DNA mutations and Inheritance
1/18
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
19 Terms
Sickle cell anemia is a WHAT disease
Sickle cell anemia is a GENETIC disease

Sickle cell is the mist common WHAT in the world with 400,000 infants a year born with the disease
Sickle cell is the mist common INHERITED DISEASE in the world with 400,000 infants a year born with the disease
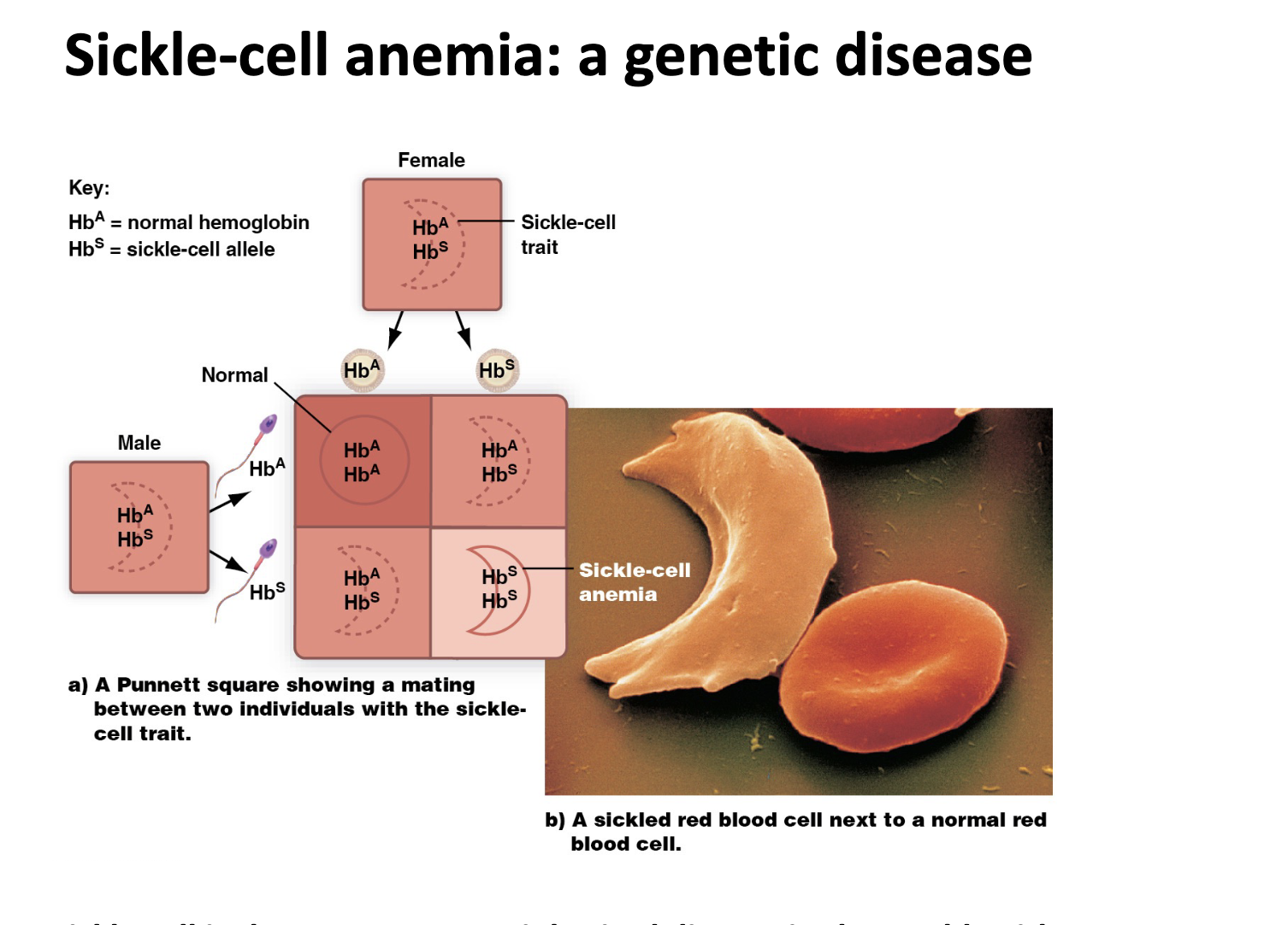
In sickle cell anemia you get painful WHAT which is cause by WHAT blockage
In sickle cell anemia you get painful VASO-OCCULSIVE CIRSIS which is cause by ARTERIOLE blockage
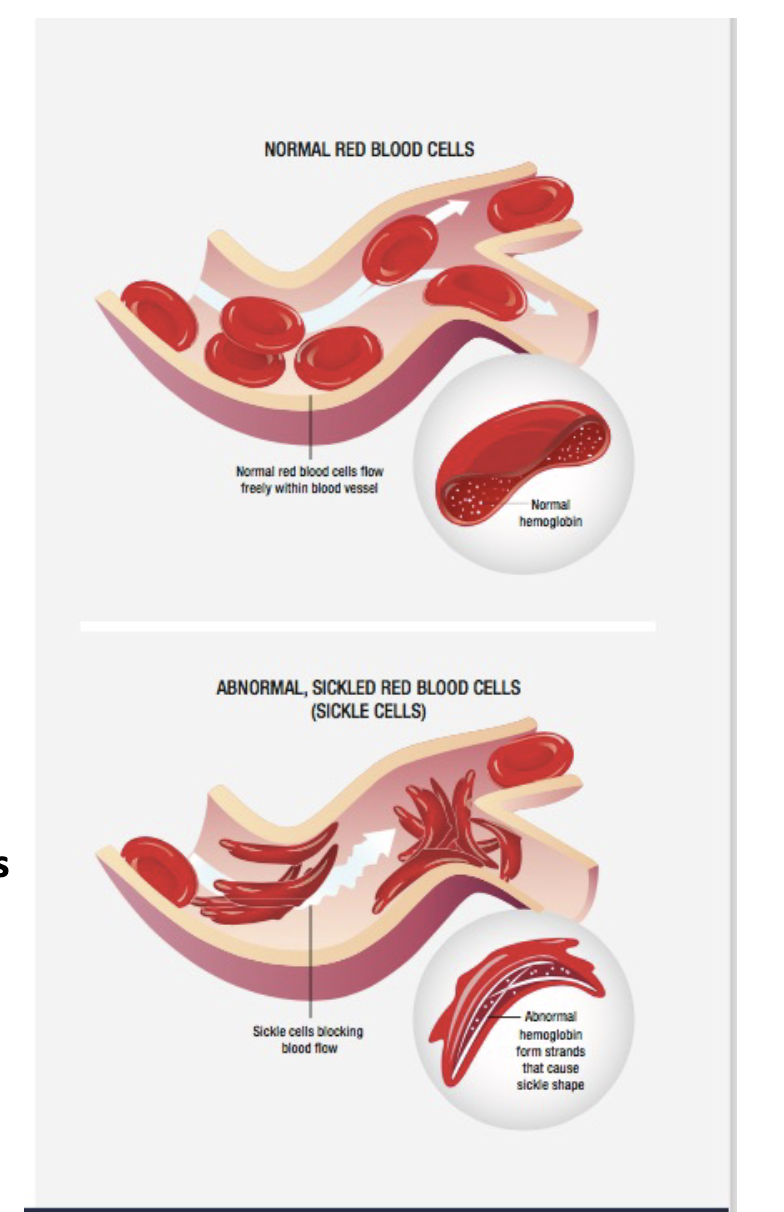
Sickle cell anemia
The WHAT gene is mutated in sickle cell anemia
A single base-pair WHAT changes a HWAT to WHAT with the WHAT codon of exon (close to the start)
The substitution of the amino acid WHAT where WHAT is normally found causes the hemoglobin to WHAT in WHAT environments
This causes the cells to have a WHAT shape making it difficult for the cells to pass through WHAT causing extreme pain and tissue damage
Sickle cell anemia
The B-GLOBIN (B-globin) gene is mutated in sickle cell anemia
A single base-pair TRANSVERSION changes a A to T with the 6th codon of exon (close to the start)
The substitution of the amino acid VALINE (Val) where GLUTAMIC ACID (Glu) is normally found causes the hemoglobin to CLUMP in LOW O2 environments
This causes the cells to have a RIGID SICKLE shape making it difficult for the cells to pass through SMALL BLOOD VESSELS (capillaries) causing extreme pain and tissue damage
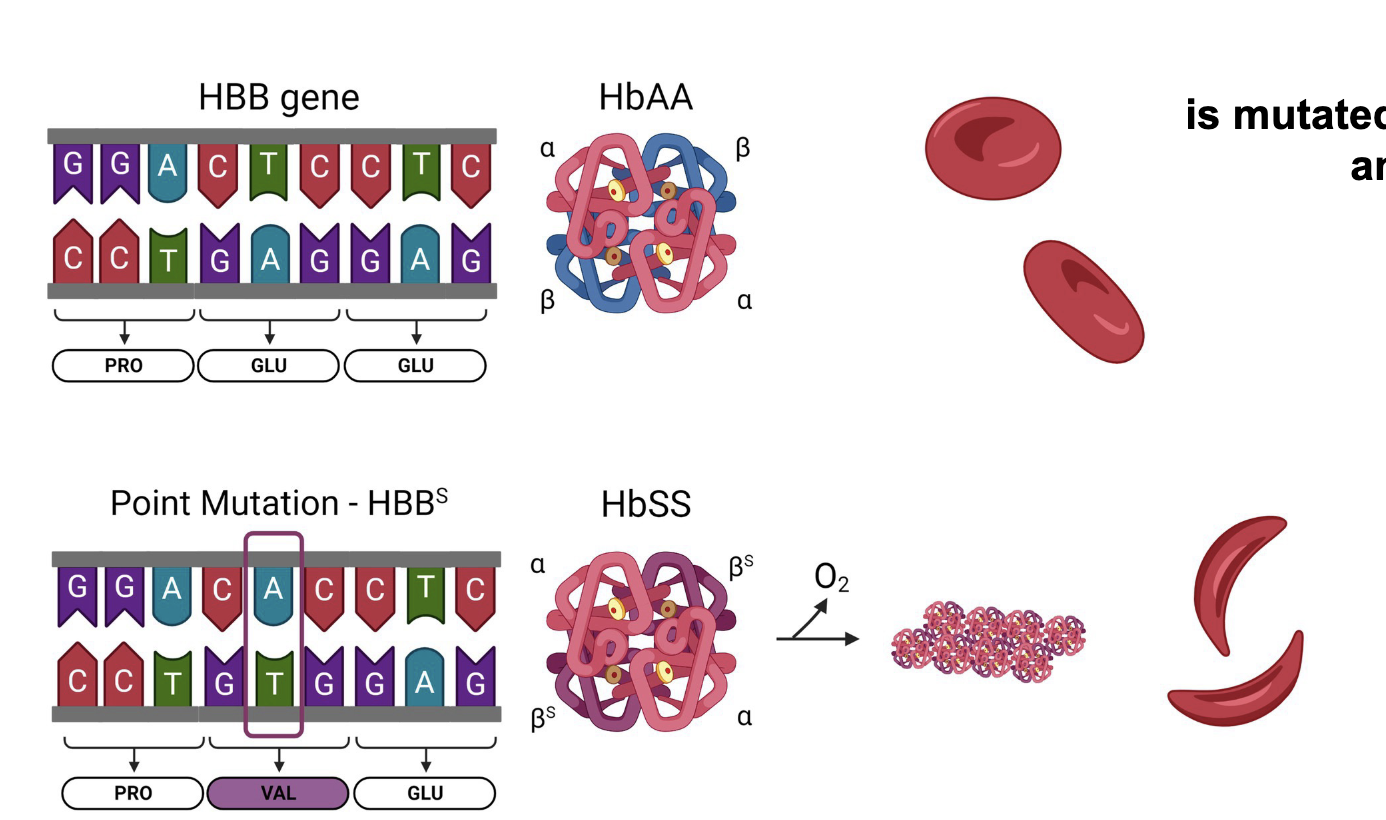
Prokaryotes have a WHAT
Microbial immune system
Microbial immune system
Prokaryotes that survive viral attack can add 40bp segment of WHAT to their genome
The WHAT array is a library of WHAT
Clustered Regularly Interspace Short Palindromic Repeat
A mix of repeated WHAT DNA sequences and WHAT DNA sequences
Microbial immune system
Prokaryotes that survive viral attack can add 40bp segment of VIRAL DNA to their genome
The CRISPR array is a library of VIRAL DNA SEQUENCES
Clustered Regularly Interspace Short Palindromic Repeat
A mix of repeated CONSTANT DNA sequences and UNIQUE (VIRAL) DNA sequences
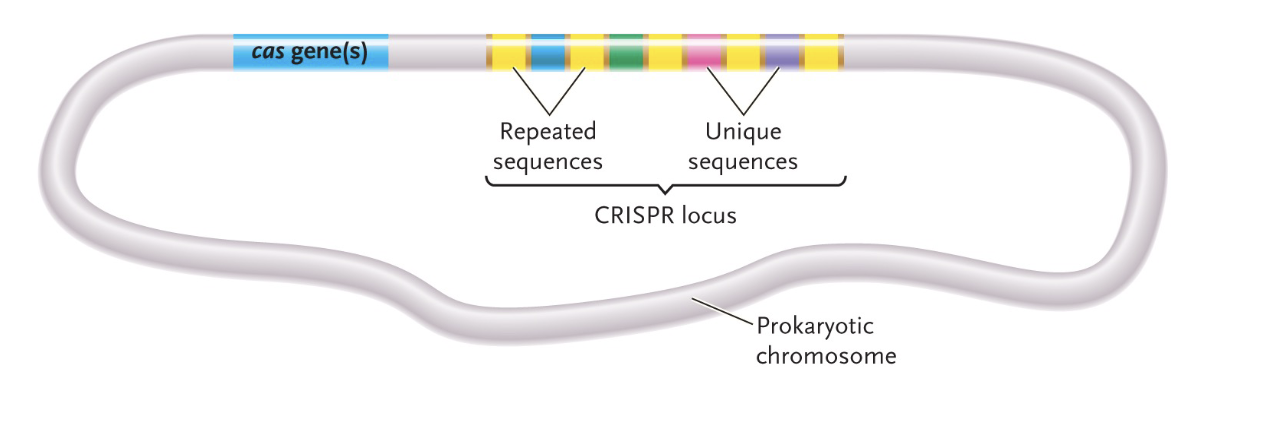
Microbial immune system: When infected. microbes will:
WHAT the CRISPR locus to generate WHAT
Express WHAT proteins
Cas are WHAT that cut WHAT
If, one of the crRNA molecules WHAT incoming viral DNA, the Cas proteins will make a WHAT, allowing the microbe to WHAT the viral DNA
Microbial immune system: When infected. microbes will:
TRANSCRIBE the CRISPR locus to generate crRNA
Express Cas (CRISPR-associated) proteins
Cas are ENDONUCLEASE that cut DOUBLE STRANDED DNA
If, one of the crRNA molecules BINDS incoming viral DNA, the Cas proteins will make a CUT, allowing the microbe to DEGRADE the viral DNA
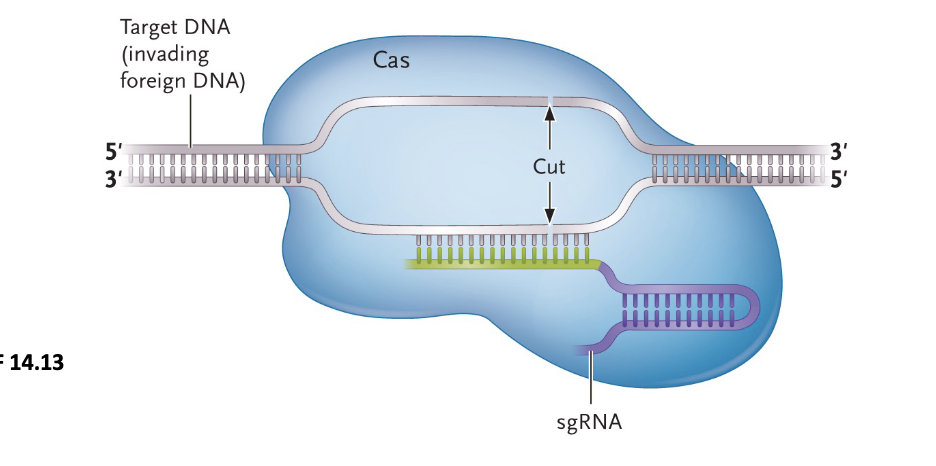
CRISPR-Cas9 Gene Editing System:
Since 2013, the understanding of an adaptive immune system from bacteria called CRISPR/Cas9 has sparked a revolution in the ability to modify or edit the genomes of variety of organisms - including humans
Compared to earlier methods, CRISPR/Cas9 is a much WHAT and WHAT method to introduce changes into genomes
CRISPR/Cas9 uses the WHAT and WHAT RNA sequence (WHAT) to target specific sites in ANY genome
CRISPR-Cas9 Gene Editing System:
Since 2013, the understanding of an adaptive immune system from bacteria called CRISPR/Cas9 has sparked a revolution in the ability to modify or edit the genomes of variety of organisms - including humans
Compared to earlier methods, CRISPR/Cas9 is a much SIMPLER and FASTER method to introduce changes into genomes
CRISPR/Cas9 uses the Cas9 ENDONUCLEASE and SYNTHETIC 20-NUCLEOTIDE RNA sequence (sgRNA (synthetic guid RNA)) to target specific sites in ANY genome
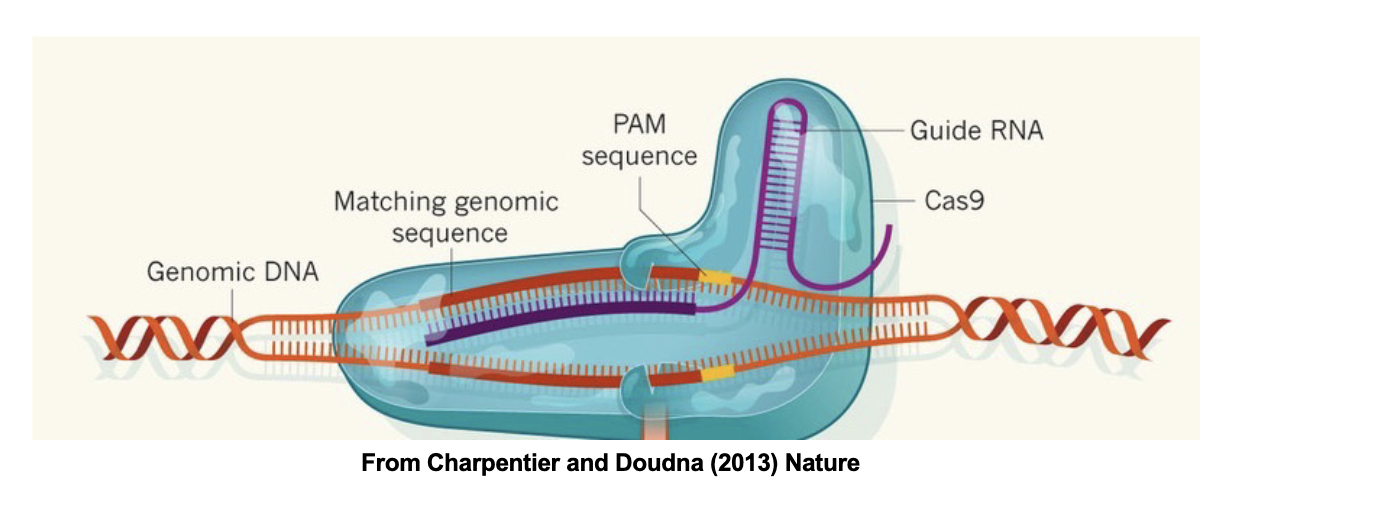
sgRNAs can be designed to bind to specific
locations in the genome and make a double
strand break (DSB) to then change or edit the
genome:
WHAT
WHAT
sgRNAs can be designed to bind to specific
locations in the genome and make a double
strand break (DSB) to then change or edit the
genome:
Non-Homologous end joining (NHEJ)
Homology-directed repair
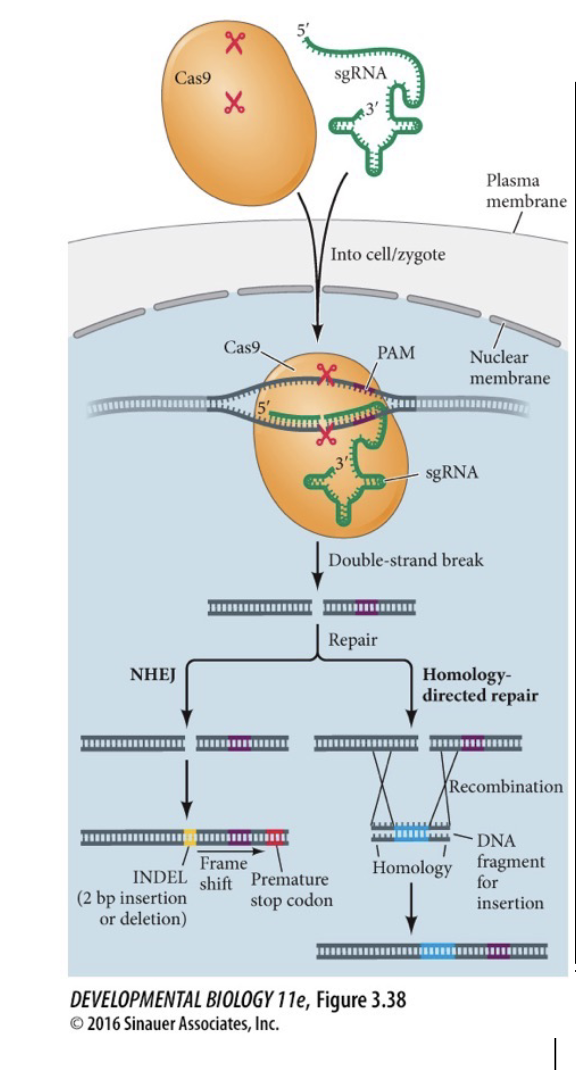
Non-Homologous end joining (NHEJ)
A natural repair mechanism to fix WHAT breaks
Error-prone – often results in WHAT that change the WHAT (insertion) + can disrupt WHAT (deletion)
Use CRISPR in genes you don’t want funtioning
Non-Homologous end joining (NHEJ)
A natural repair mechanism to fix DOUBLE-STRAND breaks
Error-prone – often results in INDELs that change the READING FRAME (insertion) + can disrupt GENES (deletion)
Use CRISPR in genes you don’t want functioning
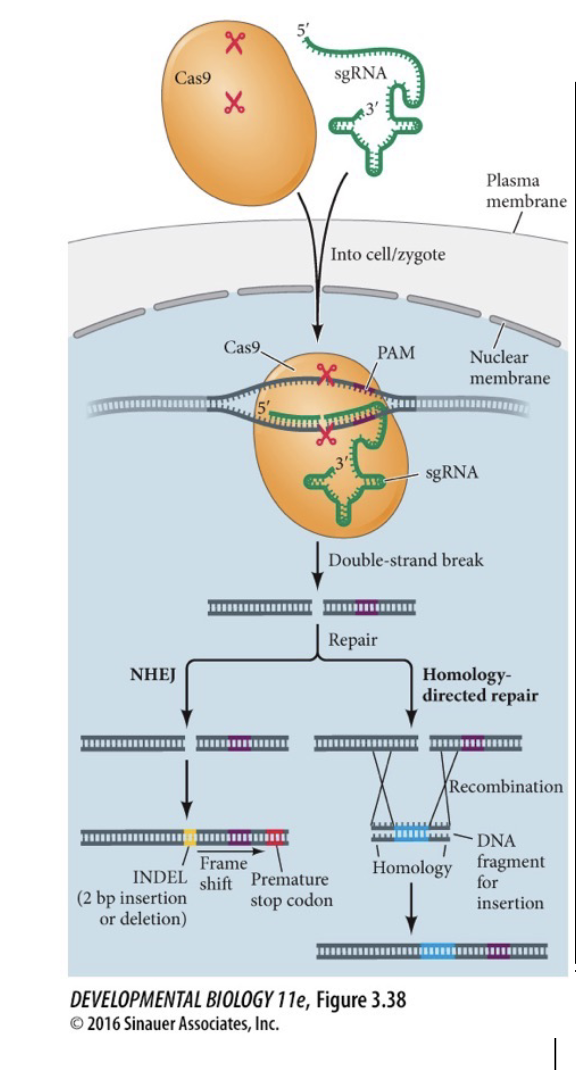
Homology-directed repair
New genes can be WHAT if flanking sequences have WHAT to the area on either side of the WHAT
Introduce genes taht were not there before
Homology-directed repair
New genes can be ADDED if flanking sequences have HOMOLOGY to the area on either side of the DSB (double-strand break)
Introduce genes taht were not there before
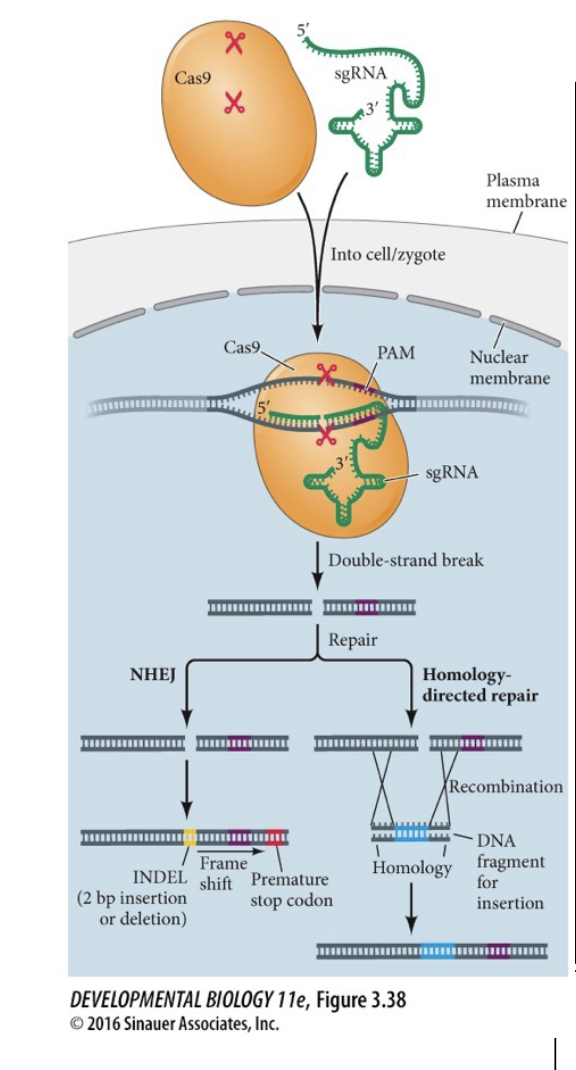
If the cell modifies is a single-celled zygote, then the whole organism has its WHAT edited
If the cell modifies is a single-celled zygote, then the whole organism has its GENOME edited
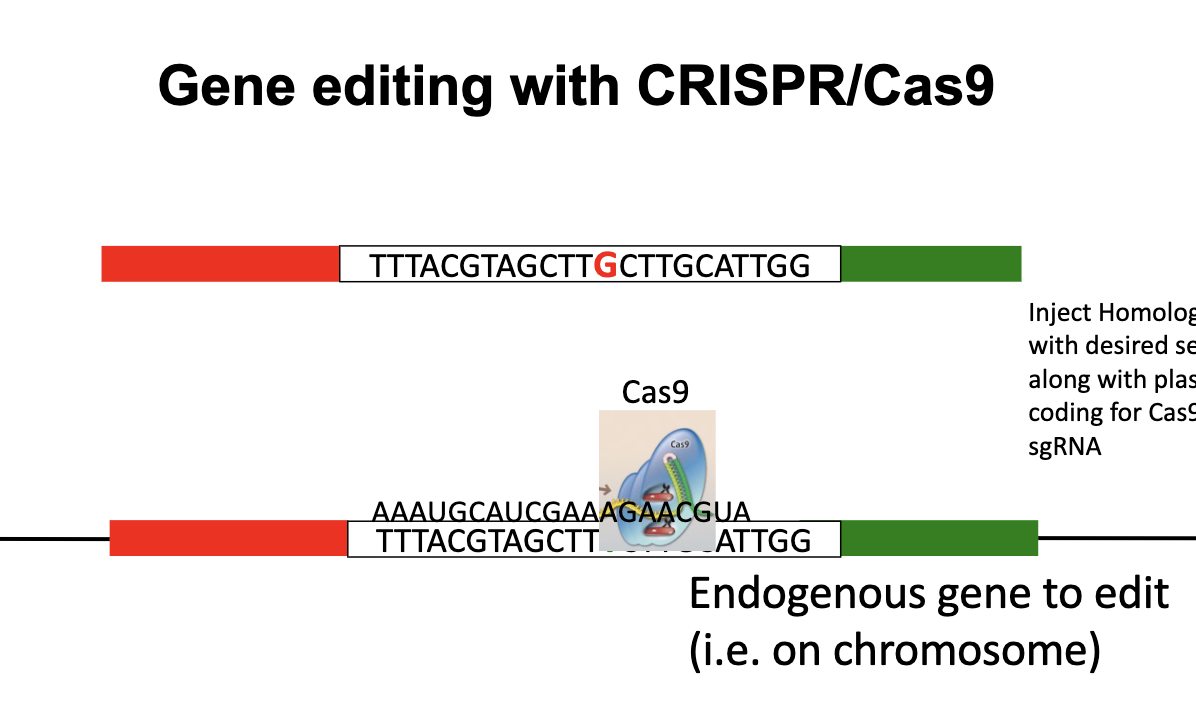
Gene editing with CRISPR/Cas9
Inject Homologous DNA with desired WHAT along with plasmid coding for WHAT and WHAT
Gene editing with CRISPR/Cas9
Inject Homologous DNA with desired SEQUENCE along with plasmid coding for Cas9 and sgRNA
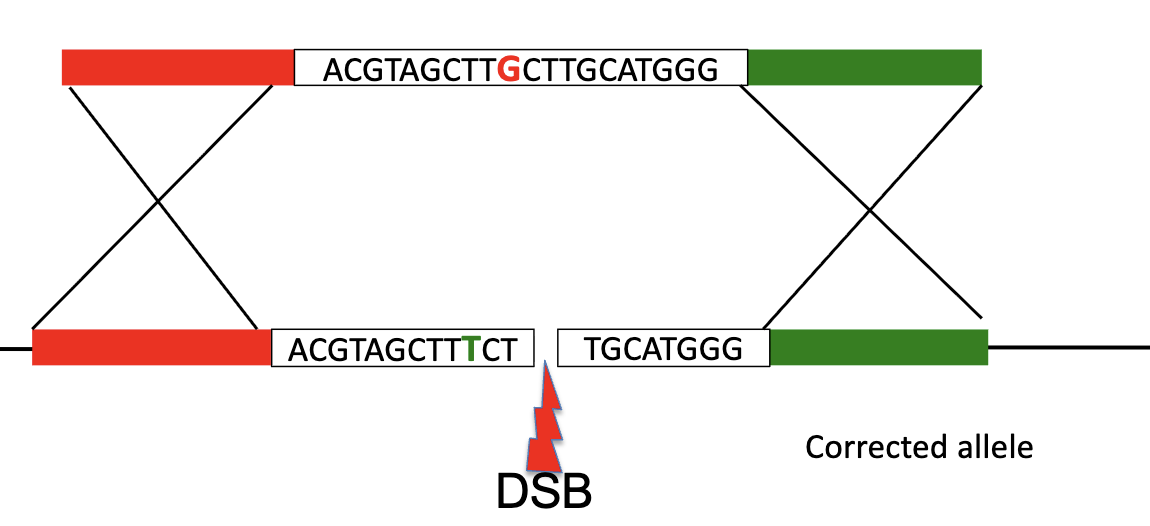
Gene editing with CRISPR/Cas9
Allow Cas9 system to cause a WHAT and then use WHAT to switch the old genes with the wanted genes
Gene editing with CRISPR/Cas9
Allow Cas9 system to cause a DOUBLE-STRAND BREAK and then use ENZYMES to switch the old genes with the wanted genes
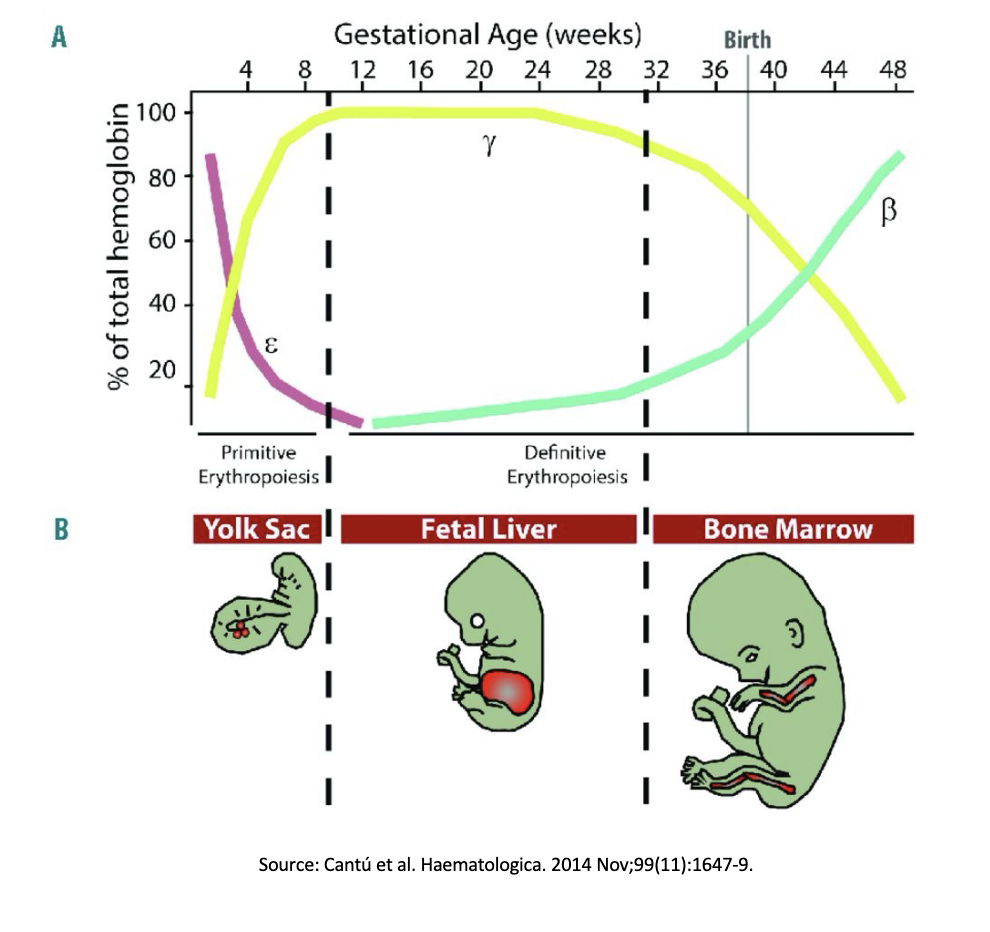
Using CRISPR-Cas9 to cure sickle cell anemia
When in utero the fetus uses WHAT hemoglobin which is more oxygen prone than the WHAT hemoglobin in adults
The WHAT hemoglobin gene mutation is what causes sickle cell anemia
Therefor to cure sickle cell anemia you want to do WHAT
Using CRISPR-Cas9 to cure sickle cell anemia
When in utero the fetus uses GAMA-hemoglobin (Y-globin)which is more oxygen prone than the BETA-hemoglobin in adults
The BETA-hemoglobin (B-globin) gene mutation is what causes sickle cell anemia
Therefor to cure sickle cell anemia you want to do REPLACE ALL B-GLOBIN WITH Y-GLOBIN
Using CRISPR to cure sickle cell anemia
Option A: Fix WHAT and WHAT fix the mutation in the adult hemoglobin gene.
Option B: Swap CRISPR reactivates the WHAT hemoglobin gene by turning off the WHAT gene.
Using CRISPR to cure sickle cell anemia
Option A: Fix CRISPR and DNA template fix the mutation in the adult hemoglobin gene.
Option B: Swap CRISPR reactivates the FETAL hemoglobin gene by turning off the BCL11A gene.
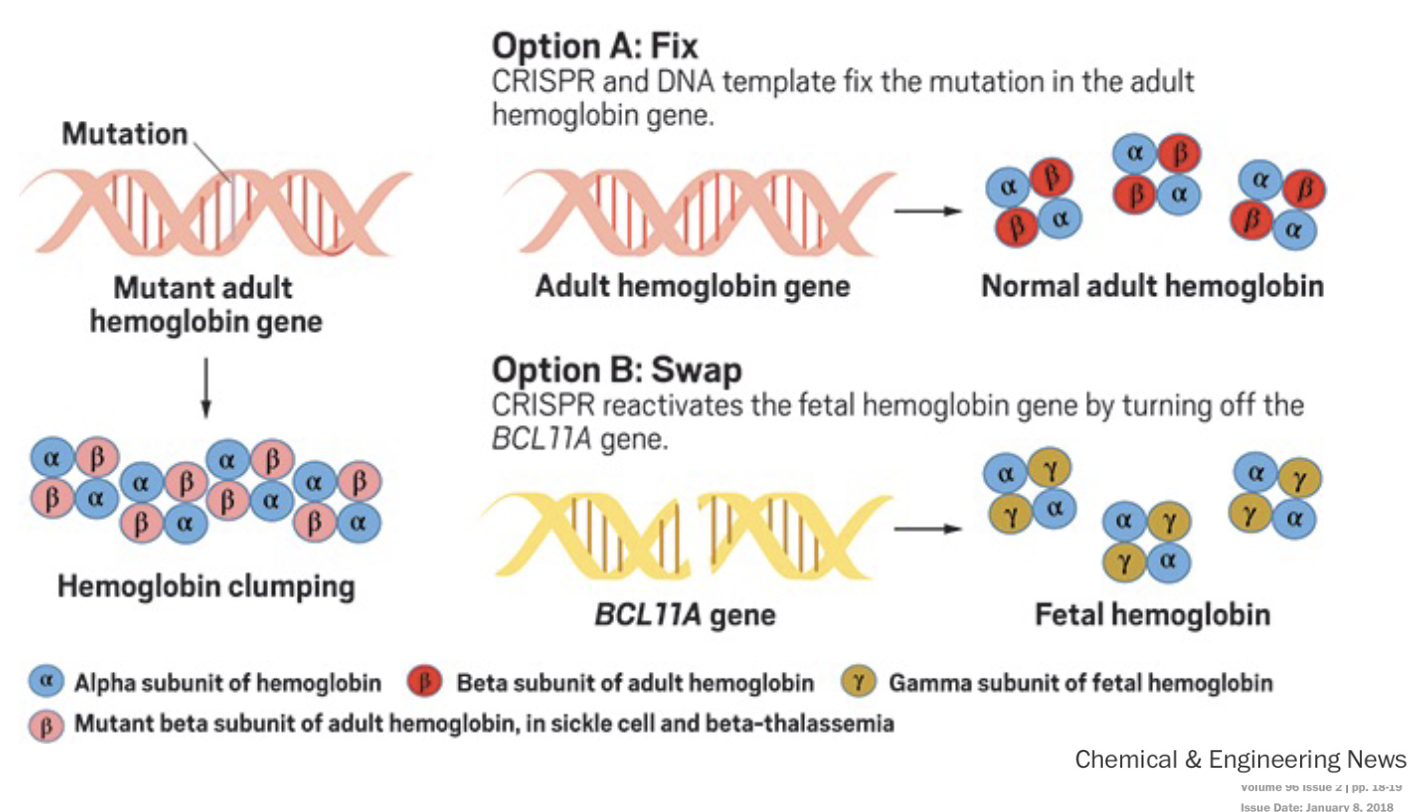
Which option was more affective
Option B
An enhancer is a WHAT DNA sequence that binds proteins called WHAT and activates transcription of a specific nearby gene on the same chromosome
An enhancer is a NON-CODING DNA sequence that binds proteins called TRANSCRIPTION FACTORS and activates transcription of a specific nearby gene on the same chromosome
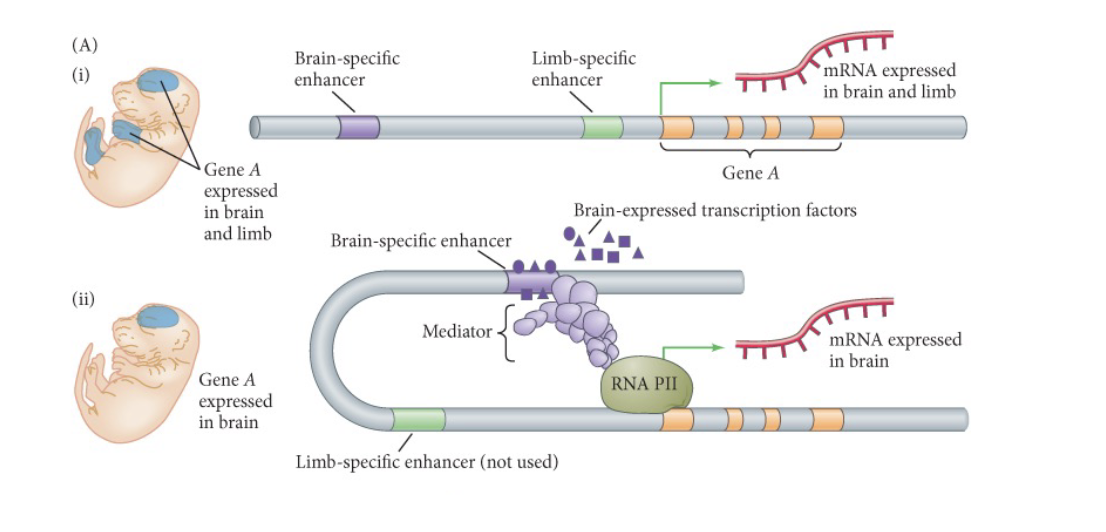
Option B: Using CRISPR-Cas9 to turn off a gene coding for a repressor of fetal hemoglobin genes
How do they inactivate this enhancer so efficiently?
Approximately 80% of CRISPR treated cells (with WHAT) have WHAT in this 2019 paper.
Not necessarily the precise WHAT used in the clinical trial.
Option B: Using CRISPR-Cas9 to turn off a gene coding for a repressor of fetal hemoglobin genes
How do they inactivate this enhancer so efficiently?
Approximately 80% of CRISPR treated cells (with sgRNA-1617) have INDELs in this 2019 paper.
Not necessarily the precise sgRNA used in the clinical trial.
