BI25M7 - 1.4-1.6 - Enzymes
1/53
Earn XP
Description and Tags
Energy for life
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
54 Terms
cofactor
non-protein chemical compound that is necessary for enzyme activity
coenzyme
an organic molecule type of cofactor that assists enzymes in catalyzing chemical reactions, usually produced from a vitamin
coenzyme examples and their derivatives
FAD from riboflavin, NAD+ from niacin, Coenzyme A from pantothenate
prosthetic group
cofactor covalently bound to the enzyme or very tightly associated with the enzyme
apoenzyme
the protein component of an enzyme that contains a cofactor
holoenzyme
whole enzyme, apoenzyme plus the cofactor(s)
six classes of enzymes
oxidoreductases, transferases, hydrolases, lyases, isomerases, ligases
oxidoreductases function
transfer electrons
transferases function
group transfer
hydrolases function
hydrolysis (transfer chemical groups to water)
lyases function
form, or add groups to double bonds
isomerases function
transfer groups within molecules (form isomers)
ligases function
formation of C-C, C-S, C-O, C-N bonds (coupled to ATP cleavage)
enzyme function
Increase rates of spontaneous reactions
Lower the activation energy of biochemical reactions
Accelerate movement towards reaction equilibrium
Gibbs free energy equation
ΔG = ΔH - ΔTS
spontaneous reactions
decrease enthalpy (H) and or increase entropy (S)
energy barrier
energy required to position chemical groups correctly, bond rearrangements, electron rearrangements, etc..
entropy reduction
Enzymes “force” the substrate(s) to be correctly orientated by binding them in the formation they need to be in for the reaction to proceed
desolvation
Weak bonds between the substrate and enzyme essentially replace most or all of the H-bonds between substrate and aqueous solution
induced fit
Conformational changes occur in the protein structure when the substrate binds
catalysis occurs when …
Active site is complementary to transition state
more substrate =
higher initial rate of reaction
low substrate concentration
almost a linear increase in V0 as ↑ [S]
michaelis-menten equation importance
crucial in enzyme kinetics to determine the rate of enzyme-catalyzed reactions and the affinity of an enzyme for its substrate.
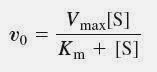
michaelis-menten equation basis
E + S ↔ ES ↔ E + P
The Michaelis constant (Km) can be defined as
the sum of the rate constants for the (part) reactions in which the enzyme-substrate complex decays, divided by the rate constant of the (part) reaction in which enzyme-substrate complex is formed.
Km
equivalent to the substrate concentration at which the initial reaction rate is half of the maximum reaction rate
Km tells us…
a clue to the affinity of the enzyme with it’s substrate
Vmax tells us
fast a reaction is proceeding when the enzyme is saturated with substrate
high Km, high Vmax
poor fit and fast enzyme
low Km, low Vmax
tight fit, slow enzyme
Lineweaver-Burk equation
linear equation
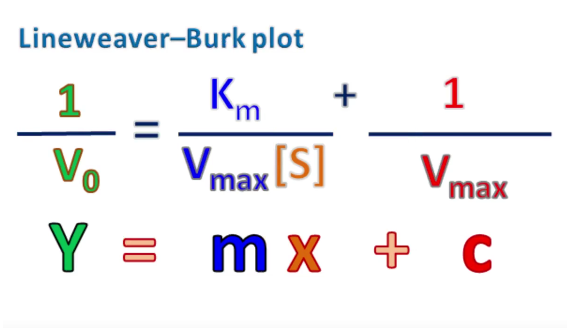
glucokinase
high Km, high Vmax, poor fit but fast enzyme. so when blood glucose goes up after a meal, the glucokinase activity increases
hexokinase
low Km, low Vmax, tight fit but slow enzyme. so when blood glucose is low, it’s active
Enzymes and two or more substrates
different Km values
Allosteric enzymes
change shape when bound to a specific molecule, impacting their activity and have a site separate from the active site
pH effect on enzymes
charge of amino acids changed, enzyme doesn’t function correctly, extremes can denature enzyme, affect the substrates of the reaction, some of which may require H+ or OH- groups to be involved in the reaction
inhibitors
competitive, non-competitive, uncompetitive
competitive inhibitors
increased Km, Vmax unchanged, reduces the affinity but doesn’t effect enzyme speed
AZT drug
competitive inhibitor of reverse transcriptase, used to treat AIDS
Transition state analouges
inhibitor to mimic transition state of enzyme
catalytic antibdies
specific to to transition state molecule
non-competitive antibodies
Km unchanged, Vmax decreased, affinity unchanged and speed of reaction decreased
ways regulatory enzyme modulate reactions
allosteric enzymes, covalently modified enzymes
feedback inhibition
product can inhibit enzymes
allosteric control
effectors that activate and inhibit enzymes
allosteric enzyme kinetics
initial low [S] sensitises the enzyme so it responds more efficiently at higher [S]
allosteric enzyme kinetics models
concerted model, sequential model
concerted model
allosteric activators and inhibitors
sequential model
substrate binding causes a change in one sub-unit which causes a change in another sub-unit allowing it to bind S more readily
reversible covalent modification
phosphorylation - Enzymes catalyse the phosphorylation of enzymes: Protein kinases (add phosphoryl groups to proteins) Protein phosphatases (remove phosphoryl groups)
multiple phosphorylation sites
These allow very fine control of enzyme function depending on the requirement of the particular enzyme at a given time - enzyme activity depends on signals
proteolytic cleavage
Enzymes can exist as an inactive precursor protein,called a proprotein or proenzyme. Proproteins can be cleaved to give active enzyme by proteases