MCDB 436 (15): Epigenetic Modifications
0.0(0)
Card Sorting
1/56
Earn XP
Description and Tags
Last updated 8:24 PM on 11/30/22
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
57 Terms
1
New cards
human genome
98% shared with chimps
- human genome encodes for 30k genes, C elegans 20k (many human analogues)
- human genome encodes for 30k genes, C elegans 20k (many human analogues)
2
New cards
organization of the genome
DNA (2nm) < nucleosome (11nm; histone core and DNA) < chromatin (30nm, 300nm, 700nm) < chromosome (1400nm)
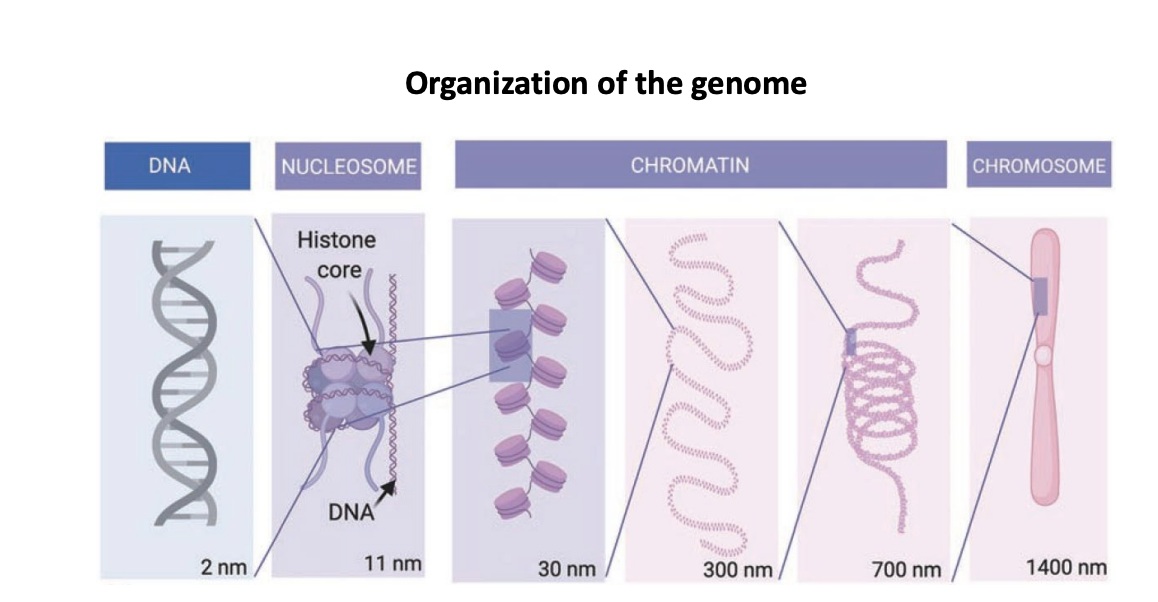
3
New cards
types of epigenetic modifications
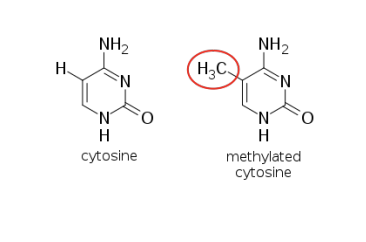
1) DNA methylation - at cytosine in CpG dinucleotide area (5mCpG)
2) histone modification - chromatin remodeling via post-translational modification of histones
2) histone modification - chromatin remodeling via post-translational modification of histones
4
New cards
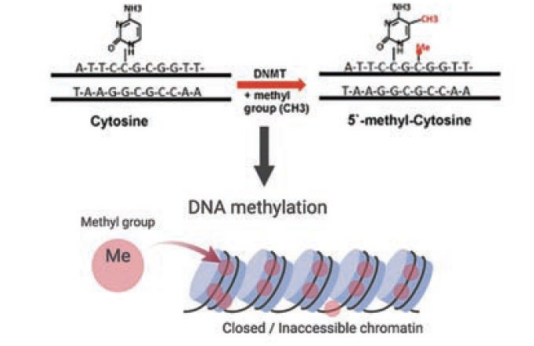
DNA methylation
(for mammals); REVERSIBLE PROCESS
DNMT enzyme catalyzes the covalent attachment of a methyl group to the C5 position residues in CpG dinuc DNA sequences
introduced methylation patterns are preserved and maintained by DNMT1 during replication
DNMT enzyme catalyzes the covalent attachment of a methyl group to the C5 position residues in CpG dinuc DNA sequences
introduced methylation patterns are preserved and maintained by DNMT1 during replication
5
New cards
DNMT1
preserve and maintain methylation patterns during replication
6
New cards
TET1-3
removes the epigenetic modification
7
New cards
DNMT3A/B
responsible for de novo methylation
- C5 position of cytosine residues in CpG dinucleotide DNA sequences
- C5 position of cytosine residues in CpG dinucleotide DNA sequences
8
New cards
effect of DNA methylation on chromatin
results in condensed chromatin, leading to transcription inactivation
9
New cards
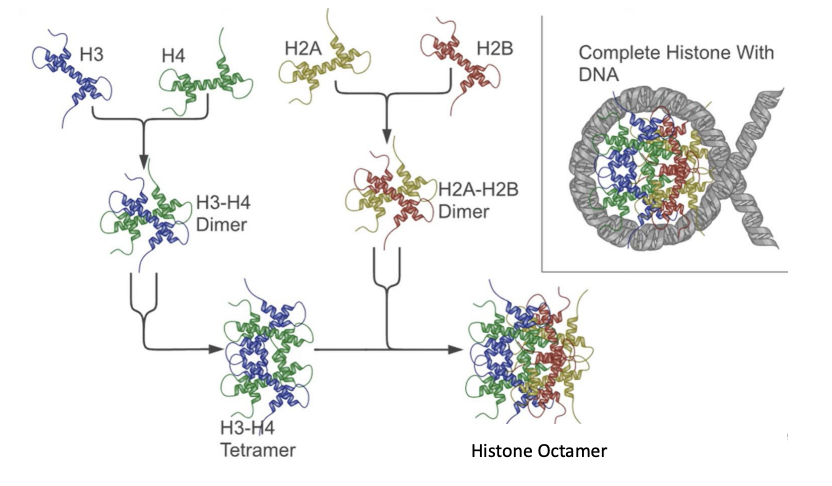
histone subunits and their products
H3 + H4 -> H3-H4 dimer -> H3-H4 tetramer
H2A + H2B -> H2A-H2B dimer ->
tetramer + dimer = histone octamer
H2A + H2B -> H2A-H2B dimer ->
tetramer + dimer = histone octamer
10
New cards
histone protein structure
histone tail: can be acetylated, methylated, phosphorylated, or ubiquitinated
histone fold
histone fold
11
New cards
four major types of histone modifications
histone acetylation - on Lys (K)
phosphorylation - on Ser/Thr (S/T)
methylation - on His/Lys/Arg (H/K/R)
ubiquitination
phosphorylation - on Ser/Thr (S/T)
methylation - on His/Lys/Arg (H/K/R)
ubiquitination
12
New cards
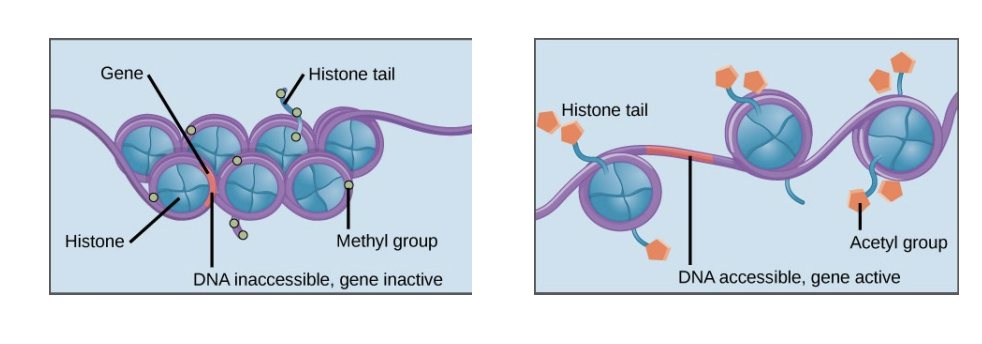
effects of histone modification on chromatin
modifications to histone tails render chromatin into open or closed conformation
- tighter wrapping = less accessible; looser wrapping = accessible DNA -> transcription
methyl = condensed
acetyl = loosened
- tighter wrapping = less accessible; looser wrapping = accessible DNA -> transcription
methyl = condensed
acetyl = loosened
13
New cards
histone methylation
catalyzed by histone methyltransferase enzyme (HMT)
- associated with activation OR repression of gene expression (depends on histone protein + AA residue)
- occurs on Arg, Lys, His
- associated with activation OR repression of gene expression (depends on histone protein + AA residue)
- occurs on Arg, Lys, His
14
New cards
Lys methylation
monomethylated (me1), dimethylated (me2), or trimethylated (me3) on epsilon-amine group
15
New cards
Arg methylation
monomethylated (me1), symmetrically dimethylated (me2s), or asymmetrically dimethylated (me2a) on guanidinyl group
16
New cards
His methylation
monomethylated (me1)
17
New cards
histone methylation abbreviations
H3K4me3
- Histidine 3 methylated
- Lysine 4 trimethylated
H4K20me1
- histidine 4 methylated
- lysine 20 monomethylated
- Histidine 3 methylated
- Lysine 4 trimethylated
H4K20me1
- histidine 4 methylated
- lysine 20 monomethylated
18
New cards
histone acetylation
dynamic process regulated by histone aceyltransferase (HAT) and histone deacetylase enzyme (HDAC)
19
New cards
HAT
RELAX chromatin -> promote interaction of RNA-polymerase and other TFs with DNA and activation of gene expression
(relax; take your hat off, stay a while)
(relax; take your hat off, stay a while)
20
New cards
HDAC
mediate CLOSED chromatin conformation -> downregulation of gene expression
(c = closed/condensed)
(c = closed/condensed)
21
New cards
cell differentiation
a biological process wherein a cell develops and acquires a more specialized form and function
- changes may include cell shape, size, membrane potential, metabolic activities, responsiveness, etc
- changes brought about by modifications in gene expressions; crucial component of the cell differentiation process
- changes may include cell shape, size, membrane potential, metabolic activities, responsiveness, etc
- changes brought about by modifications in gene expressions; crucial component of the cell differentiation process
22
New cards
differentiated cell
a cell that has changed in form and matured from being generalized into being more specific in terms of function
23
New cards
undifferentiated cell
a progenitor cell that is yet to undergo cellular differentiation
24
New cards
T cell differentiation
CD4 and CD8 T cells leave the thymus and enter circulation as resting cells in the G0 stage
- 2x as many CD4 than CD8
- 2x as many CD4 than CD8
25
New cards
naive T cell activation/proliferation
characteristics: condensed chromatin, very little cytoplasm, and little transcriptional activity
can activate by recognizing Ag-MHC complex on APC/target cell
- IL-2 -> IL2R -> proliferation -> 48hrs, enlarges into blast cell and undergoes rounds of cell division -> effector or memory cells
can activate by recognizing Ag-MHC complex on APC/target cell
- IL-2 -> IL2R -> proliferation -> 48hrs, enlarges into blast cell and undergoes rounds of cell division -> effector or memory cells
26
New cards
transformation
conversion of a normal cell into a tumor cell
- changes at cellular, genetic, and epigenetic levels and abnormal cell division
- changes at cellular, genetic, and epigenetic levels and abnormal cell division
27
New cards
6 hallmarks of cancer
1) sustaining proliferative signaling
2) evading growth suppressors
3) activating invasion and metastasis
4) enabling replicative immorality
5) inducing angiogenesis
6) resisting cell death
2) evading growth suppressors
3) activating invasion and metastasis
4) enabling replicative immorality
5) inducing angiogenesis
6) resisting cell death
28
New cards
sustaining proliferative signaling
- the accelerator signals instruct cells to grow and divide chronically
- formation of oncogenes
- formation of oncogenes
29
New cards
oncogenes
genes that transform normal cells into cancer cells
30
New cards
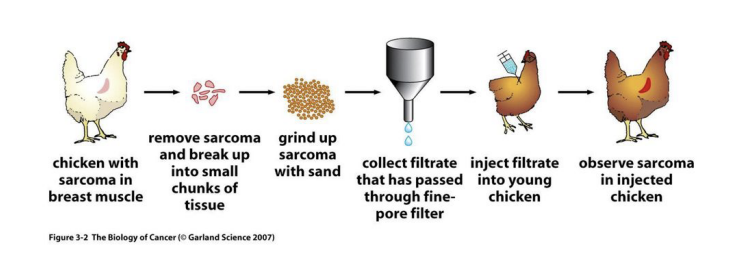
discovery of Rous Sarcoma Virus
by peyton rous, nobel prize/physiology or medicine
chicken with sarcoma in breast muscle ->
remove sarcoma and break up into small chunks of tissue ->
grind up sarcoma with sand ->
collect filtrate that has passed through fine-pore filter ->
inject filtrate into young chicken ->
observe sarcoma in injected chicken
chicken with sarcoma in breast muscle ->
remove sarcoma and break up into small chunks of tissue ->
grind up sarcoma with sand ->
collect filtrate that has passed through fine-pore filter ->
inject filtrate into young chicken ->
observe sarcoma in injected chicken
31
New cards
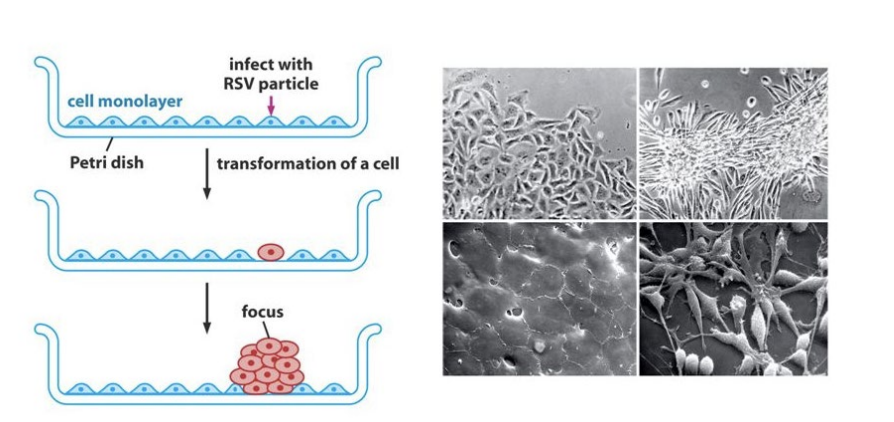
result of RSV discovery
RSV can transform infected cells into tumor cells
- with sand, the cell membrane broke -> no individual cells
result: tumor is not caused by another tumor cells. trigger of sarcoma = RSV, a single viral particle
- with sand, the cell membrane broke -> no individual cells
result: tumor is not caused by another tumor cells. trigger of sarcoma = RSV, a single viral particle
32
New cards
RSV genome
contains core proteins, reverse transcriptase, envelope protein, and Src
33
New cards
first human oncogene
Ras oncogene
34
New cards
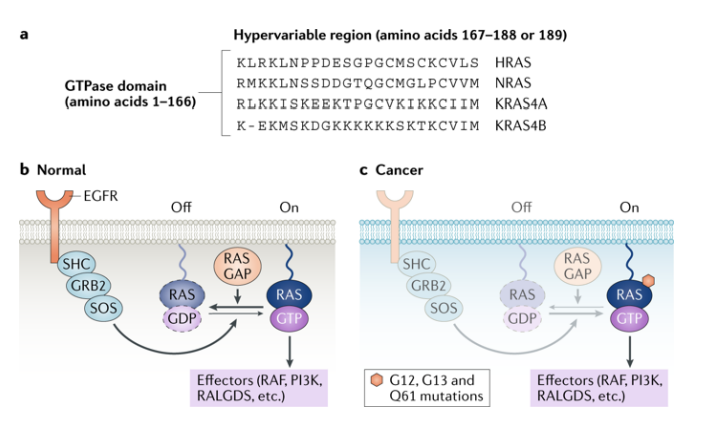
Ras oncogene
a constitutively activated form of RAS (KRAS, HRAS, NRAS). can stimulate cell growth without any growth factor
- proto-oncogene = ras, oncogene = mutated ras
- proto-oncogene = ras, oncogene = mutated ras
35
New cards
how proto-oncogenes beome oncogenes
1) mutation (point/delete/inser) -> hyperactive protein
2) gene amplification -> greatly overproduced
3) chromosome translocation (promoter/gene fusion) -> abnormal production of normal protein or novel protein
2) gene amplification -> greatly overproduced
3) chromosome translocation (promoter/gene fusion) -> abnormal production of normal protein or novel protein
36
New cards
classification of oncogenes
1) growth factors (PDGF)
2) growth factor receptors (EGFR, PDGFR)
3) signal transducers (Ras, Src, Abl)
4) cell cycle regulators (cyclin D, CDK4)
5) transcription factors (Myc, MITF)
6) anti-apoptotic regulators
2) growth factor receptors (EGFR, PDGFR)
3) signal transducers (Ras, Src, Abl)
4) cell cycle regulators (cyclin D, CDK4)
5) transcription factors (Myc, MITF)
6) anti-apoptotic regulators
37
New cards
BCL-2
oncogene; translocations lead to over-expression -> favors prolonged survival of the cell and confers resistance to the cytotoxic effect of chemotherapeutic agents
38
New cards
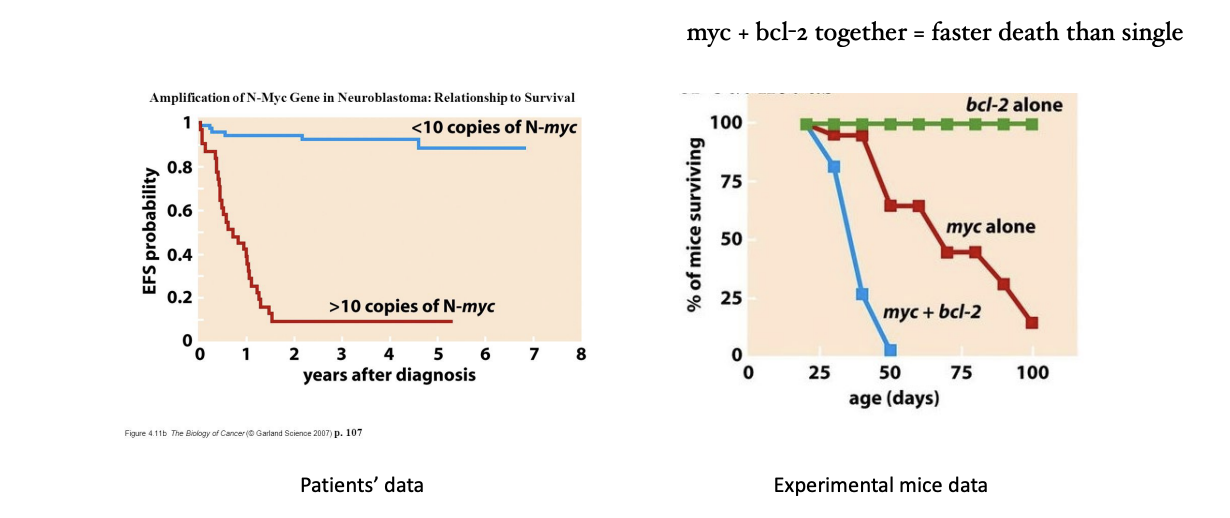
MYC expression
transcription factor oncogene
patients: > 10 copies of N-myc = lower rate of survival
mice: bcl-2 alone = lived
myc alone = less likely to live
myc + bcl-2 = guaranteed death
patients: > 10 copies of N-myc = lower rate of survival
mice: bcl-2 alone = lived
myc alone = less likely to live
myc + bcl-2 = guaranteed death
39
New cards
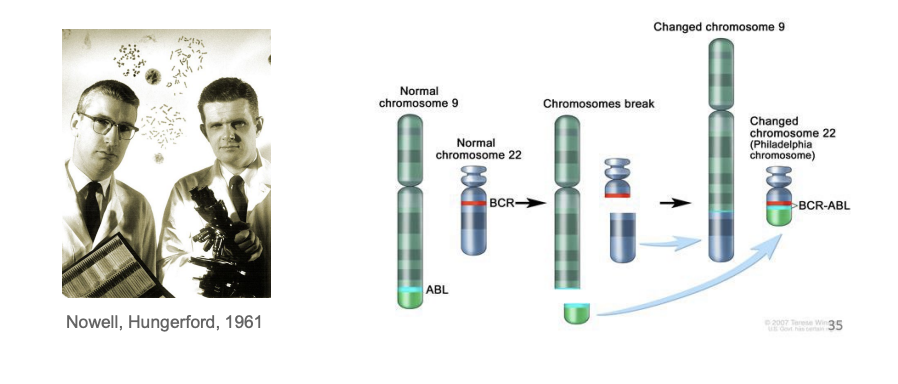
Philadelphia chromosome
ABL translocation from normal chromosome 9 onto chromosome 22
- found in 90% of chronic myeloid leukemia (CML)
- gives rise to a BCR-ABL fusion protein (p210) with constitutive tyrosine kinase activity
- inhibited by Gleevec
- found in 90% of chronic myeloid leukemia (CML)
- gives rise to a BCR-ABL fusion protein (p210) with constitutive tyrosine kinase activity
- inhibited by Gleevec
40
New cards
Gleevec/imatinib
competitive inhibitor of BCR-ABL
blocks ATP binding to enzyme -> block chromosome formation -> block downstream signaling cascade
blocks ATP binding to enzyme -> block chromosome formation -> block downstream signaling cascade
41
New cards
tumor suppressor genes
encoded proteins that negatively regulate cell proliferation in their normal state
- lost or inactivated in many tumors, contributing to abnormal proliferation of tumor cells (P53, PTEN, etc)
- lost or inactivated in many tumors, contributing to abnormal proliferation of tumor cells (P53, PTEN, etc)
42
New cards
Rb
cell cycle regulator; first human tumor suppressor gene ID'd
- retinoblastoma requires loss of both functional copies of Rb gene
- retinoblastoma requires loss of both functional copies of Rb gene
43
New cards
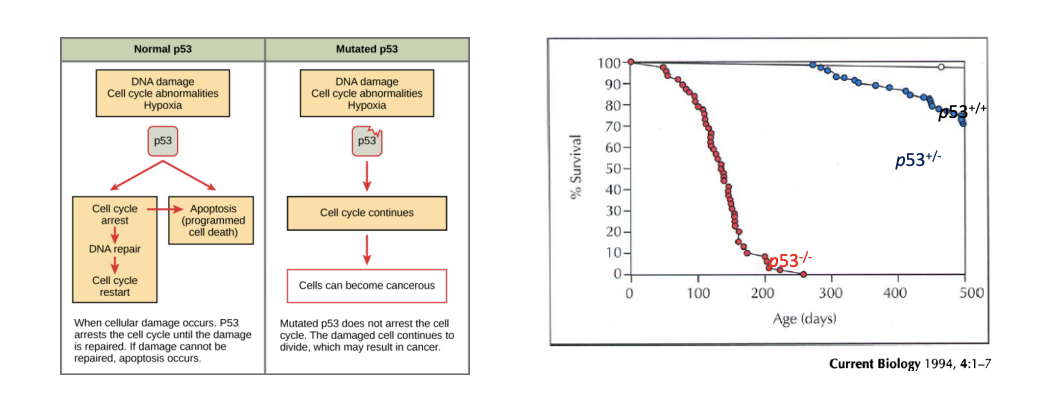
p53
inactivated in many cancers; mutations of p53 in about 50% of all cancers
- normally responsible for cell cycle arrest and apoptosis
- normally responsible for cell cycle arrest and apoptosis
44
New cards
PTEN
lipid a protein phosphatase mutated in many cancers
45
New cards
oncogenes vs tumor suppressor genes
- oncogenes: activation increases cancer risk. mutated form of proto-onco gene. gain-of-function mutation can overactivate a proto-oncogene to make it an oncogene
- tumor suppressor genes (TSG): activation decreases cancer risk; loss-of-funcion can lead to loss of activity, allowing for cancer to occur
- tumor suppressor genes (TSG): activation decreases cancer risk; loss-of-funcion can lead to loss of activity, allowing for cancer to occur
46
New cards
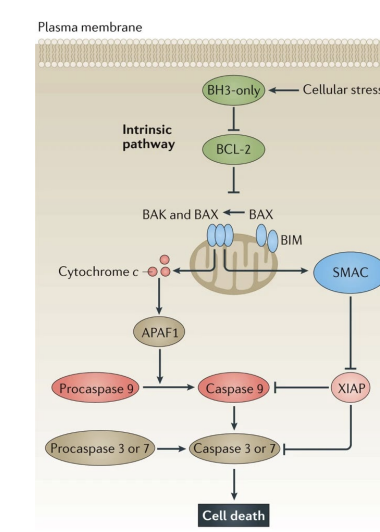
resisting cell death
avoiding assisted suicide of outlaw cells; abrogation of the inborn willingness of cells to die for the benefit of the organism
- increases expression of anti-apoptotic Bcl-2 family proteins
- down regulating pro-apoptotic Bcl-2 family members
- increases expression of anti-apoptotic Bcl-2 family proteins
- down regulating pro-apoptotic Bcl-2 family members
47
New cards
Bcl-2 and B cell lymphoma
alterations in the Bcl-2 family of proteins (increased expression -> anti-apoptosis)
48
New cards
enabling replicative immorality
circumventing a counting mechanism that disrupts continuing cell division when a set limit is reached
- telomere activity related
- telomere activity related
49
New cards
telomere
a region of repetitive nucleotide sequences at each end of a chromosome, which protects the end of the chromosome from deterioration or from fusion with neighboring chromosomes
50
New cards
telomeres and cancer
- cancer cells maintain their telomeres (90% by increasing production of telomerase)
- telomerase: adds telomeric DNA to the ends of chromosomes
- telomerase: adds telomeric DNA to the ends of chromosomes
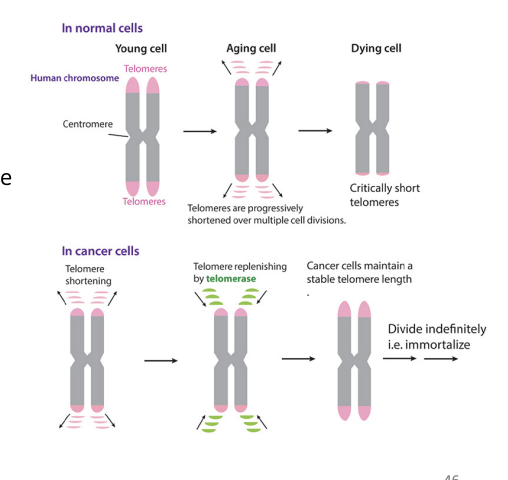
51
New cards
hTERT
telomerase reverse transcriptise
- in HEK cells: without hTERT, eventually die. with hTERT, HEK cells keep growing
- in HEK cells: without hTERT, eventually die. with hTERT, HEK cells keep growing
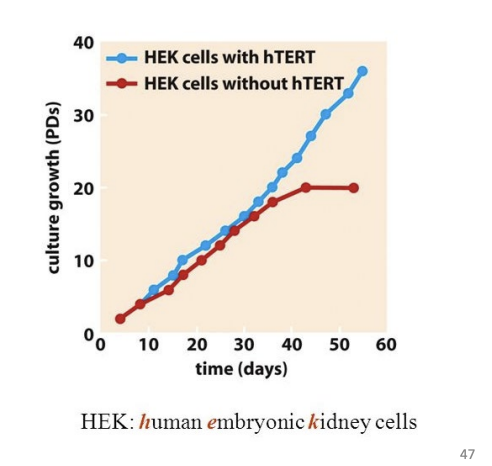
52
New cards
inducing angiogenesis
turning on new blood vessel growth to feed and nurture the growing mass of cancer cells
- hypoxia-inducible transcription factor (HIF) system
- hypoxia-inducible transcription factor (HIF) system
53
New cards
hypoxia-inducible transcription factor (HIF) system
genes that indirectly/directly induce angiogenesis and other stress-adaptive capabilities of cancer cells
1) tumor secretes VEGF
2) VEGF increases blood vessel expression and movement to tumor
3) tumor has increased blood supply
1) tumor secretes VEGF
2) VEGF increases blood vessel expression and movement to tumor
3) tumor has increased blood supply
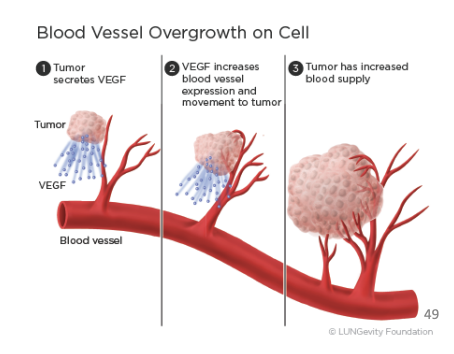
54
New cards
activating invasion and metastasis
- malignant tumor cells tend to invade locally and metastasize to other organs
- EMT and seed and soil hypothesis
- EMT and seed and soil hypothesis
55
New cards
epithelia mesenchymal transition (EMT)
epithelia cells acquire mesenchymal traits
- loss of adherent junctions, change in cellular morphology, increased motility
- roughly 90% of cancers arise as carcinomas in epithelial (surface) tissue
- loss of adherent junctions, change in cellular morphology, increased motility
- roughly 90% of cancers arise as carcinomas in epithelial (surface) tissue
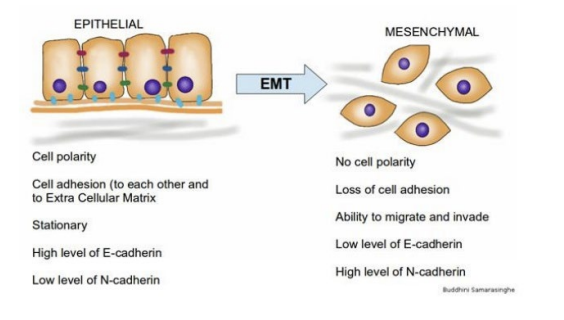
56
New cards
tumor cell invasion and metastasis
tumor splits into mesenchymal-like cancer cells of a collective invasion of epithelial-like + mesenchymal-like cancer cells -> intravasation -> metastasis -> extravasation
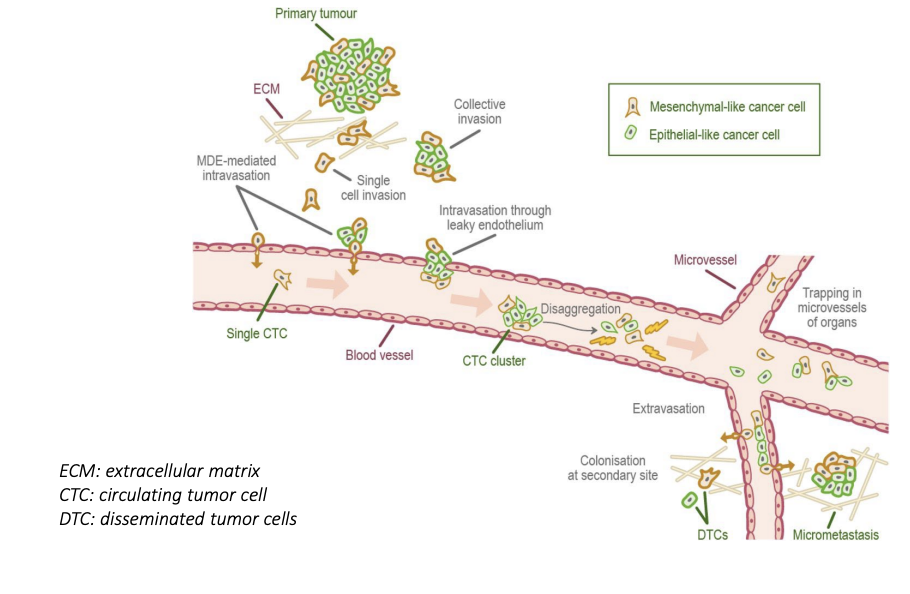
57
New cards
seed and soil hypothesis
tumor cells grow preferentially in the microenvironment of select organs
- metastases resulted only when the appropriate seed was implanted in its suitable soil
- 50% related to brain; might be because brain is a hospitable environment
- metastases resulted only when the appropriate seed was implanted in its suitable soil
- 50% related to brain; might be because brain is a hospitable environment