DNA: Structure + Replication
1/163
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No study sessions yet.
164 Terms
What does DNA stand for and why is it called a nucleic acid?
Deoxyribonucleic acid
It is called a nucleic acid because it is found in the nucleus and possesses many acidic phosphate groups.
Why is DNA considered semiconservative?
DNA replication is called "semiconservative" because each new DNA molecule consists of one old strand (from the original DNA) and one new strand. This means half of the original DNA is conserved in each new DNA molecule.
What is the building block of DNA and what are its components?
The building block of DNA is deoxyribonucleoside 5'-triphosphate (dNTP). It consists of three components:
a monosaccharide (deoxyribose),
a nitrogenous base (adenine, guanine, cytosine, or thymine),
and a phosphate group.
What makes ribose special in the context of DNA?
In DNA, ribose is modified to deoxyribose, where the 2' carbon only has a hydrogen atom instead of a hydroxyl group (-OH), making it "deoxy."
What are the nitrogenous bases in DNA and how are they categorized?
The nitrogenous bases in DNA are adenine (A), guanine (G), cytosine (C), and thymine (T).
Adenine and guanine are purines, while cytosine uracil thymine are pyrimidines.
What is a nucleoside and how is it different from a nucleotide?
A nucleoside is composed of a ribose sugar linked to a nitrogenous base via a β-N-glycosidic linkage.
A nucleotide is a nucleoside with one, two, or three phosphate groups attached to the ribose's 5' carbon.
What is β-N-glycosidic linkage?
A β-N-glycosidic linkage is a type of chemical bond that connects a sugar molecule to a nitrogen-containing group (often an amine) in another molecule.
In simpler terms, it's a specific way sugars can attach to other molecules through a nitrogen atom, forming a stable connection that's important in various biological processes.

How is DNA repaired during replication? (2)
During DNA replication, the cell copies its DNA to pass on to a new cell. This process needs to be highly accurate, but sometimes mistakes happen, like the wrong nucleotide being added to the growing DNA strand. To fix these errors, the cell uses several mechanisms:
Proofreading by DNA Polymerase: DNA polymerase is the enzyme responsible for adding nucleotides to the new DNA strand. As it adds each nucleotide, it checks to make sure it's the correct one. If it detects an error, the enzyme can remove the incorrect nucleotide and replace it with the right one. This process is like a built-in spell-checker that helps reduce the number of mistakes during replication.
Mismatch Repair: If a mistake escapes the proofreading step, the mismatch repair system can catch it after replication. This system scans the newly made DNA for any mismatched bases (pairs that don't match according to the base-pairing rules). When a mismatch is found, the incorrect nucleotide is removed, and the correct one is inserted in its place.
How does DNA repair mutations? (3)
Base Excision Repair (BER): This mechanism fixes small, non-bulky damage to the DNA, like when a single base is altered (for example, due to oxidation). The damaged base is removed by a special enzyme, and the resulting gap is filled with the correct nucleotide by DNA polymerase and sealed by DNA ligase.
Nucleotide Excision Repair (NER): This process repairs bulky damage that distorts the DNA helix, such as damage caused by UV light, which can create thymine dimers (where two thymine bases bond together incorrectly). The damaged section of DNA is cut out by enzymes, and the gap is filled in with the correct nucleotides.
Homologous Recombination and Non-Homologous End Joining (HR and NHEJ): These mechanisms repair more severe types of damage, like double-strand breaks (where both strands of the DNA are broken). In homologous recombination, the cell uses a sister chromatid (an identical copy of the DNA) as a template to accurately repair the break. In non-homologous end joining, the cell directly joins the broken ends together, which is quicker but can sometimes lead to small errors.
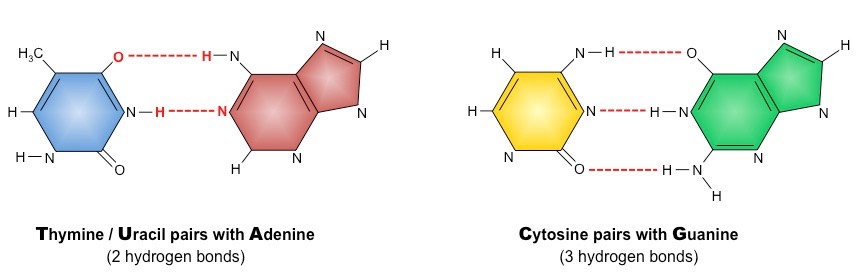
Why can adenine and thymine form hydrogen bonds with each other?
Adenine and thymine can form hydrogen bonds with each other due to the presence of complementary hydrogen bonding sites, which facilitate base pairing in the DNA double helix.
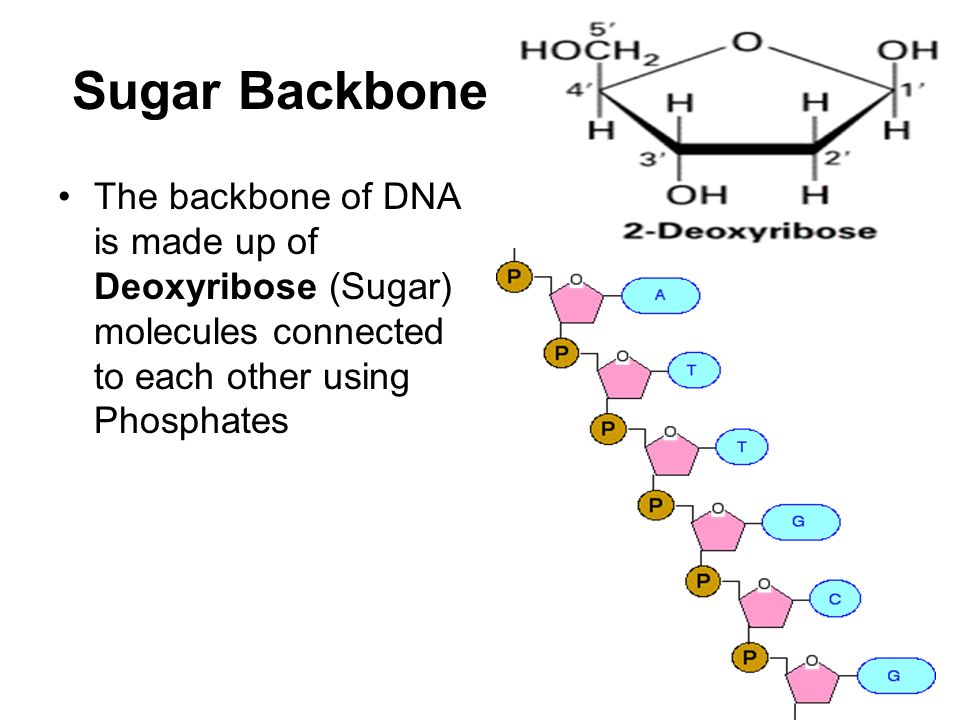
What is the significance of the phosphate group in nucleotides?
The phosphate group in nucleotides contributes to the formation of the DNA backbone by linking the 5' carbon of one nucleotide to the 3' carbon of the next nucleotide, forming a sugar-phosphate backbone.

What are the two types of nucleic acid?
DNA and RNA
What is the precursor to nucleotides?
How do they transform?
Nucleosides
Kinase enzymes add a phosphate group to turn them into nucleotides
What are the building blocks of nucleic acid?
Nucleotides
Nitrogenous bases are made of what? How does this help bonding?
Nitrogen
Helps bonding because bases form hydrogen bonds with each other to pair up. Hydrogen bonds are weak attractions between a hydrogen atom bonded to an electronegative atom like oxygen or nitrogen.
(Electronegative meaning measure of atom’s ability to attract and hold onto electrons in a chemical bond — oxygen is highly electronegative — strong tendency to attract electrons towards itself in a chemical bond)
What is referred to as the backbone of DNA and why?
The ribose + phosphate portion of the nucleotide is referred to as the backbone of DNA because it is invariant, meaning it does not change.
The base is the variable portion of the building block.
How many different dNTPs are there and what makes them different?
There are four different deoxyribonucleoside triphosphates (dNTPs) in DNA: dATP, dGTP, dCTP, and dTTP.
They differ only in the aromatic base they contain (adenine, guanine, cytosine, and thymine).

What is a hydroxyl group?
Functional group consisting of an oxygen atom bonded to a hydrogen atom (-OH), commonly found in alcohols and many other organic compounds.

What is a phosphodiester bond?
Covalent bonds that form the backbone of DNA and RNA molecules, linking the 3' carbon atom of one sugar molecule to the 5' carbon atom of another
Creating a strong connection crucial for building the long chains of genetic material that carry information in living cells
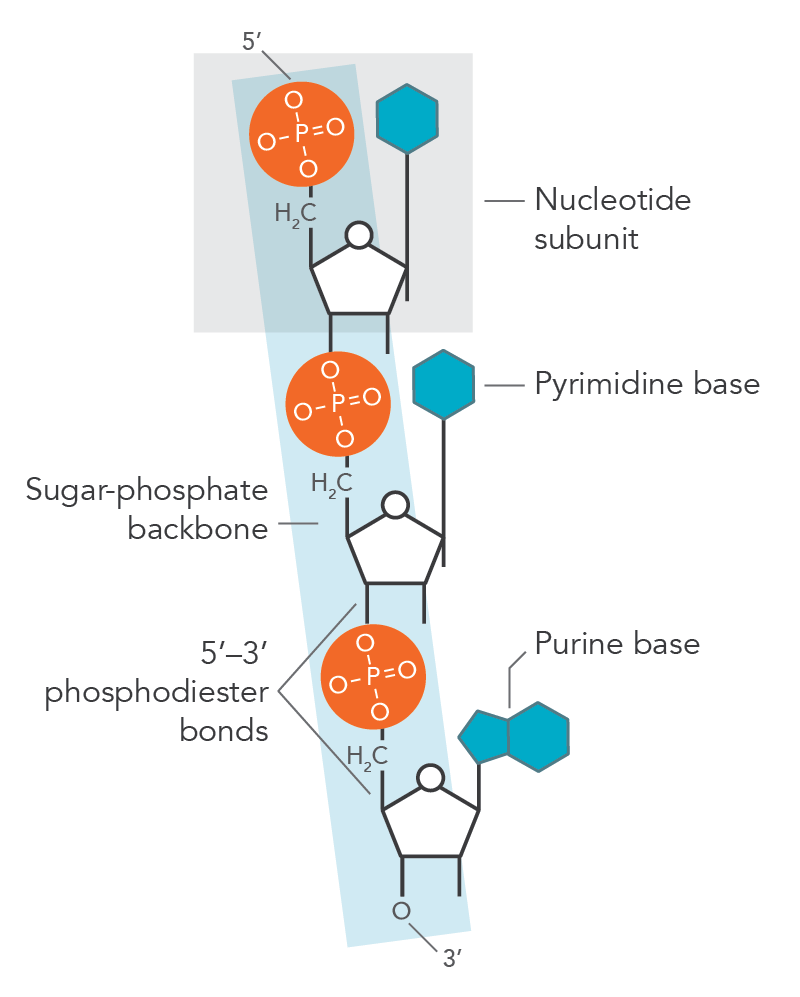
What type of bond links nucleotides in the DNA chain?
Nucleotides in the DNA chain are covalently linked by phosphodiester bonds between the 3' hydroxyl group of one deoxyribose and the 5' phosphate group of the next deoxyribose.

What is the difference between an oligonucleotide and a polynucleotide?
An oligonucleotide is a polymer of several nucleotides linked together, whereas a polynucleotide is a polymer of many nucleotides.
How is the sequence of a polynucleotide abbreviated and written?
The sequence of a polynucleotide is abbreviated by listing the bases attached to each nucleotide in the chain. The end of the chain with a free 5' phosphate group is written first, with the sequence indicated in the 5' to 3' direction.
For example, a polynucleotide sequence might be written as pGpApC or guanyladenylcytosine (GAC)
Why is the 5' to 3' direction significant in DNA sequencing?
The 5' to 3' direction is significant in DNA sequencing because it represents the orientation in which nucleotides are added during DNA synthesis, with the 5' phosphate group of one nucleotide connecting to the 3' hydroxyl group of the next.
What determines the variability in the DNA structure?
By the nitrogenous bases (adenine, guanine, cytosine, and thymine) that are attached to the deoxyribose sugars in the nucleotide chain.
Who developed the Watson-Crick model of DNA structure, and what does it propose?
James Watson and Francis Crick, with contributions from Maurice Wilkins and Rosalind Franklin, developed the Watson-Crick model of DNA.
It proposes that DNA is a right-handed double helix structure held together by hydrogen bonds between complementary bases.
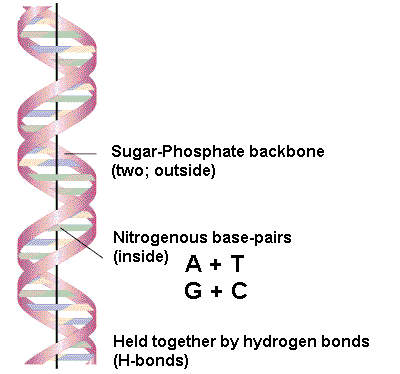
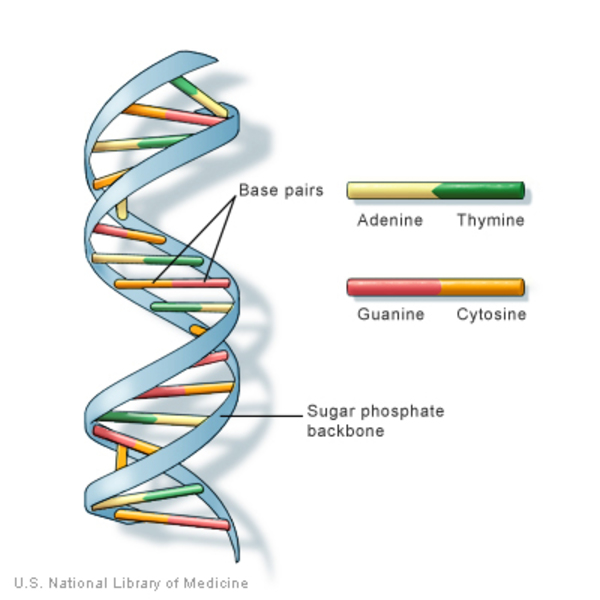
Describe the structure of double-stranded DNA (ds-DNA) according to the Watson-Crick model.
In ds-DNA, two long polynucleotide chains are hydrogen-bonded together in an antiparallel orientation, meaning the 5' end of one chain is paired with the 3' end of the other.

What is the specificity of base pairing in DNA according to the Watson-Crick model?
According to the Watson-Crick model, adenine (A) always pairs with thymine (T) via two hydrogen bonds, and guanine (G) always pairs with cytosine (C) via three hydrogen bonds.
** A - T = 2 hydrogen bonds
** G - C = 3 hydrogen bonds

What is meant by the terms annealing, hybridization, melting, and denaturation in the context of DNA?
Annealing or hybridization refers to the binding of two complementary DNA strands into a double-stranded structure.
Melting or denaturation refers to the separation of the two DNA strands due to high temperature or other denaturing conditions.

What is the significance of Tm in DNA studies?
Tm, or melting temperature, is the temperature at which 50% of the DNA molecules in a solution are denatured (melted). It is a critical parameter in various DNA-related experiments.

How is the stability of the DNA double helix maintained?
The stability of the DNA double helix is maintained by several interactions:
hydrogen bonds between complementary bases: A/T 2 hydrogen bonds, G/C 3 hydrogen bonds
van der Waals: weak attractions between molecules that arise due to temporary fluctuations in electron distribution around atoms
and hydrophobic interactions between bases in the interior of the helix — their chemical structure tends to repel water molecules.
Explain the term "right-handed double helix" in the context of DNA structure.
A right-handed double helix refers to the clockwise spiral arrangement of the two DNA strands around each other. This helical structure is stabilized by the interactions between bases and the sugar-phosphate backbone.

How does the term "complementary strands" apply to DNA structure?
Complementary strands of DNA are those that can hydrogen bond with each other when oriented in an antiparallel fashion.
This complementary base pairing is essential for maintaining the integrity and function of DNA molecules.

What is a genome, and how is it structured in eukaryotes?
A genome is the total genetic information of an organism.
In eukaryotes, the genome consists of several large pieces of linear double-stranded DNA, each called a chromosome.

Describe the difference between eukaryotic and prokaryotic genomes.
Eukaryotic genomes are composed of multiple linear chromosomes, whereas prokaryotic genomes typically consist of a single circular chromosome.

What is the role of DNA gyrase in prokaryotes?
DNA gyrase is an enzyme in prokaryotes that uses ATP to induce supercoiling in the circular DNA molecule, making it more compact and stable.
How is eukaryotic DNA packaged to fit within the cell nucleus?
Eukaryotic DNA is wrapped around globular proteins called histones to form nucleosomes. These nucleosomes are connected by linker DNA and further compacted to form chromatin, which is tightly packed DNA.
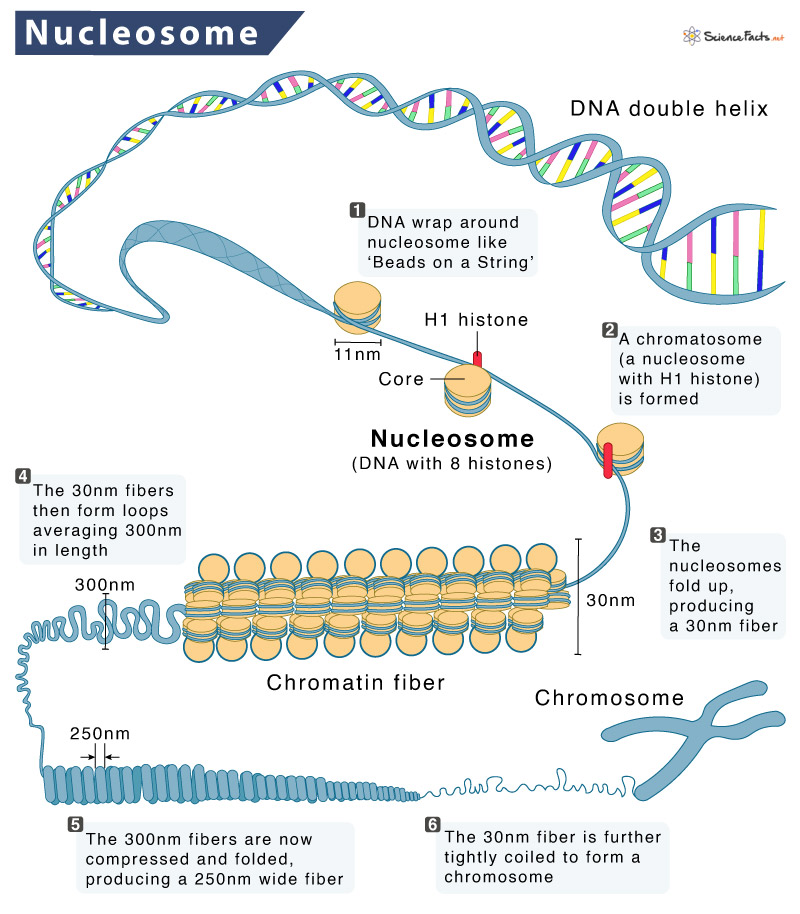
What are nucleosomes, and what is their composition?
Nucleosomes are the basic unit of chromatin structure, consisting of DNA wrapped around an octamer of histone proteins
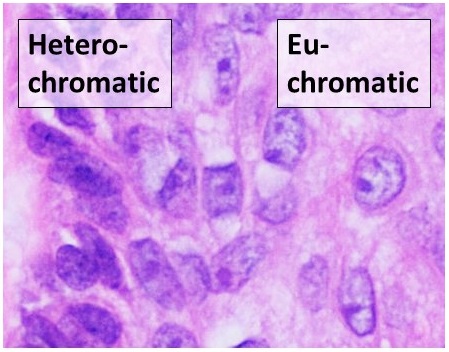
Define heterochromatin and euchromatin in terms of DNA packing and activity.
Heterochromatin is densely packed, transcriptionally inactive DNA, often found in darker-staining regions of chromosomes.
Euchromatin is less dense and transcriptionally active, located in lighter-staining regions.

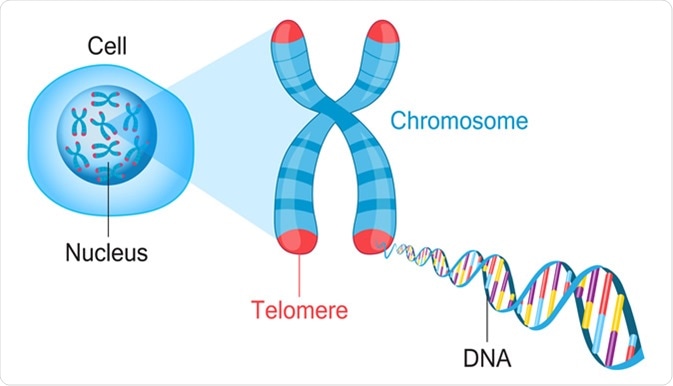
What are telomeres, and what is their function?
Telomeres are repetitive DNA sequences (e.g., TTAGGG in humans) at the ends of linear chromosomes.
They protect chromosome ends from degradation and prevent fusion with neighboring chromosomes during DNA replication.

Explain the structure and function of centromeres
Centromeres are specialized regions of chromosomes where spindle fibers attach during cell division.
They contain repetitive DNA sequences and serve as attachment sites for kinetochores, which are protein complexes involved in chromosome segregation.
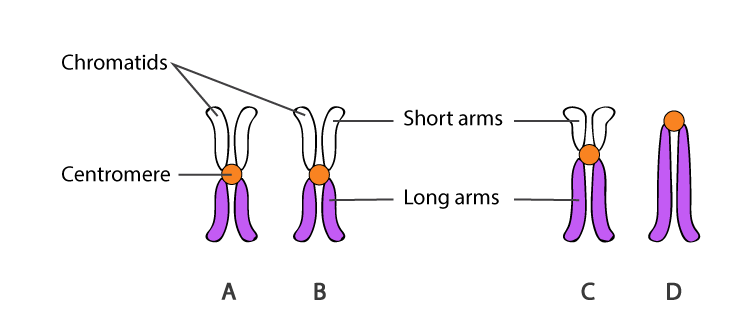
How do chromosome staining techniques like Giemsa stain provide insight into chromosome structure?
Giemsa staining reveals distinct banding patterns on chromosomes, where darker bands (heterochromatin) indicate more compact DNA and lighter bands (euchromatin) indicate less compact, more accessible DNA regions.

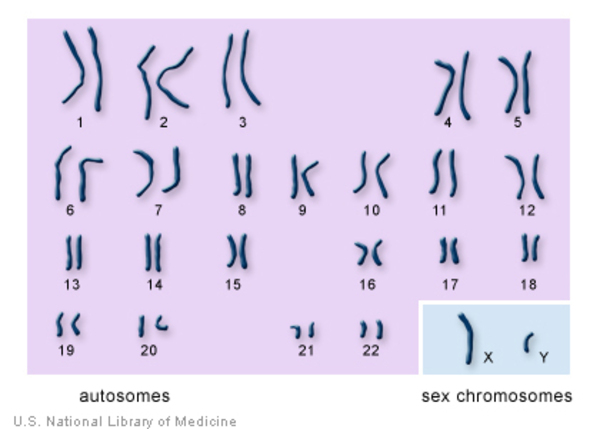
How many chromosomes are in the human genome?
The human genome contains 24 different chromosomes (22 autosomes — any chromosome that is not a sex chromosome and 2 sex chromosomes), 3.2 billion base pairs, and codes for about 21,000 genes.
What are intergenic regions and what are its major components?
Intergenic regions are segments of DNA located between genes that do not code for proteins.
They can influence chromatin structure and regulate nearby genes, although many have no known function
Major components of intergenic regions include:
tandem repeats, which are short, repeated DNA sequences,
and transposons, which are DNA sequences that can move around the genome.


What is a gene and what does it include?
A gene is a specific sequence of DNA that contains the instructions for making a gene product, which can be either a protein or a noncoding RNA. This sequence includes:
Regulatory Regions: Sections such as promoters and transcription stop sites that control when and how much the gene is expressed.
Coding Region: The part of the gene that is transcribed into RNA and then translated into a protein, or that functions directly as a noncoding RNA.
These components work together to ensure the proper production and regulation of the gene product.

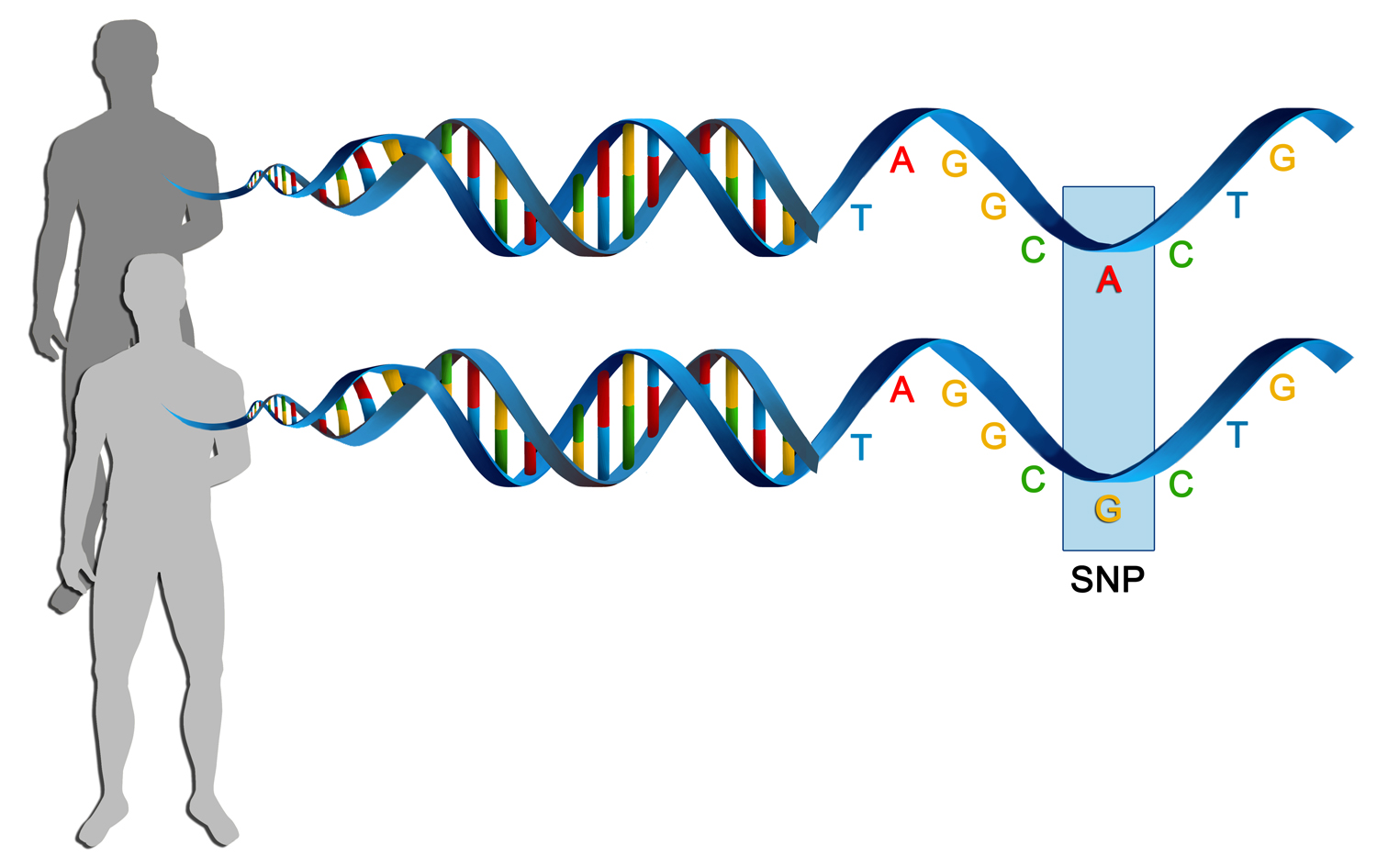
What are SNPs and how frequent are they in the human genome?
SNPs, or single nucleotide polymorphisms, are variations where one nucleotide in the genome differs between individuals. They occur approximately once every 1,000 base pairs in the human genome.
Most SNPs have no noticeable effect on health or development, and many are found in noncoding regions of DNA or in regions of genes that do not affect gene function.
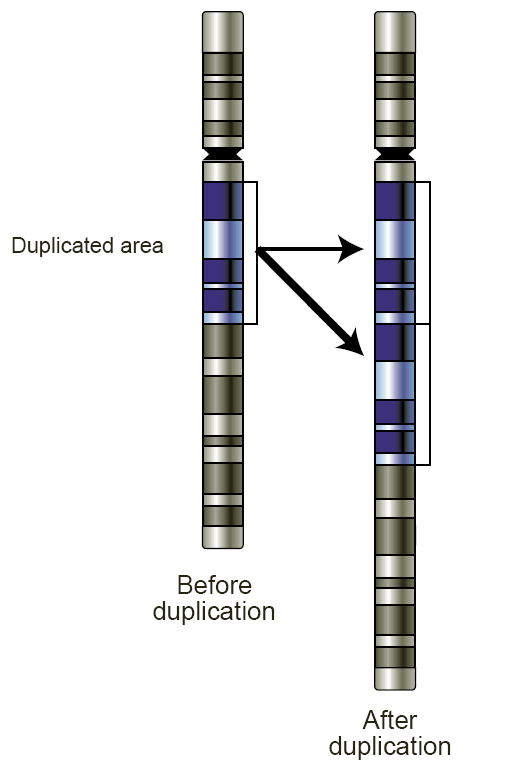
What are CNVs and what can they cause?
CNVs, or copy-number variations, are structural variations in the genome where large sections (103 to 106 base pairs) can be duplicated or deleted — duplications or deletions
They are associated with diseases such as cancer and affect genes involved in immune function and brain activity.
Ex: Autism, breast cancer, schizophrenia, epilepsy, lung cancer
What are tandem repeats and why are they important?
Tandem repeats are regions where short sequences of nucleotides are repeated one after another.
They can be unstable and lead to chromosome breaks, but are also useful in DNA fingerprinting — DNA fingerprinting is a technique used to identify individuals by analyzing unique patterns in their DNA.

What are transposons and how do they function?
Transposons are mobile genetic elements that can jump around the genome, causing mutations and chromosome changes.
They contain a gene for transposase, an enzyme that facilitates their movement.
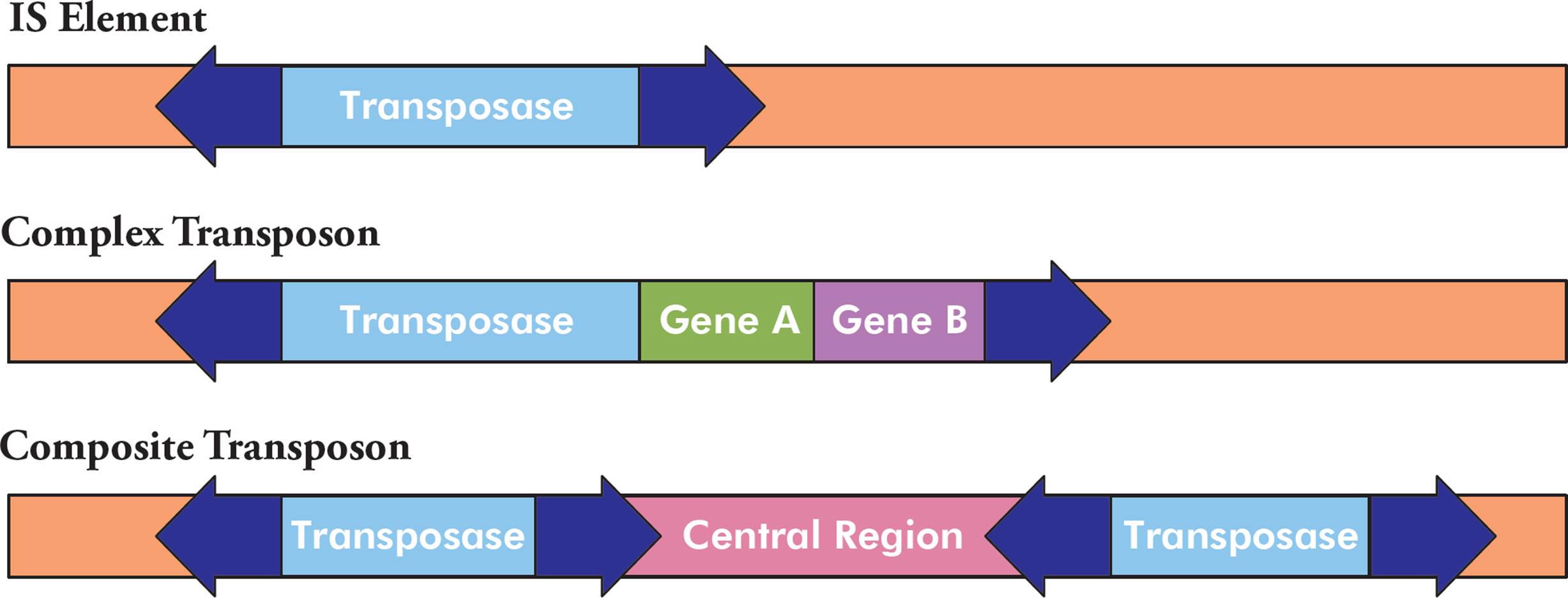
What are the three common types of transposons?
IS Elements (Insertion Sequences):
Structure: Simple elements with inverted repeats and a transposase gene.
Function: Move within the genome, potentially disrupting genes or regulatory regions. Can affect gene expression and lead to gene duplication during meiosis.
Complex Transposons:
Structure: Larger elements with extra genes, such as antibiotic resistance genes.
Function: Move around and carry additional genes, disrupting gene function and influencing gene expression.
Composite Transposons:
Structure: Two IS elements flanking a central region with additional genes.
Function: Move as a unit, carrying new combinations of genetic material, which can lead to changes in gene function or expression.
Key Difference:
IS Elements: Simple and do not carry extra genes.
Complex Transposons: Larger and carry additional genes within the same unit.
Composite Transposons: Consist of two IS elements surrounding a central region with extra genes, moving as a combined unit.
What is the function of the transposase enzyme?
Transposase catalyzes the mobilization of the transposon by cutting and pasting it from the donor site to a new acceptor site in the genome.
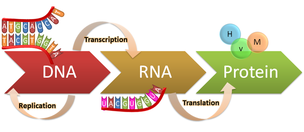
What does the Central Dogma of molecular biology state?
The Central Dogma states that genetic information flows from DNA to RNA to protein.
What did Oswald Avery demonstrate regarding DNA in 1944?
Oswald Avery showed that DNA, not protein, carries genetic information, by proving that pure DNA from one type of bacteria could transform another type.
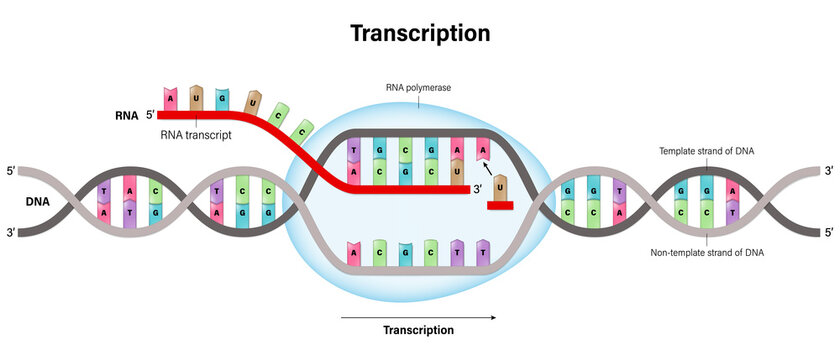
What is transcription in molecular biology? Where does it occur in eukaryotes and prokaryotes?
Transcription is the process where DNA is used as a template to synthesize RNA molecules
Occurs in the: nucleus/eukaryotic & cytoplasm/prokaryotic
What is translation and where does it occur?
Translation is the process where mRNA is decoded by ribosomes to produce proteins.
It occurs in the cytoplasm of cells.
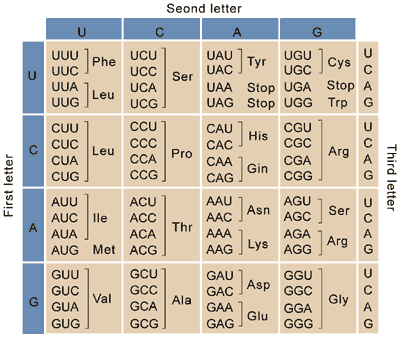
How does the genetic code specify the building blocks of proteins?
The genetic code uses three-letter words (codons) to specify each of the 20 amino acids. These codons are read by ribosomes during translation
What is the relationship between codons and anticodons?
In translation, messenger RNA (mRNA) is used to build proteins in ribosomes.
A type of RNA that brings amino acids to the ribosome. Each tRNA has a specific region called an anticodon.During translation, the anticodon on a tRNA molecule pairs with its complementary codon on the mRNA.
Anticodons on tRNA molecules base pair with codons on mRNA during translation, ensuring the correct amino acid sequence in the synthesized protein.

Why is the genetic code considered degenerate?
The genetic code is degenerate because multiple codons can code for the same amino acid, due to the redundancy in the third nucleotide of the codon (wobble pairing).
Ex: Alanine can be coded for by the codons GCU, GCC, GCA AND GCG

What are start and stop codons, and what do they signify?
Start codons (e.g., AUG) initiate protein synthesis, while stop codons (e.g., UAA, UAG, UGA) signal the termination of translation.
Are there variations in the genetic code used by different organisms?
Yes, some organisms use alternative genetic codes different from the standard code, and mitochondria use a slightly modified code.
What processes go beyond the Central Dogma of molecular biology?
Processes like reverse transcription in retroviruses (RNA to DNA), DNA methylation (epigenetic modification of DNA), and post-translational modifications of proteins (chemical changes after protein synthesis) are not explicitly covered because they involve additional layers of regulation and information transfer beyond the simple linear sequence flow described by the Central Dogma.
What is meant by semiconservative replication of DNA?
Each new DNA molecule consists of one strand from the original DNA molecule and one newly synthesized strand.
![]()
Name an enzyme responsible for unwinding the double helix during DNA replication.
Helicase unwinds the double helix and separates the DNA strands during replication.

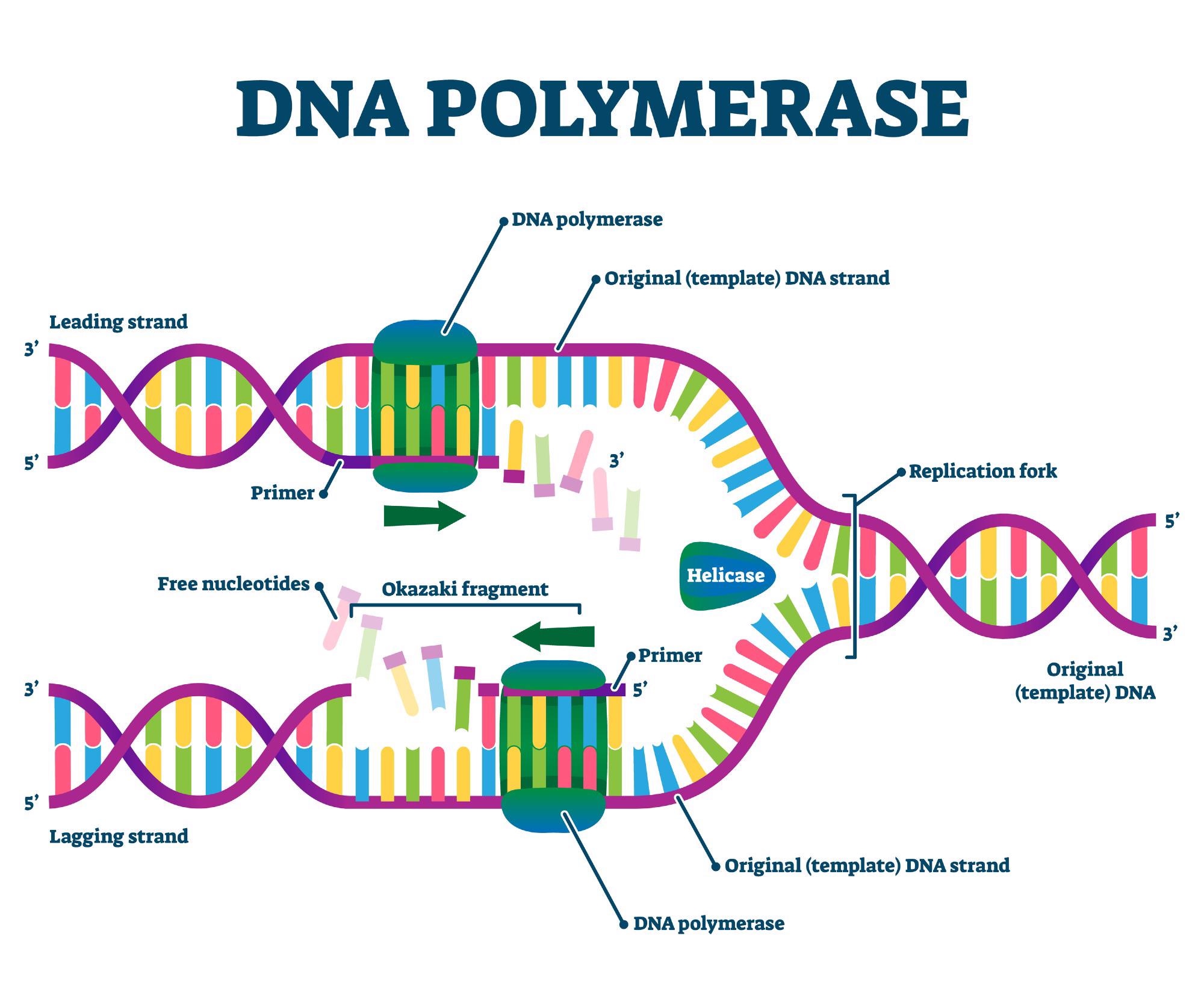
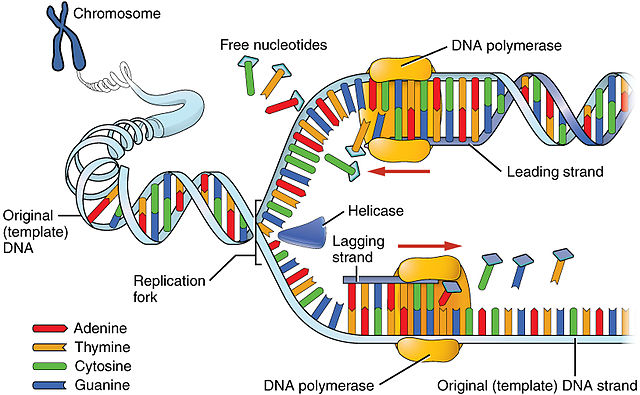
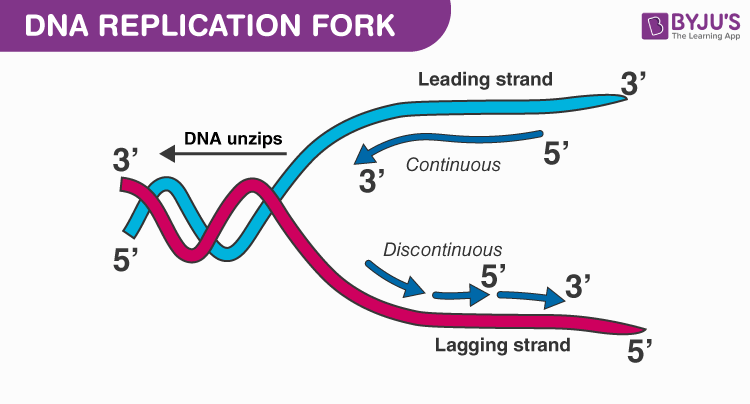
What are leading and lagging strands in DNA replication?
The leading strand is synthesized continuously in the 5' to 3' direction, while the lagging strand is synthesized discontinuously in Okazaki fragments.

What are Okazaki fragments?
Okazaki fragments are short DNA fragments on the lagging strand synthesized discontinuously during DNA replication.
What are the primary functions of DNA polymerase during DNA replication?
DNA polymerase synthesizes new DNA strands using a template, performs proofreading by removing incorrect nucleotides, and repairs mistakes during replication.
How does DNA replication differ between eukaryotes and prokaryotes?
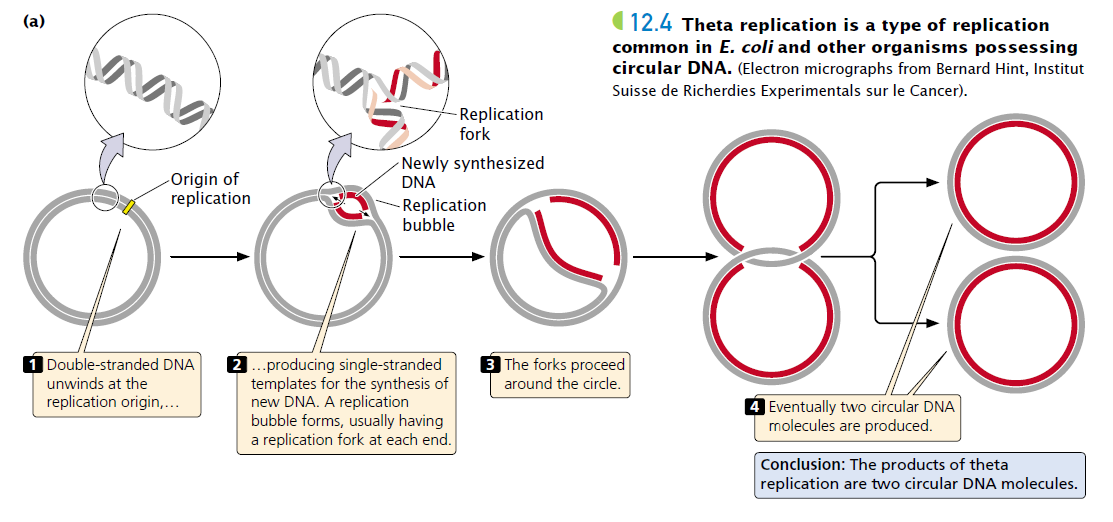
Eukaryotes have multiple origins of replication per chromosome, while prokaryotes have a single origin due to their circular chromosome.
Eukaryotic replication creates replication bubbles, while prokaryotic replication follows a theta mechanism.
What are telomeres, and what role does telomerase play in their maintenance?
Telomeres are repetitive DNA sequences at the ends of chromosomes that protect them from degradation.
Telomerase is an enzyme that adds repetitive nucleotide sequences to telomeres, counteracting their shortening during each round of cell division.
What is the advantage of DNA polymerase's proofreading activity during replication?
DNA polymerase's proofreading activity increases the fidelity of DNA replication by correcting errors in nucleotide incorporation.

What are replication bubbles, and how are they formed during DNA replication?
Replication bubbles are regions of DNA where replication forks have initiated and DNA is being synthesized.
They are formed as replication forks progress bidirectionally from origins of replication.
What is the role of helicase in DNA replication?
Helicase unwinds the double-stranded DNA helix ahead of the replication fork by breaking hydrogen bonds between complementary bases.

What is the function of topoisomerase during DNA replication?
Topoisomerase cuts one or both DNA strands to relieve the tension created by helicase unwinding the DNA helix.
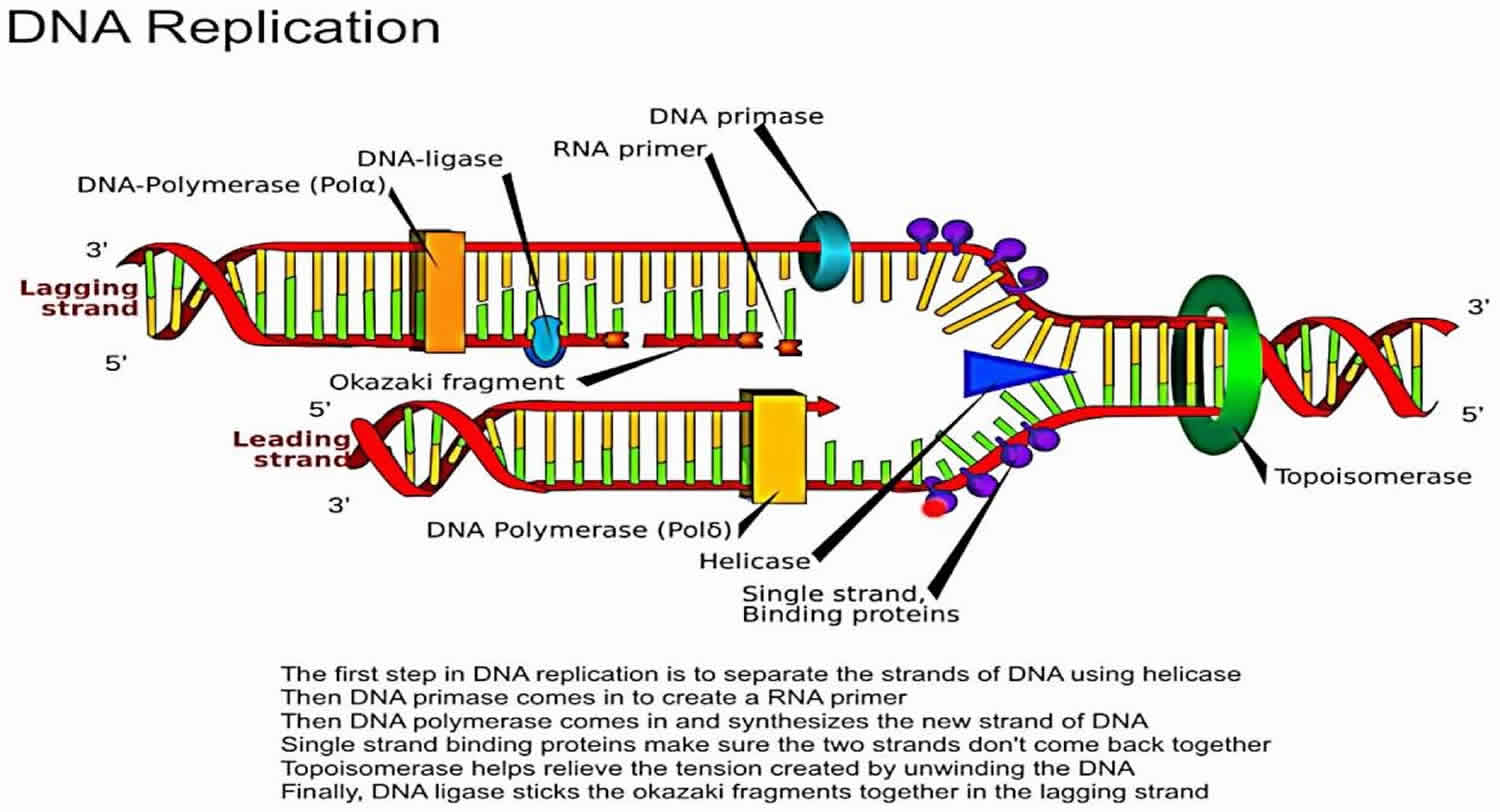
What is the role of single-strand binding proteins in DNA replication?
Single-strand binding proteins stabilize and protect single-stranded DNA during replication, preventing it from reannealing or forming secondary structures.

What is the origin of replication (ORI), and how is it recognized?
The ORI is a specific sequence where replication begins.
Proteins recognize this sequence and recruit helicase and other enzymes to initiate replication.

What is the role of primase in DNA replication?
Primase synthesizes a short RNA primer to provide a starting point for DNA polymerase to begin DNA synthesis.
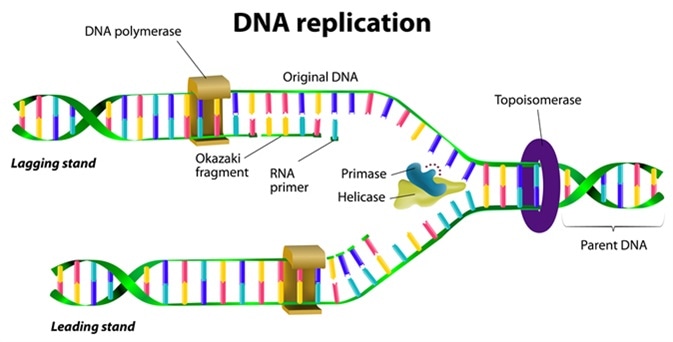
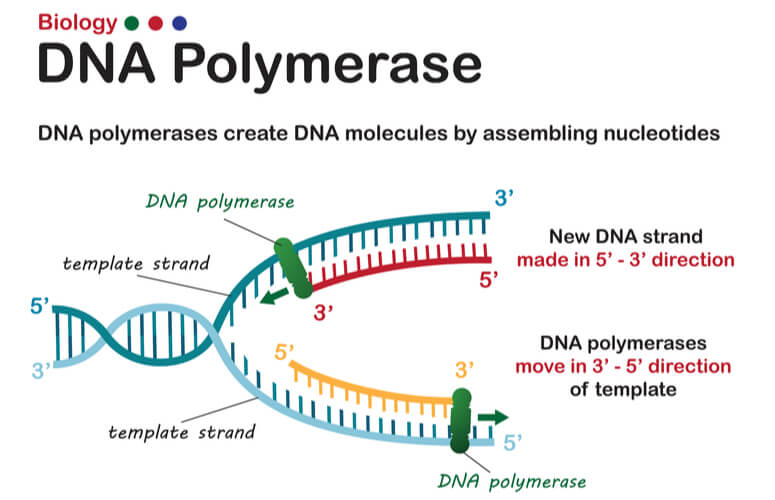
What is the primary function of DNA polymerase in replication?
DNA polymerase synthesizes the new DNA strand by adding nucleotides to the 3' end of the RNA primer, using the parental DNA as a template.

In which direction does DNA polymerization occur?
DNA polymerization occurs in the 5' to 3' direction, meaning new nucleotides are added to the 3' end of the growing strand.
How do the leading and lagging strands differ during DNA replication?
The leading strand is synthesized continuously towards the replication fork, while the lagging strand is synthesized discontinuously away from the fork in short segments called Okazaki fragments.

What is the role of DNA ligase in DNA replication?
DNA ligase joins Okazaki fragments on the lagging strand, forming a continuous DNA strand by sealing the nicks between fragments.

Compare the functions of DNA polymerase I and DNA polymerase III in prokaryotes.
DNA Polymerase ΙΙΙ: the primary enzyme involved in DNA replication.
DNA Polymerase Ι: this enzyme is involved in the processing of Okazaki fragments generated by lagging strand synthesis.
Why is proofreading by DNA polymerase important during replication?
Reduces errors in DNA replication by removing incorrectly paired nucleotides and increasing the fidelity of DNA synthesis.
Why do eukaryotic chromosomes have multiple origins of replication?
Eukaryotic chromosomes have multiple origins of replication because they are large, and multiple origins allow for faster replication of the entire chromosome.
Describe theta replication in prokaryotes.
In prokaryotes, theta replication involves the circular DNA forming a theta-shaped structure with replication forks proceeding bidirectionally from a single origin.

What are replication forks, and how do they function in DNA replication?
Replication forks are the areas where the DNA double helix is unwound and new strands are being synthesized.
They move away from the origin of replication as replication proceeds.
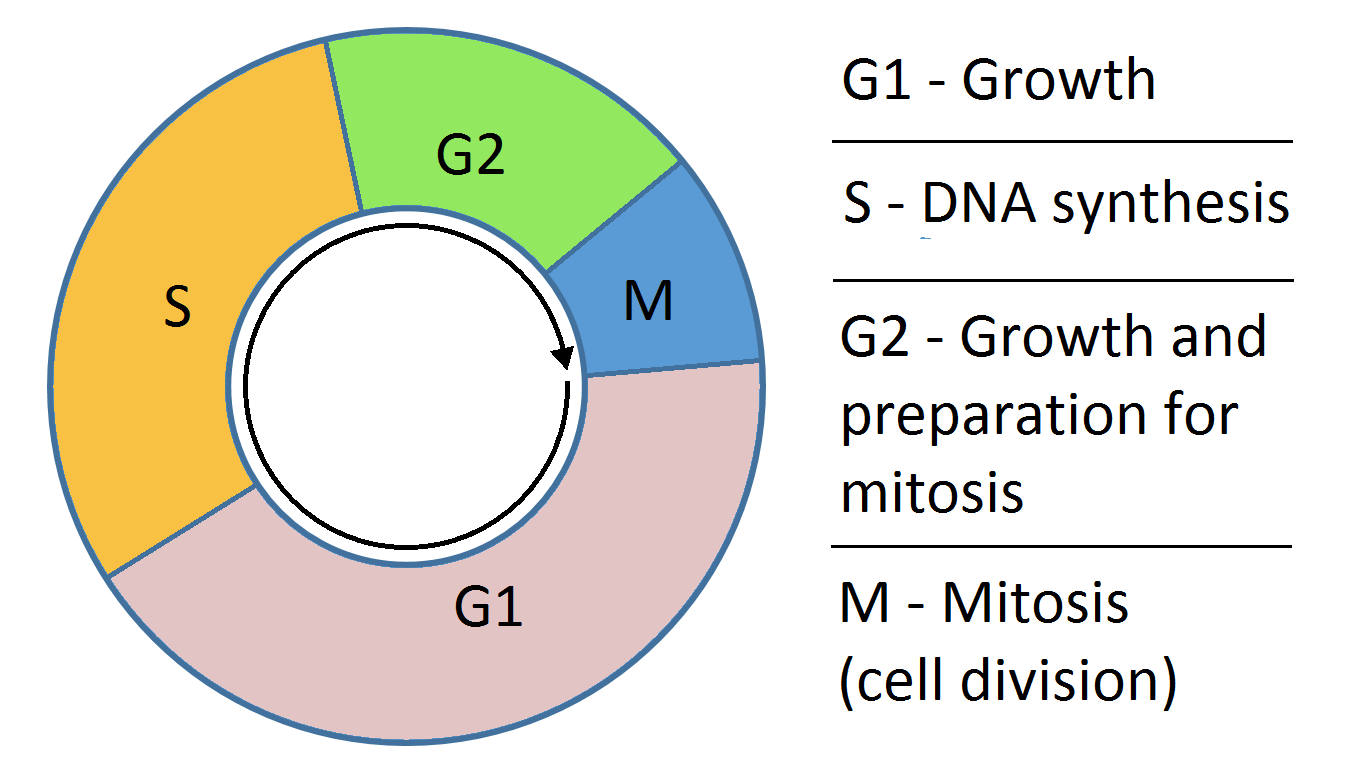
During which phase of the cell cycle does DNA replication occur?
DNA replication occurs during the S (synthesis) phase of interphase in the cell cycle.
What is a primosome, and what is its role in DNA replication?
A primosome is a complex of proteins, including primase, that synthesizes RNA primers necessary for DNA polymerase to begin DNA synthesis.
Why are RNA primers necessary for DNA replication?
RNA primers provide a starting point with a free 3' hydroxyl group for DNA polymerase to begin adding DNA nucleotides.
What are the functions of DNA polymerase II, IV, and V in prokaryotes?
DNA Pol II participates in DNA repair and serves as a backup for Pol III.
DNA Pol IV and V stall replication at forks during DNA repair and have additional repair functions.
What happens when telomeres become too short? How does cancer bypass this?
When telomeres become too short, they can trigger cellular senescence, a state where cells stop dividing and enter a state of growth arrest.
If the cell continues to divide despite critically short telomeres, it can lead to genomic instability.
This instability can trigger a DNA damage response, which can lead to apoptosis, the programmed cell death mechanism.
Telomere shortening is a natural part of aging and contributes to the aging process by limiting the replicative potential of cells.
In contrast, cancer cells often activate the enzyme telomerase, which extends telomeres and allows cells to divide indefinitely, bypassing apoptosis and contributing to tumor growth.

What is the Hayflick limit?
Number of times a normal human cell can divide before telomere shortening prevents further division.
What is the function of reverse transcriptase in telomerase?
Reverse transcriptase (opposite of transcription) in telomerase reads the RNA template and synthesizes DNA to extend telomeres.

Why do prokaryotes typically have a single origin of replication?
Prokaryotes have a single origin of replication because their circular chromosome is small enough to be replicated efficiently from one starting point.
What is the chromosome end replication problem, and how is it solved?
The chromosome end replication problem arises because DNA polymerase cannot replicate the very ends of chromosomes.
Telomerase solves this by extending the telomeres.
What are the 5 replication rules?
Polymerization occurs in the 5′ to 3′ direction, without exception. This means the existing chain is always lengthened by the addition of a nucleotide to the 3′ end of the chain. There is never 3′ to 5′ polymerase activity.
DNA pol requires a template. It cannot make a DNA chain from scratch but must copy an old chain. This makes sense because it would be pretty useless if DNA pol just made a strand of DNA randomly, without copying a template.
DNA pol requires a primer. It cannot start a new nucleotide chain.
Replication forks grow away from the origin in both directions. Each replication fork contains a leading strand and a lagging strand.
Eventually all RNA primers are replaced by DNA, and the fragments are joined by an enzyme called DNA ligase.
List the steps of DNA Replication:
We start at the origin recognition complex (ORC)
Helicase unwinds the DNA double helix to create a replication fork — topoisomerases helps prevent supercoiling (1: single strand cut / 2: double strand cut)
Single-strand binding proteins (SSBs) stabilize the unwound DNA strand
Primase synthesizes short RNA primers to provide starting points for DNA
DNA Pol I starts DNA synthesis by extending RNA primer with short stretch of DNA
DNA Pol II/III takes over to synthesize leading (II) / lagging strand (III)
DNA Pol II fills the gaps left by RNA primer
DNA Ligase joins the Okazaki fragments on lagging strand
Telomerase extends the telomeres

Where does DNA replication occur in prokaryotes/eukaryotes?
Prokaryotes: DNA replication occurs in the cytoplasm since they lack a nucleus.
Eukaryotes: DNA replication occurs in the nucleus.
What is the effect of ionizing radiation on DNA?
Ionizing radiation can cause single-strand breaks or double-strand breaks in DNA.
Double-strand breaks split the chromosome into two pieces, making it harder to repair.
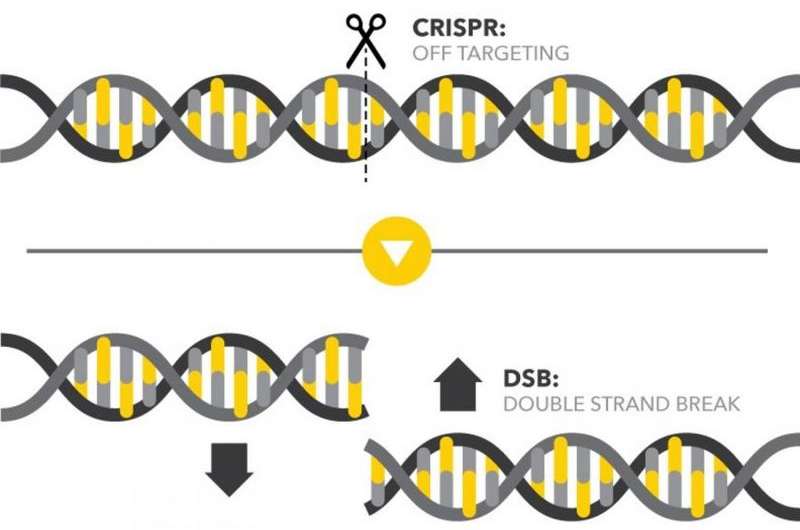
What are double-strand breaks (DSBs) in DNA?
Double-strand breaks occur when both backbones of the DNA helix are broken close to each other, splitting the chromosome into two pieces.

Why have cells evolved repair pathways?
To help deal with mutations and maintain genetic integrity.
Besides physical mutagens, what other types of mutagens can cause genetic mutations?
Can also be induced by chemical mutagens and spontaneous errors during DNA replication.
How does UV light cause damage to DNA? What are the most common occurances?
UV light causes pyrimidine dimers by directly affecting the chemical bonds between adjacent pyrimidine bases in DNA, leading to structural distortions and potential genetic mutations if not repaired.
The absorbed UV energy induces a chemical reaction that forms covalent bonds between adjacent pyrimidine bases on the same DNA strand.
This most commonly occurs between two adjacent thymine (T) bases or less frequently between two cytosine (C) bases.