gene 3200: unit 3 exam
1/108
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
109 Terms
why are different genes expressed in different places?
even though the chromosomes and genes in each cell should be the same, different tissues and cell types transcribe different genes
what are genes?
sequences of DNA that are transcribed into RNA
includes the transcribed sequences as well as the sequences that control transcription
is all DNA transcribed into RNA?
no
is all RNA translated into protein?
no
what are the 5 important characteristics of genetic material?
replication (must be able to make exact copies)
stability (structural and chemical integrity)
mutability (potential for mutations for genetic variability)
expression (encodes phenotype)
heritability (passed from parent to offspring)
what are the repeating units of DNA and RNA? (3)
= nucleotides
pentose sugar
phosphate group
nitrogenous base
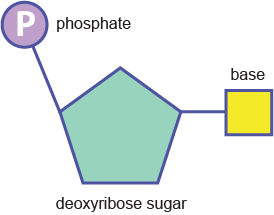
what sets RNA and DNA apart?
RNA has an extra oxygen, making DNA much more stable than RNA
what makes up the phosphodiester backbone of DNA and RNA?
pentose sugars and phosphate groups
what are the purines and their basic shape?
adenine and guanine
2 ringed nitrogenous base
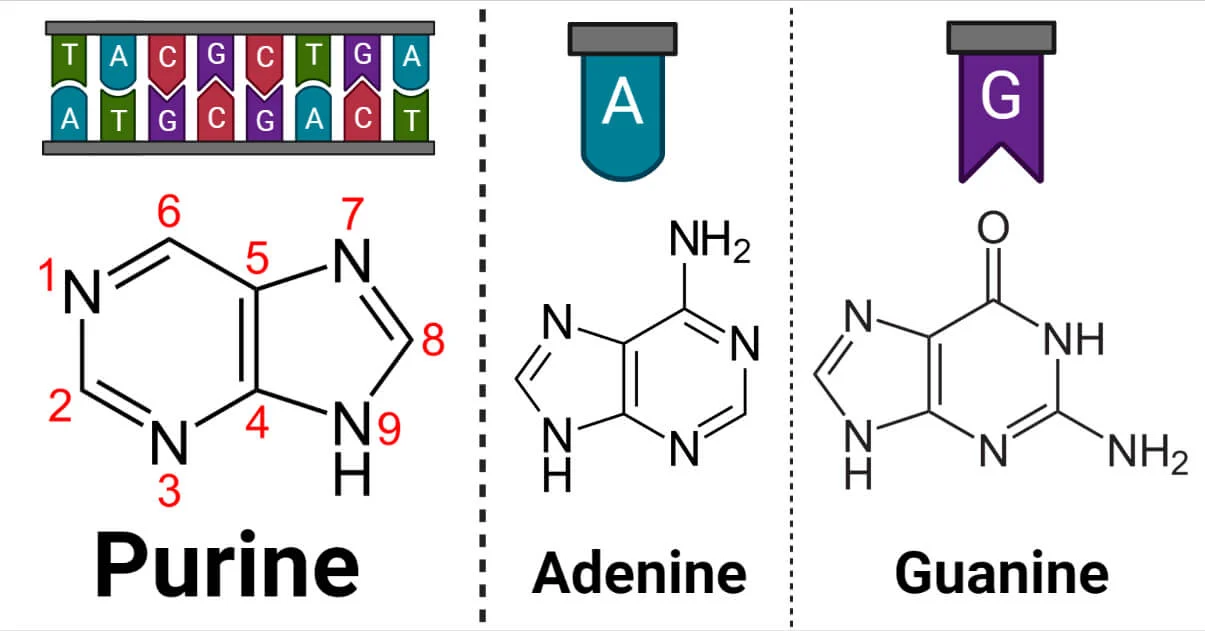
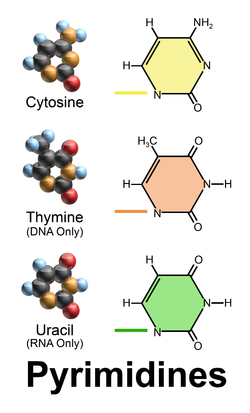
what are the pyrimidines and their basic shape?
cytosine, thymine, uracil
1 ringed nitrogenous base
what nitrogenous bases go together?
A : T = 1
G : C = 1
A + G = T + C
what does it mean to be antiparallel?
the orientation of the two strands of DNA run parallel to each other but in opposite directions
in DNA, one strand runs in the 5' to 3' direction while the other runs 3' to 5'
where is the phosphodiester bond?
bond between the 5’-PO4 and 3’-OH groups of adjoining nucleotides on each strand
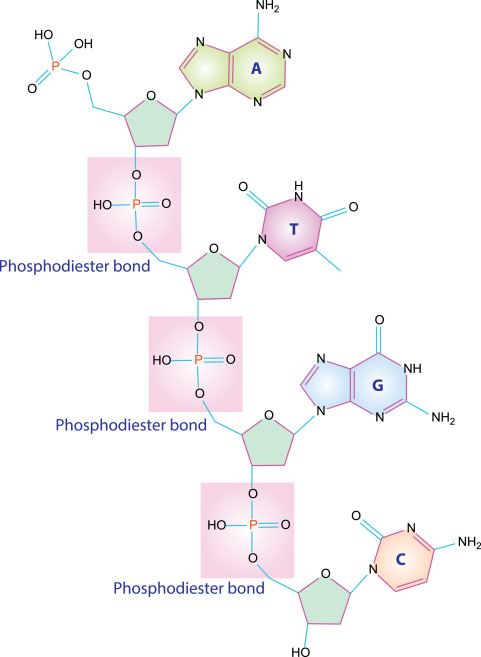
where is the hydrogen bond?
bond between complementary bases of opposite strands (intermolecular)
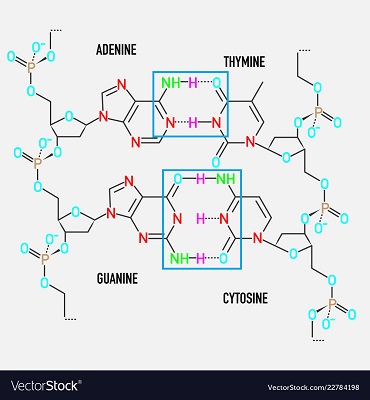
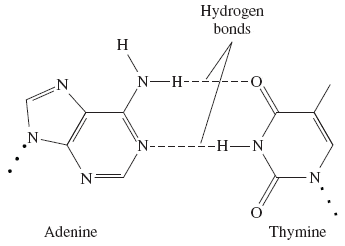
how many H bonds are between A-T base pairs?
2
how many H bonds are between G-C base pairs?
3
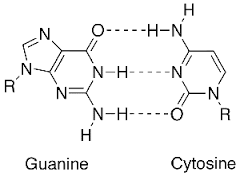
is more energy required to denature G-C pairs or A-T pairs?
G-C since its 3 H bonds
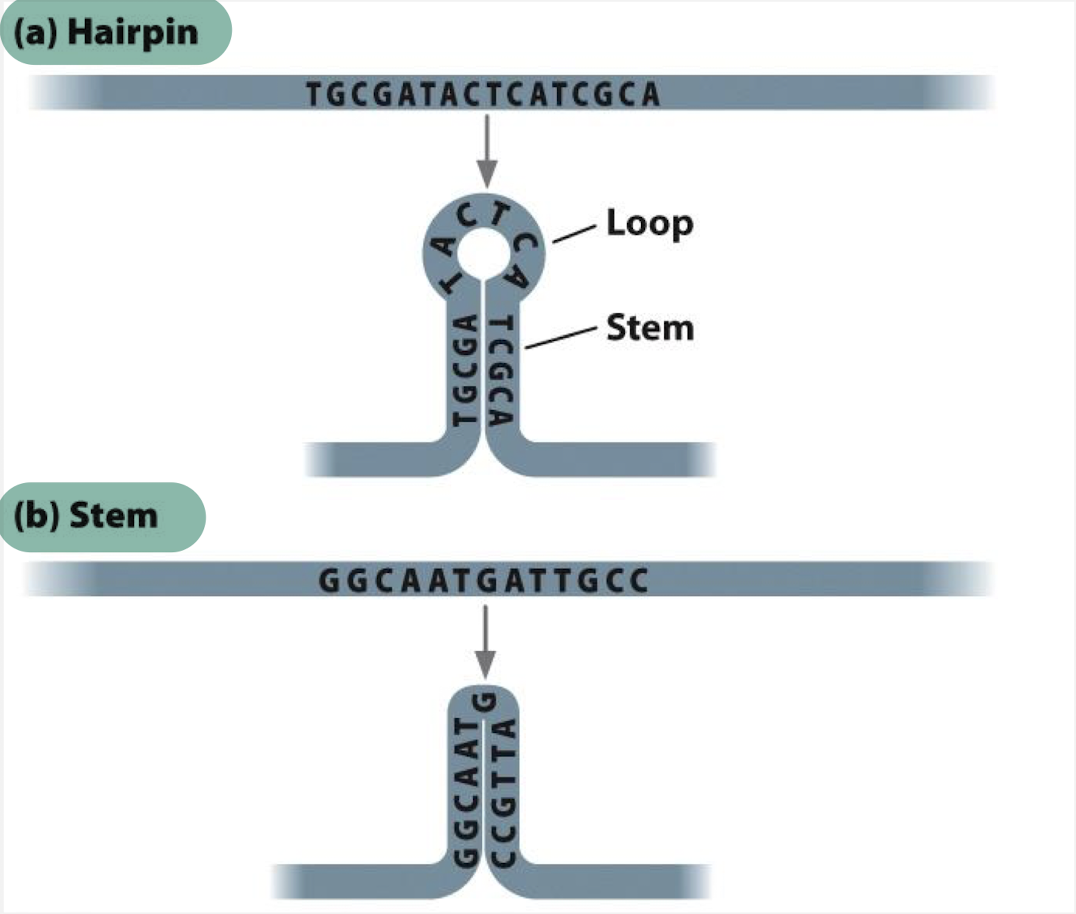
what is a hairpin in DNA?
a specific structure formed when a single strand of DNA folds back upon itself, creating a loop which is stabilized by base pairing within the same strand
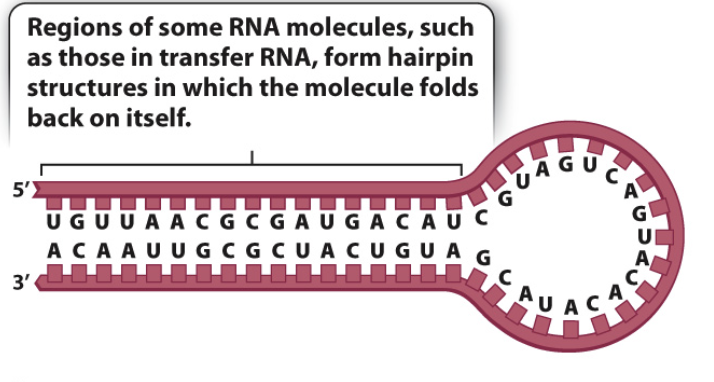
what is a stem in DNA?
a double-stranded region formed by intramolecular base pairing within a single-stranded nucleic acid molecule
how do prokaryotic vs eukaryotic genetic material differ?
location:
prokaryotic is found in cytoplasm
eukaryotic is found in the nucleus
nucleus: prokaryotic don’t have nuclei
structure:
prokaryotic is a single circular chromosome
eukaryotic is multiple linear chromosomes
packaging:
prokaryotic aren’t wrapped in histones
eukaryotic are wrapped in histones (chromatin)
replication + transcription:
prokaryotic have transcription and translation occurring simultaneously
eukaryotic have transcription in the nucleus and translation in the cytoplasm

what is supercoiled DNA?
additional winding/twisting of double-helix DNA, allowing more information to fit into a small space
what is positive supercoiling?
clockwise direction of winding, overrotated
what is negative supercoiling?
counterclockwise direction of winding, underrotated
is most DNA positively or negatively supercoiled?
negatively for easier separation of strands for replication and transcription
what are topisomerases?
add or remove rotations in DNA by breaking and then rejoining DNA strands
what is chromatin?
DNA and associated proteins of a chromosome
protein component is essential for condensation, segregation, and organization of chromosomes
what are histones?
small, positively-charged, highly conserved proteins found in all eukaryotes to bind to and neutralize negatively charged DNA
what is the composition of a chromosome?
about ½ DNA, ½ protein (1/2 of this is histone proteins)
what are the 5 types of histone proteins?
H1, H2A, H2B, H3, H4
what is euchromatin?
relaxed chromatin, usually transcriptionally active
lightly stained regions of chromosomes
what is constitutive heterochromatin?
tightly condensed, usually transcriptionally inactive (silenced)
darkly stained regions of chromosomes
highly compacted even during interphase
what is facultative heterochromatin?
condensed or relaxed under certain conditions
what is a restriction enzyme?
recognizes a specific sequence of bases anywhere within the DNA
a protein produced by bacteria that cuts DNA at specific sequences, creating fragments with predictable ends
what are recognition sites?
place where restriction enzyme cuts
each enzyme cuts at the same place relative to its specific recognition sequence
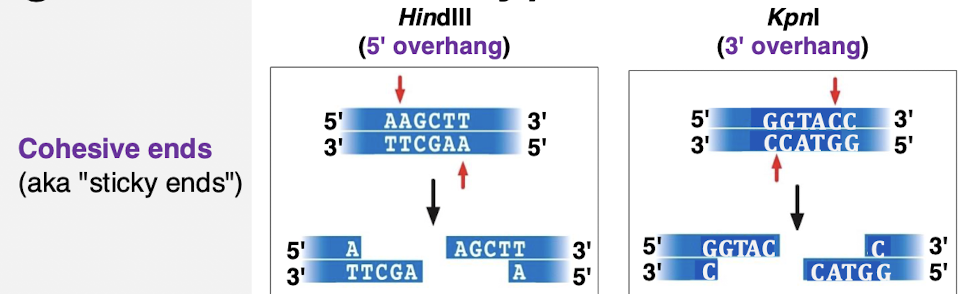
what are cohesive ends?
5’ and 3’ overhangs
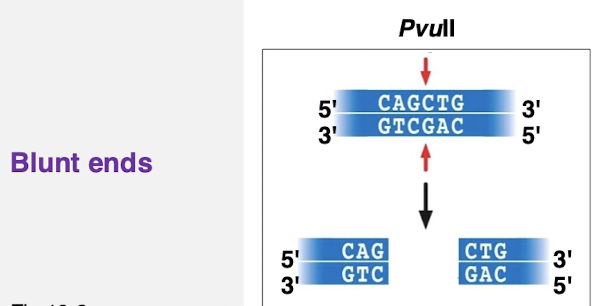
what are blunt ends?
no overhang
what is gel electrophoresis?
different types of gels can be used to separate macromolecules based on size; generates resolving fragments
what factors can affect the migration or final position of a macromolecule after gel electrophoresis?
size
molecular charge
molecular shape
what is a southern blot?
membrane that contains transferred DNA
what is a northern blot?
membrane that contains transferred RNA
what is a western blot?
membrane that contains transferred proteins
when do errors in DNA occur?
whenever information is copied—the more times it is copied, the greater the potential number of errors
why is DNA polymerase so accurate?
it has a proofreading mechanism (3’ → 5’ exonuclease)
what were the 3 proposed mechanisms for DNA replication?
semiconservative replication
conservative replication
dispersive replication
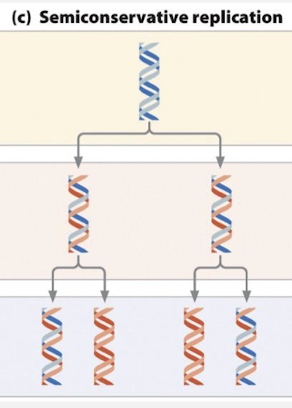
what is the semiconservative model of DNA replication?
each daughter duplex contains one parental and one daughter strand
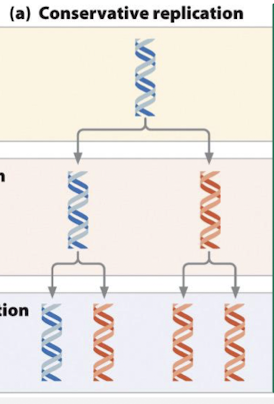
what is the conservative model of DNA replication?
one daughter duplex contains both parental strands and the other contains both daughter strants
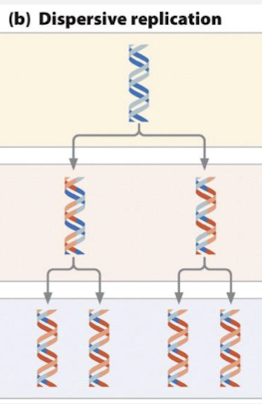
what is the dispersive model of DNA replication?
each daughter duplex contains interspersed parental and daughter segments
what was the meselson-stahl experiment?
confirmed DNA was semiconservative
grew e. coli with a heavy and a light nitrogen isotope
in the first generation, DNA only showed one intermediate band and not 2 light and heavy bands, ruling out the conservative model
in the second generation, DNA showed both intermediate and light bands, ruling out dispersive model
where does theta replication occur?
in E. coli and other bacteria
single origin of replication
bidirectional replication from the origin
where does linear replication occur?
in all eukaryotes
where is replication initiated?
at origins (Ori) and is initiated with an RNA primer
how does DNA polymerase execute replication?
is template driven through H-bonding, occurs in the 5’ → 3’ direction, and requires free 3’-OH
catalyzes the formation of phosphodiester bonds with the 3’-OH of the new strand and the 5’-PO4 of the next nucleotide
part of the strand is synthesized continuously while other parts of the same strand are synthesized semi-discontinously
what are the 4 stages of DNA replication in E. coli?
initiation: OriC
unwinding: single strand binding proteins, helicase, DNA gyrase
elongation: primase, RNA primer, DNA pol III, leading strand, lagging strand, okazaki fragments, DNA pol I, ligase
termination
what happens in the initiation phase of E. coli DNA replication?
an initiator protein binds to the a single replication origin and causes a short section of DNA to unwind
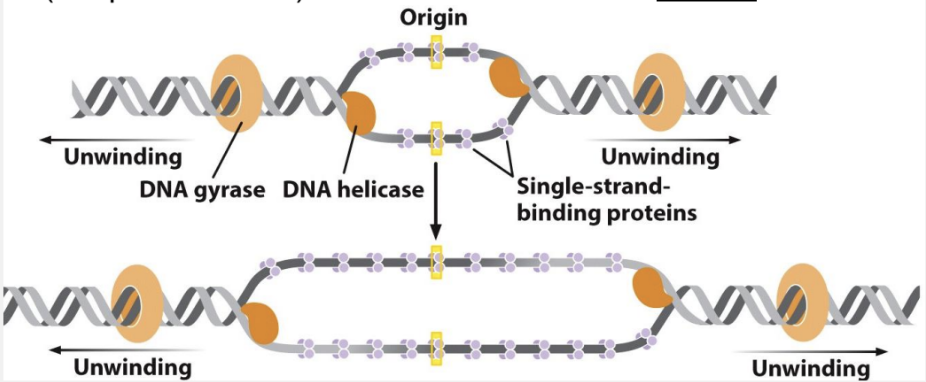
what happens in the unwinding phase of E. coli DNA replication?
helicase binds to the lagging strand and moves in the 5’ → 3’ direction to break H bonds, single stranded binding protein stabilize single stranded regions, DNA gyrase relieves torsional strain in front of each fork
what happens in the elongation phase of E. coli DNA replication?
DNA synthesis is initiated at RNA primers and synthesized by primase
a new primer is needed for each leading strand and each okazaki fragment
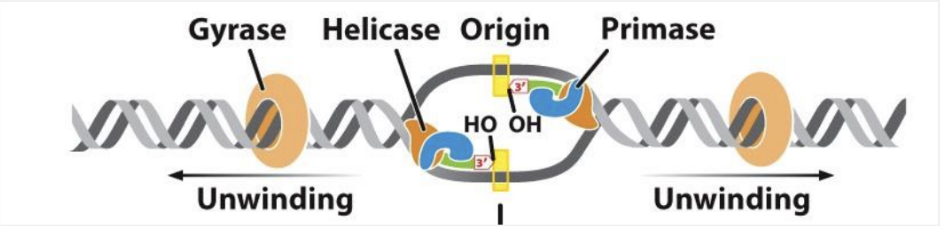
what are the characteristics of RNA primers?
complementary and antiparallel to each template strand
what is the leading strand?
synthesis of one daughter strand occurs continuously in the same direction as the replication fork progresses
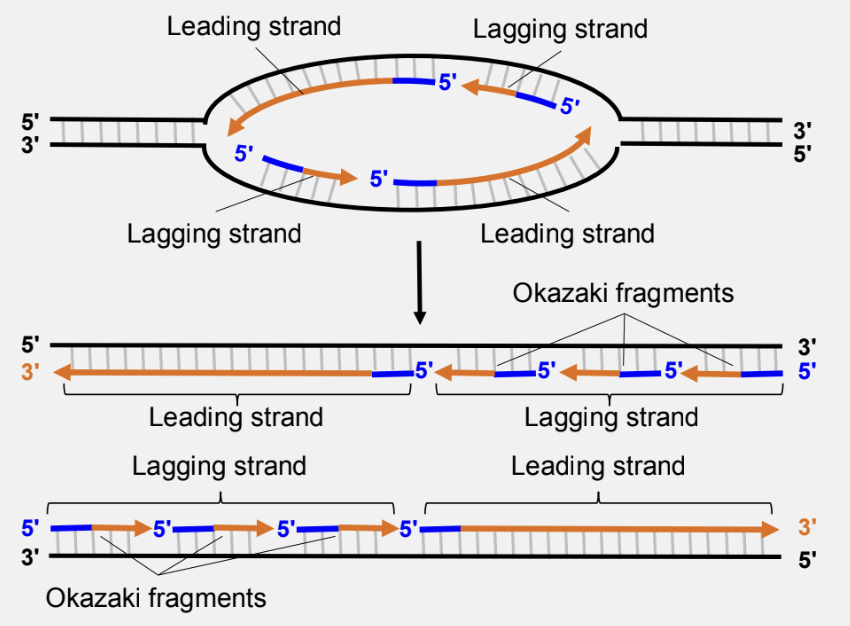
what is the lagging strand?
synthesis of the daughter strand occurs discontinuously, in the opposing direction to fork progression, via short segments called okazaki fragments
what does DNA polymerase I do?
removes and replaces primers
completes replication by using:
5’ to 3’ exonuclease to remove the RNA primers
5’ to 3’ polymerase activity to add DNA nucleotides to the 3’ end of the DNA segment preceding the primer
what does DNA ligase do?
catalyzes formation of a phosphodiester bond between adjacent DNA segments (3’-OH and 5’-PO4)
what does DNA polymerase III do?
elongates DNA
what is the origin-recognition complex (ORC)?
binds to origins to initiate DNA replication during G1 of interphase (replication occurs only during S phase)
what are telomeres?
consist of specific repetitive DNA sequences and don’t contain genes in order to maintain integrity of chromosomal ends and protect the ends of chromosomes to prevent chromosome fusion
what is the end-replication problem?
in linear chromosomes, the lagging strand cannot fully replicate its 5’ end after primer removal, causing gradual shortening of chromosomes after each replication cycle
alleviated by telomeres and telomerases
telomeres contain a 3’ G-rich overhang that folds over and forms a t-loop (a lasso-like structure formed at the ends of chromosomes) by H-bonding with the complementary strand
a reverse transcriptase of telomerase (TERT) copies the DNA template on htR to add DNA repeats onto the telomere end
hTR is the RNA component of telomerase that contains the sequence that is complementary to the tandem repeat on the telomere end
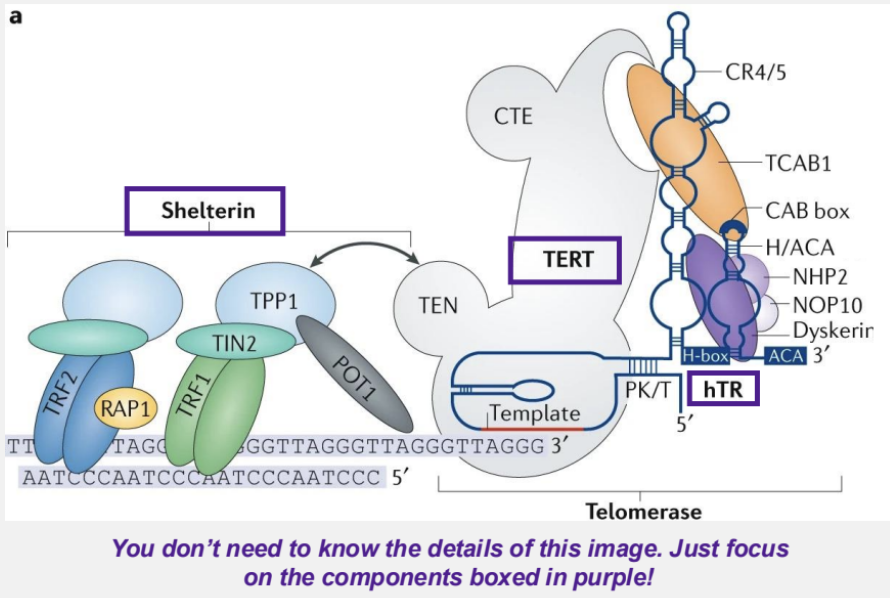
what is the shelterin complex?
6 proteins that bind the t-loop and help to:
mediate t-loop formation
protect the end sequences
recruit and regulate telomerase
what is the TERT gene?
provides instructions for making telomerase reverse transcriptase but also can give you cancer
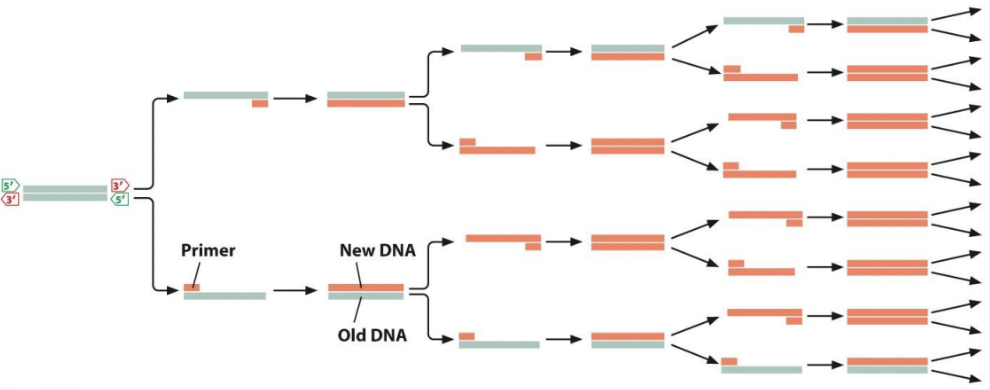
what is the polymerase chain reaction (PCR)?
a lab technique used to make many copies of a specific DNA segment
what is required for PCR?
a dsDNA template containing the target sequence to be amplified
a mixture of all 4 dNTPs
2 primers for opposite strands of the target DNA
an aqueous buffer solution
thermocycler
a thermo-stable DNA polymerase (most common is Taq polymerase, which lacks 3’ → 5’ exonuclease)
what are the steps in each PCR cycle?
denaturation of DNA into single strands
hybridization of primers to their complementary sequence
DNA synthesis
amount of DNA is doubled with each cycle
what are some of the limitations of PCR?
need prior knowledge of part of the sequence of target DNA
very susceptible to contamination
Taq polymerase lacks 3’ → 5’ exonuclease (proofreading)
size limitation of DNA fragments
why is RNA’s secondary structure important?
inverted repeats in RNA sequence can form intramolecular H-bonding called hairpins or stem-loops
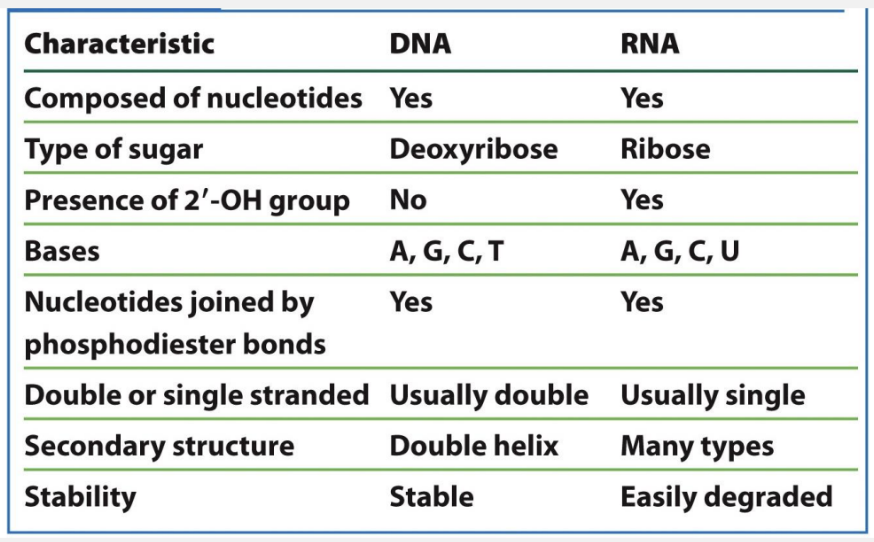
what are some comparisons between DNA and RNA structures?
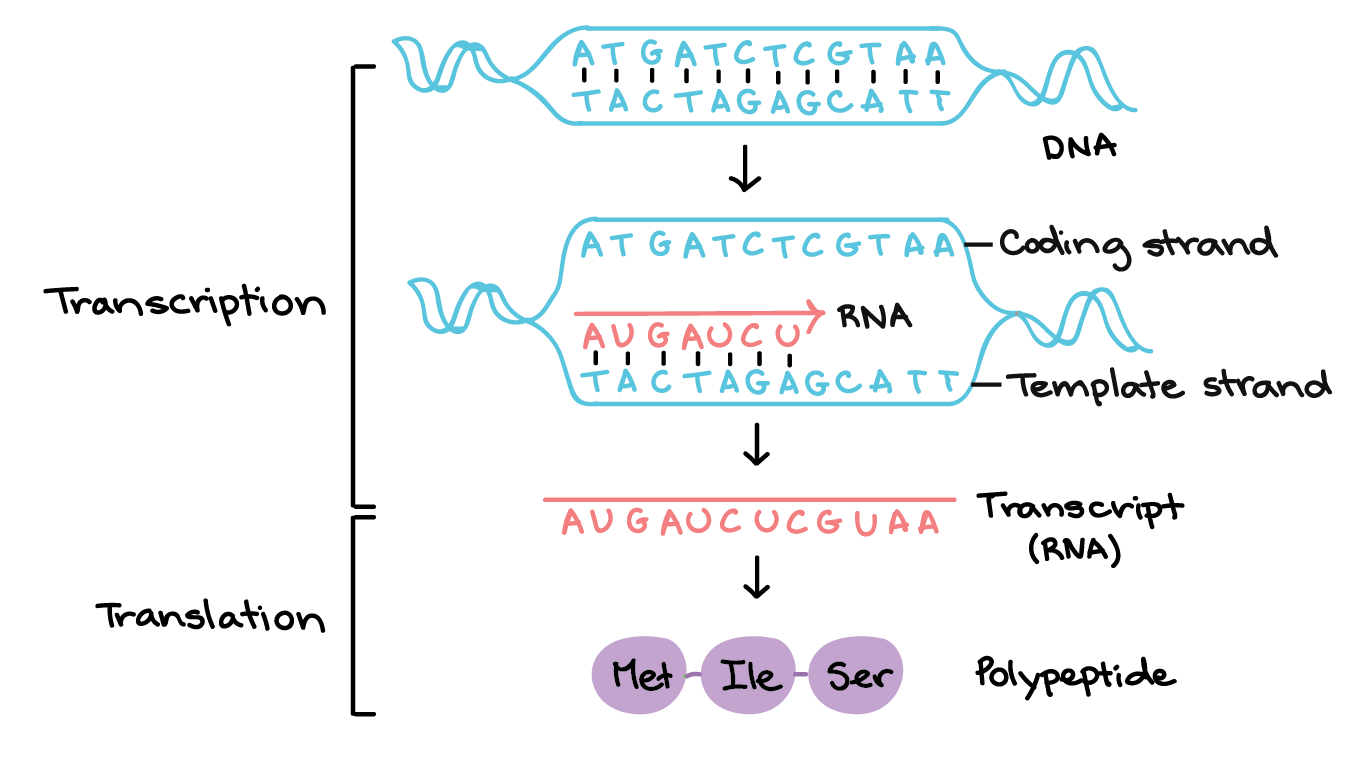
what is transcription?
the synthesis of RNA from the template strand of DNA
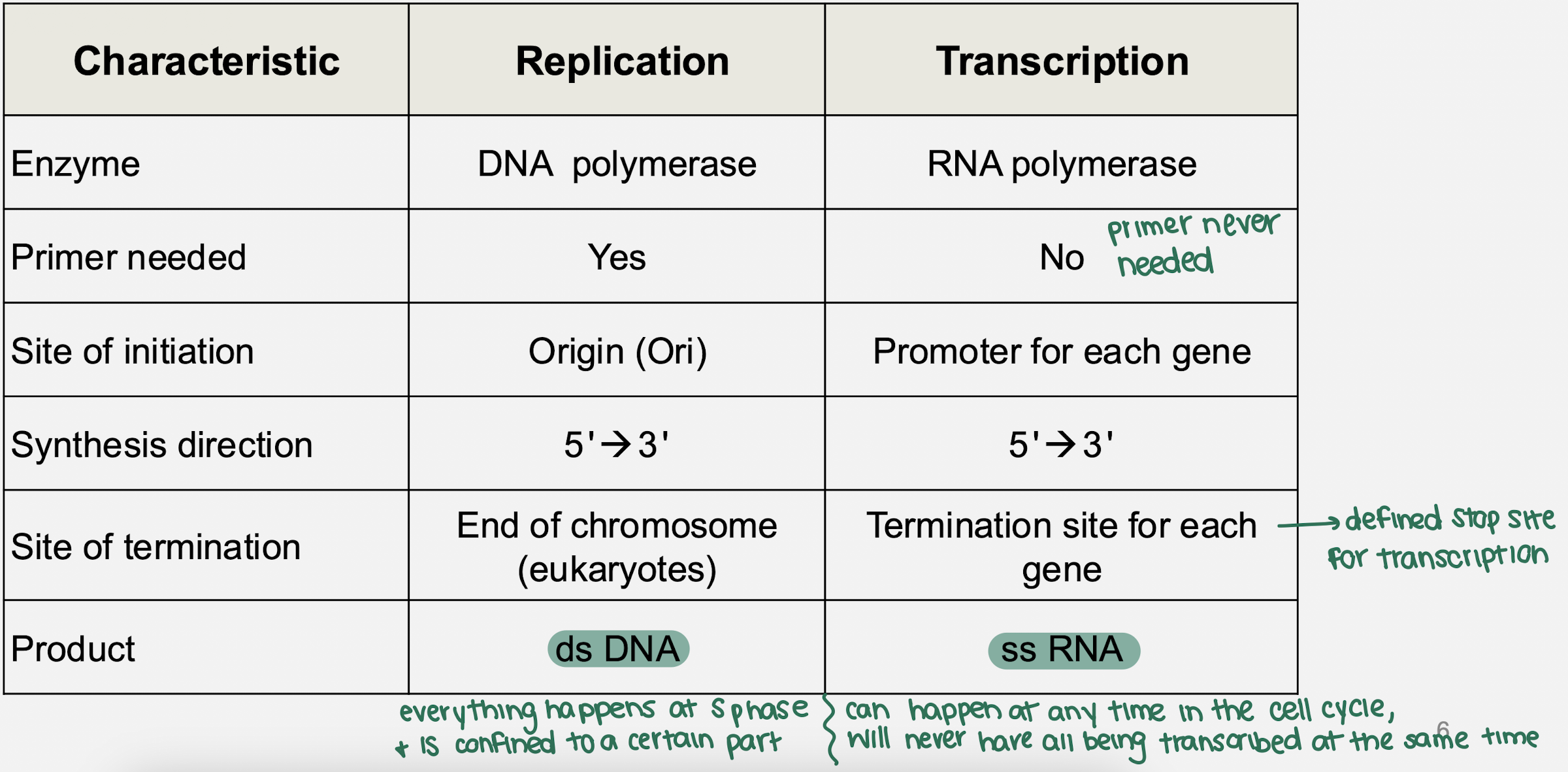
what are some comparisons between DNA replication and transcription?
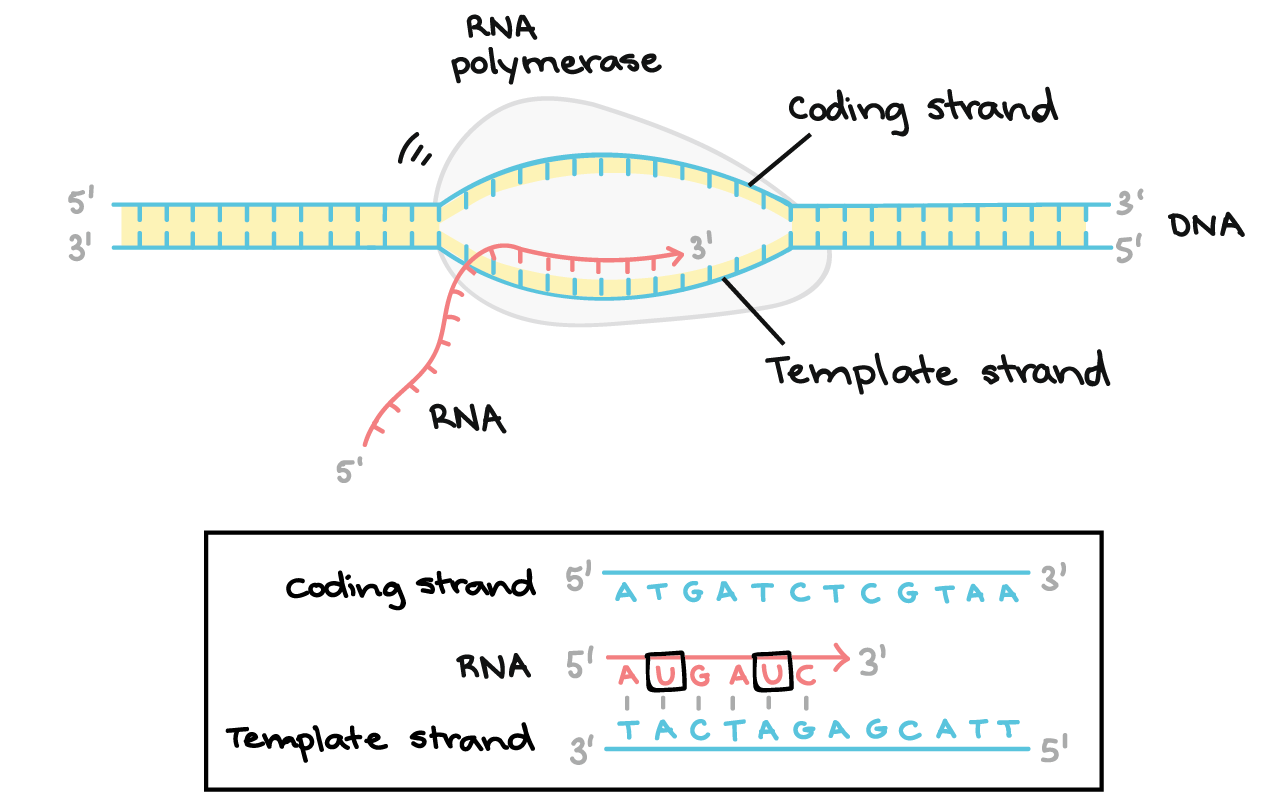
what is the coding strand?
non-template strand → 5’ to 3’
the RNA base sequence in transcription is identical to the base sequence of the coding strand (except for U/T)
what is the non-coding strand?
template strand
antiparallel and complementary to the RNA base sequence of transcription
what is the transcription bubble?
a localized region of unwound DNA that forms during transcription, allowing RNA polymerase to access and transcribe the template strand of DNA into RNA
moves 3’ → 5’ with respect to the template DNA but the synthesis occurs 5’ → 3’
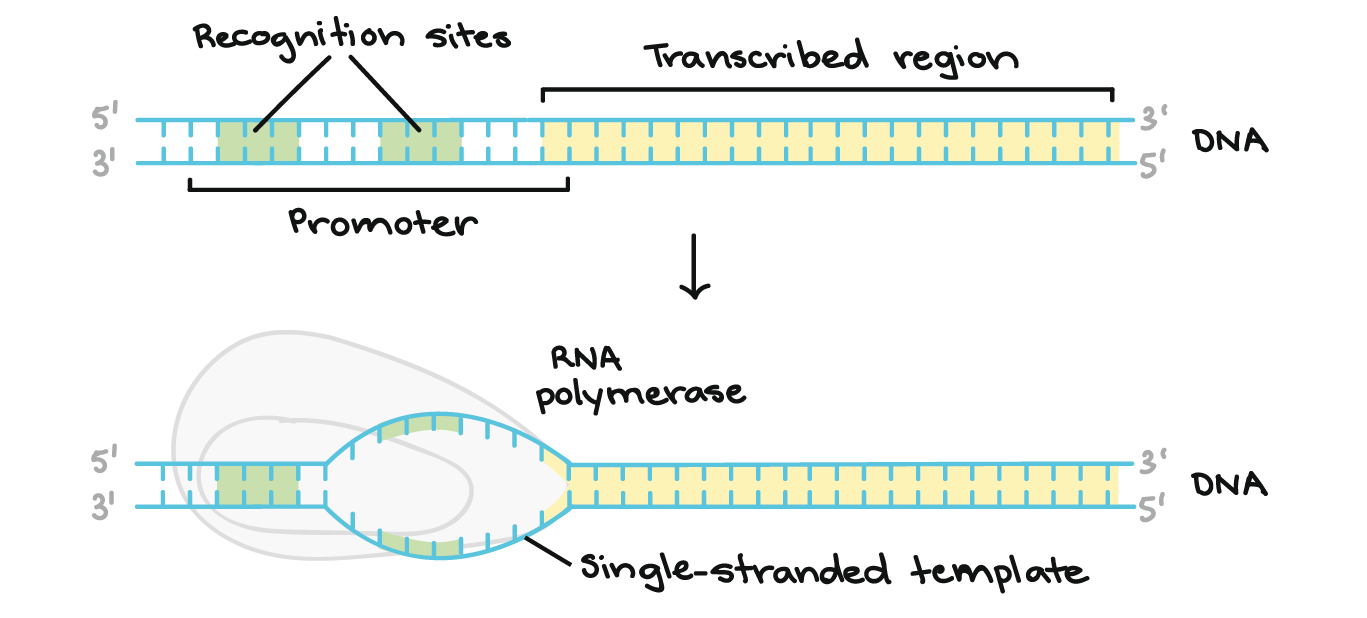
what is the promoter in transcription?
determines where transcription initiates but is NOT part of the transcribe sequence
RNA polymerase binds here and transcription starts at a defined distance from the promoter (+1)
what is the consensus sequence?
the most common base sequence found at a specific location or alignment in DNA
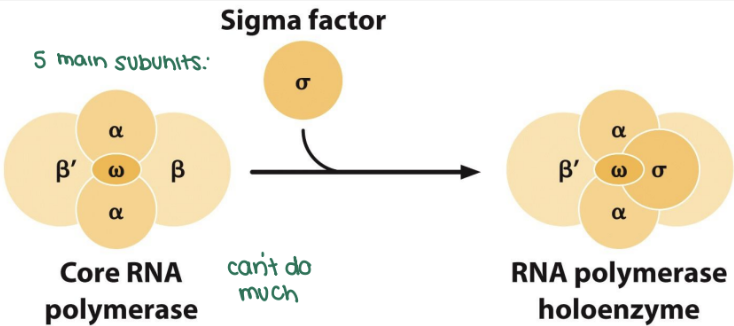
what is sigma factor?
the core RNA polymerase cannot bind the promoter or initiate RNA synthesis without a sigma factor
what are the 3 steps of bacterial transcription?
initiation
elongation
termination
what happens during initiation of bacterial transcription?
the holoenzyme starts unwinding at the -10 position of the promoter
needs the sigma factor to bind which then dissociates from the core polymerase after intiation
what happens during elongation of bacterial transcription?
transcription bubble forms, newly synthesized RNA forms complementary base pairs with template DNA
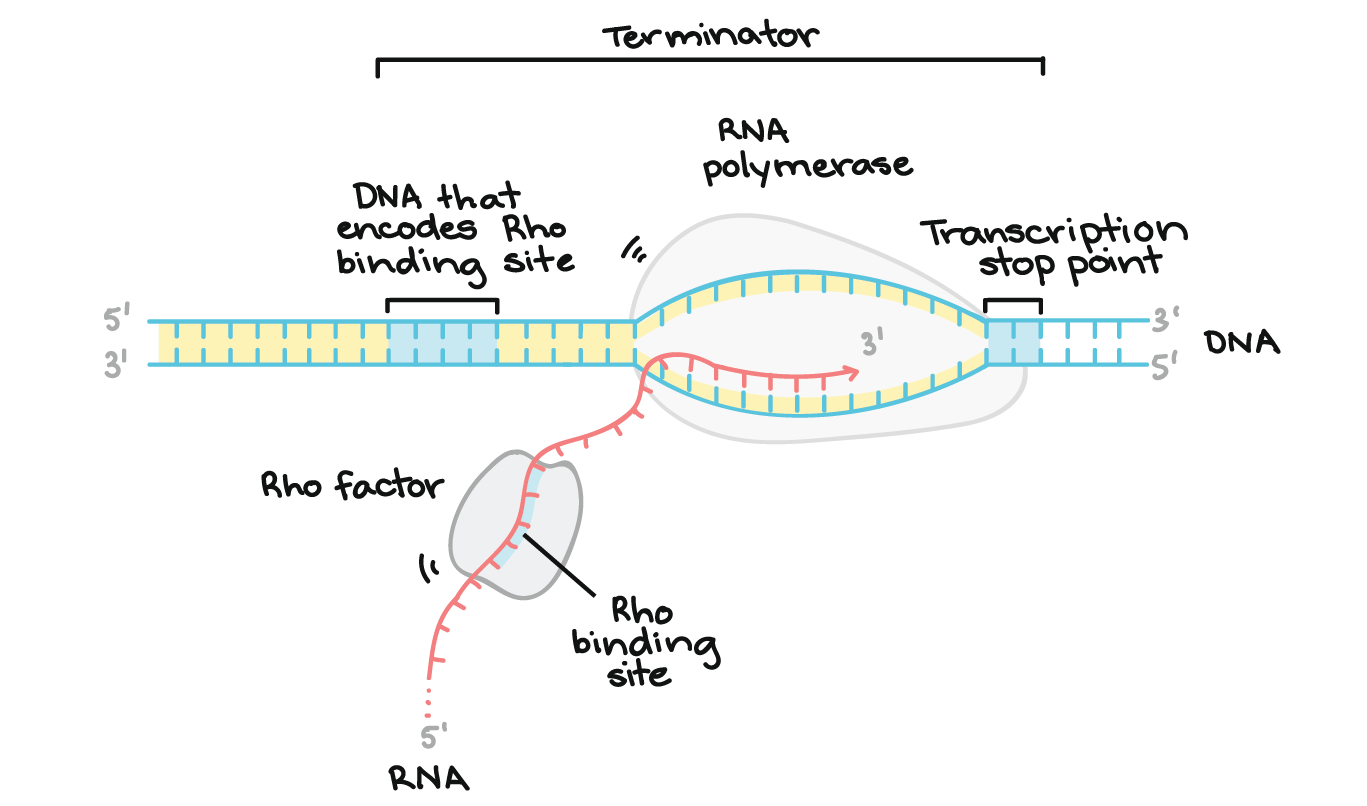
what is Rho-dependent termination?
Rho: an ATP-dependent helicase that unzips DNA by breaking H bonds
recognizes and binds to a Rut site in the RNA (C-rich region that lacks secondary structure) and moves 5’ to 3’ until it encounters RNA polymerase, which has paused near the 3’ end of transcription
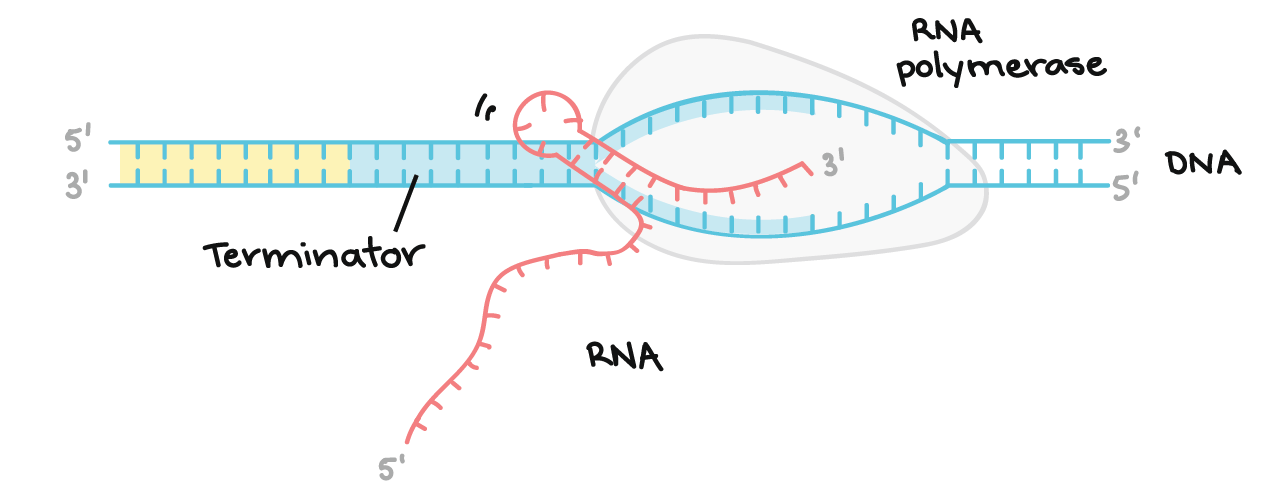
what is Rho-independent termination?
RNA polymerase pauses at strings of U’s allowing the hairpins to form at the inverted repeats which causes pairing between DNA and RNA to be destabilized, leading to release of RNA
are eukaryotic or prokaryotic promoters more diverse?
eukaryotic - eukaryotic transcription is a lot more complex than prokaryotic
in eukaryotes, how many different RNA polymerases recognize different promoters and transcribe different types of RNA?
3
what are low levels of transcription initiation mediated by?
basal transcription apparatus that assembles at the core promoter
what are high levels of transcription initiation mediated by?
mediated when specific transcription factors bind to regulatory promoter sequences and/or more distant enhancers
what is the core promoter?
site for assembly of basal transcription apparatus, contains atleast one consensus sequence
RNA pol II promoter sequence
what is the TATA box?
a common consensus sequence that marks the beginning of transcription for a gene
most common sequence, at -25
RNA pol II promoter sequence
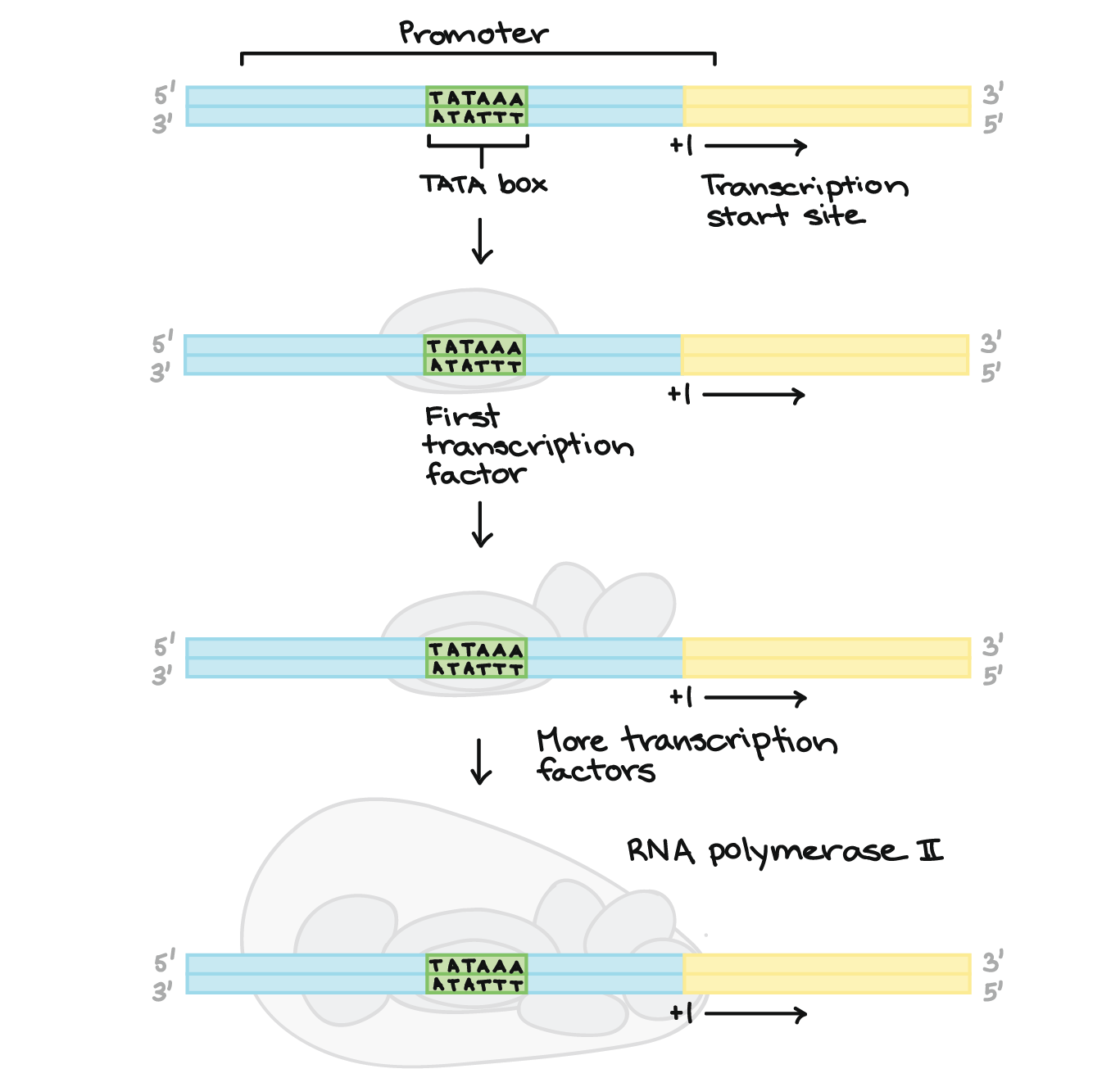
what is a regulatory promoter?
binding sites for specific transcription factors
RNA pol II promoter sequence
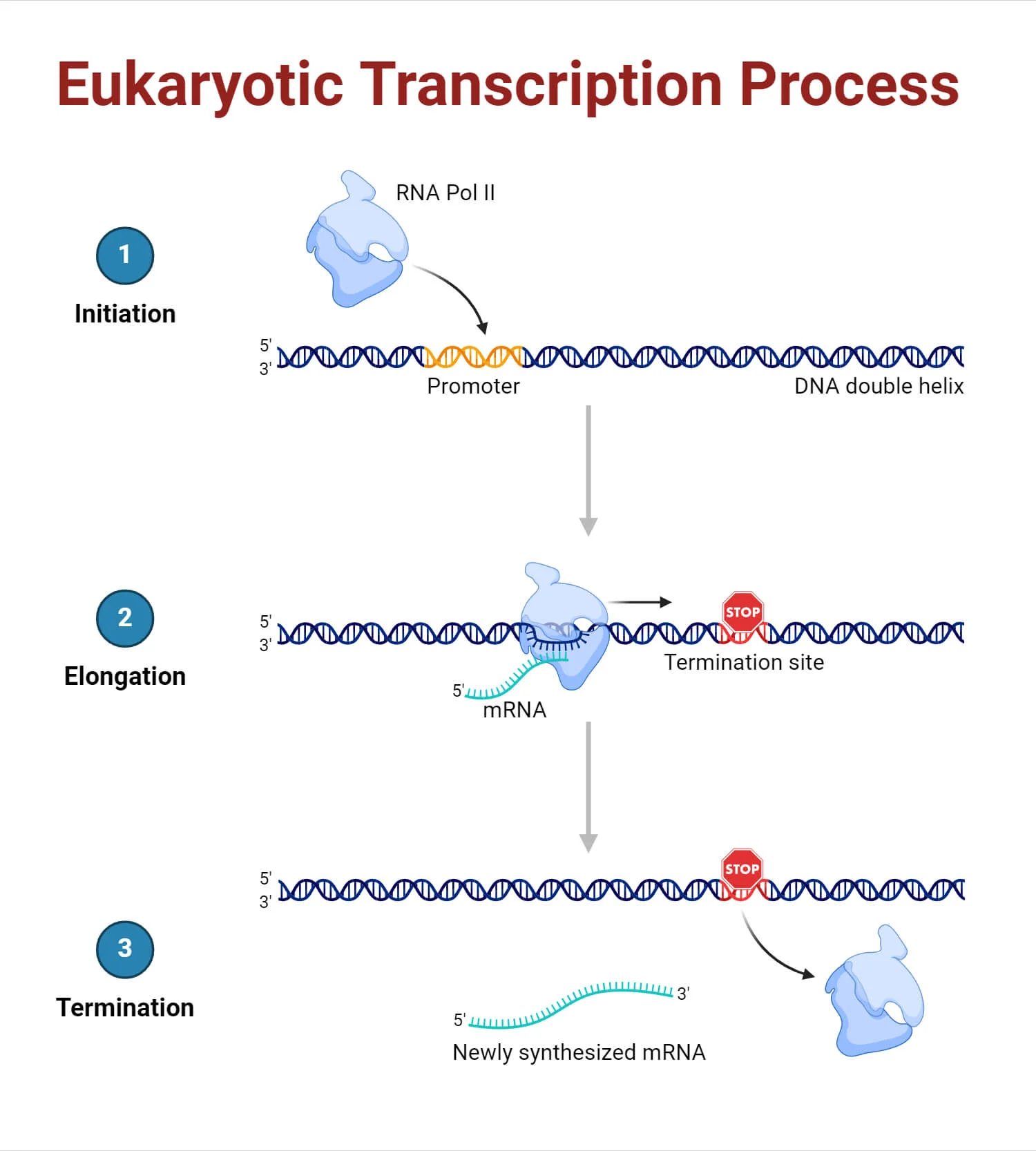
what are the steps of eukaryotic transcription?
intiation
elongation
termination
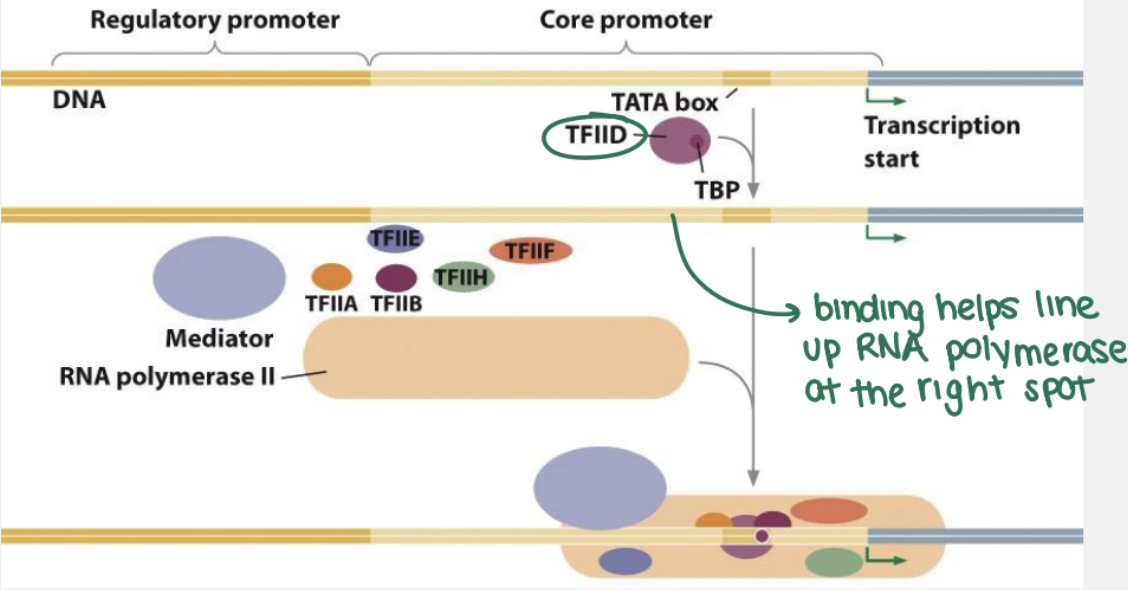
what happens in the initiation phase of eukaryotic transcription?
transcription factors and RNA polymerase II assemble at the promoter region, forming a pre-initiation complex (PIC), which then unwinds the DNA, allowing the template strand to be read and transcription to begin
RNA pol II and general transcription factors assemble on the core promoter
TFIID (general TF) binds to the TATA box
specific transcription factors binds to enhancers and regulatory promoters and interact with the basal transcription apparatus through mediator and coactivator proteins
11-15 bp of DNA is unwound and positioned within the active site of RNA polymerase
what happens in the elongation phase of eukaryotic transcription?
RNA polymerase, after initiating transcription, moves along the DNA template strand, adding nucleotides to the growing RNA transcript in a 5' to 3' direction, forming the mRNA molecule
RNA pol II leaves promoter and general TFs available to form another open complex with another RNA pol II
what happens in the termination phase of eukaryotic transcription?
a poly(A) signal sequence in the transcribed RNA triggers cleavage and polyadenylation, followed by the release of the RNA transcript and the RNA polymerase from the DNA template
cleavage occurs while transcription is still taking place
polyadenylation takes place at 3’ end of cleaved mRNA
an exonuclease (Rat1) degrades RNA in 5’-3’ direction
transcription terminates when Rat1 reaches RNA pol II
what are some posttranscriptional modifications to eukaryotic precursor mRNA? (3)
addition of 5’ cap
3’ cleavage and addition of polyA tail
RNA splicing
what is the significance of 5’ cap?
facilitates binding of ribosome to 5’ end of mRNA, increases mRNA stability, enhances RNA splicing
what is the significance of the 3’ cleavage and addition of polyA tail?
increases stability of mRNA, aids in export of mRNA from the nucleus, and facilitates binding of ribosome to mRNA