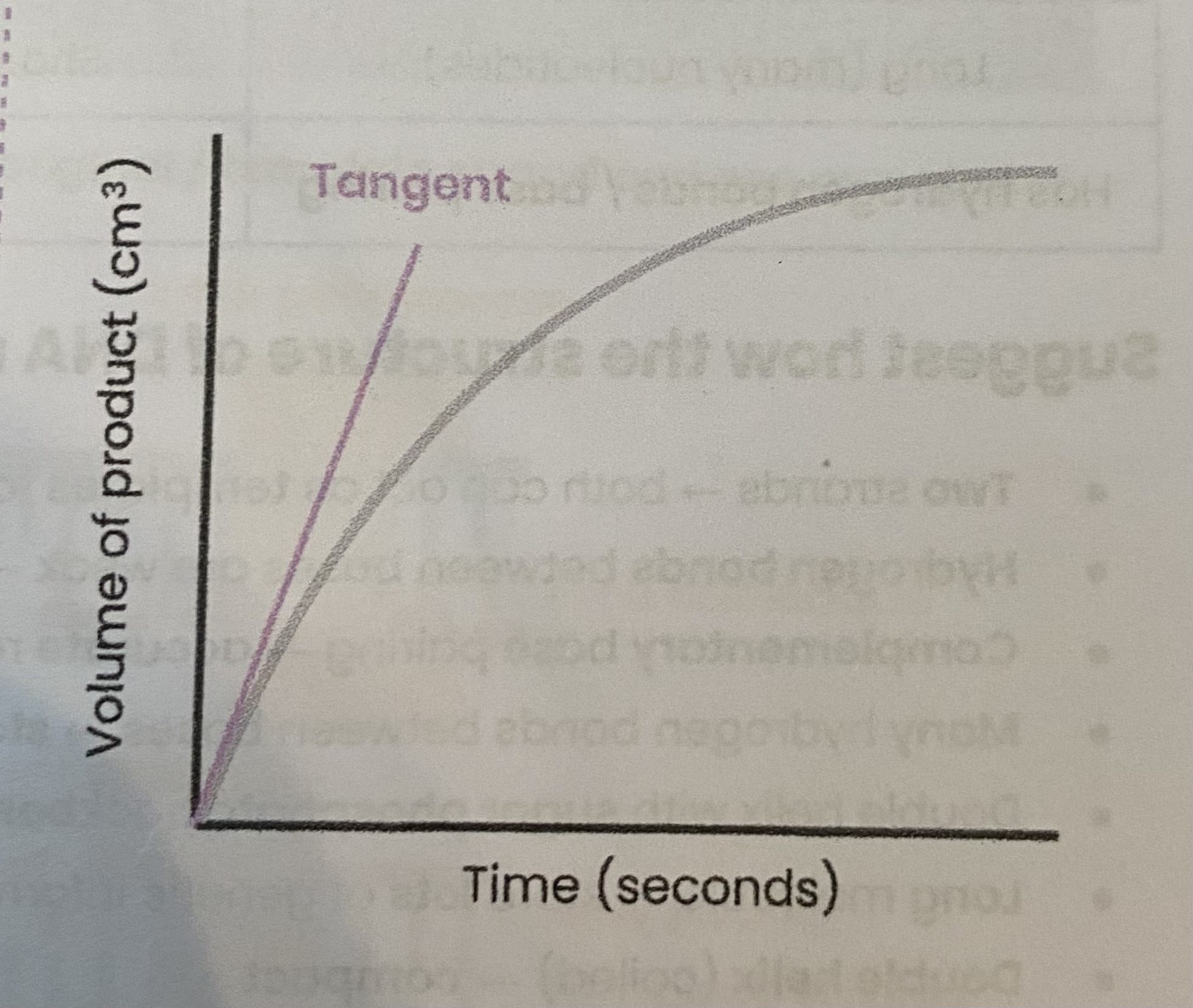
RP1: Effect of a named variable on the rate of an enzyme-controlled reaction
Suggest a safety risk and explain how to reduce this risk
handling enzymes may cause an allergic reaction
avoid contact with skin by wearing gloves and eye protection
RP1: Effect of a named variable on the rate of an enzyme-controlled reaction
Explain why using a colourimeter to measure colour change is better than comparison to colour standards
not subjective
more accurate
RP1: Effect of a named variable on the rate of an enzyme-controlled reaction
Explain a procedure that could be used to stop each reaction
boil/ add strong acid/ alkali= denature enzyme
put in ice= lower kinetic energy so no E-S complexes form
add high conc of inhibitor= no E-S complexes form
RP1: Effect of a named variable on the rate of an enzyme-controlled reaction
Describe how processed data can be presented in a graph
independent variable on x axis, rate of reaction on y axis including units
linear number sequence on axis, appropriate scale
plot coordinates accurately as crosses
join point to point with straight lines if cannot be certain of intermediate values OR draw a smooth curve
RP1: Effect of a named variable on the rate of an enzyme-controlled reaction
Explain why the rate of reaction decreases over time throughout each experiment
initial rate is highest as substrate conc not limiting/ many E-S complexes form
reaction slows as substrate used up and often stops as there is no substrate left