Unit 4: DNA Replication, Transcription, Translation, and Protein synthesis.
1/57
Earn XP
Description and Tags
Laura Mozingo BIO 135 At JCCC
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
58 Terms
Hershey and Chase
DNA is the genetic material.
Chargaff's Rule
DNA has equal amounts of A=T and G=C.
Chemical Structure of DNA
(-) including a nitrogenous base, pentose sugar, and phosphate group.
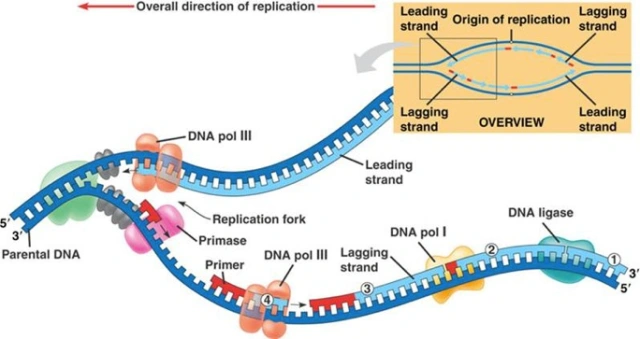
DNA REPLCIATION
Semiconservative Model of Replication
Type of DNA replication where double helix has parent and daughter strands.
Leading Strand Replication
Nucleotides continuously added in 5' to 3' direction
TOWARDS replication fork.
Lagging Strand Replication
Nucleotides discontinuously added in Okazaki fragments in 5' to 3' direction
AWAY from replication fork.
DNA Polymerase I
Removes RNA primer and replaces them with DNA.
DNA Polymerase III
Adds nucleotides to RNA primer or DNA strand
Builds new DNA strands.
Uses parental DNA as a template
Helicase
Unzips DNA at replication forks
Ligase ‘gluer’
Glues bonds between Okazaki fragments to form one continuous DNA strand.
Nuclease
Cuts damaged DNA.
Primase
Makes RNA primer by adding RNA nucleotides.
RNA Primer
Short nucleotide chain
Starting point for DNA synthesis.
Single Strand Binding Protein
Binds DNA strands during replication
Stabilizes and keeps separated.
Telomerase
Adds DNA to telomeres in eukaryotic germ cells, healing their length after replication.
Topoisomerase
Breaks and rejoins DNA strands after trauma.
Telomere
Nucleotide sequences at the end of chromosomes
Act as buffer zones and delay shortening of genes
Histone
Protein molecules that pack DNA into chromatin in interphase.
Origins of Replication
Sites where DNA replication occurs
Prokaryotic Origin of Rep.
ONE circular chromosome
Eukaryotic Origin of Rep.
INFINITE number of linear chromosomes
Replication Fork
Y-shaped region
Parental strands unwind, and new strands are created.
Antiparallel Elongation
Two new strands must run ANTIPARALLEL to parent strands.
Okazaki fragment
Short strands of DNA that make up lagging strand.
DNA proofreading and repair
DNA polymerases check each nucleotide against its template as it's added. If incorrect, it's removed and corrected.
Mismatch repair
Enzymes fix DNA mistakes by removing and replacing incorrectly paired nucleotides.
Nucleotide excision repair
A repair system where damaged DNA is removed and replaced, using undamaged strand as template.
NUCLASE, DNA POLYMERASE, LIGASE
Damage can pass into replication accidentally.
Telomere function
Acts as a buffer zone, delays gene shortening/loss
Telomerase function
Adds DNA to telomeres, helping restore length.
Histones
Proteins that pack DNA in Interphase chromatin.
Eukaryotic DNA
Nucelus
Long
Linear
Many proteins (histones)
No correlation with gene #
Prokaryotic DNA
No Nucleus, uses cytoplasm
Short
Circular
Fewer proteins
Correlation with gene #
How is DNA packaged in the cell? How does this change throughout the cell cycle?
Prophase: Condensin II (Large Loops), Condensin I (Small Loops)
Metaphase: chromosomesare fully condensed, MOST dense
Euchromatin
Less condensed chromatin
Available for transcription (gene expression)
Heterochromatin
Very compacted chromatin that stays condensed during Interphase
Not used for gene expression
Central Dogma of Molecular biology
DNA —> RNA —> Protein
Prokaryotic Transcription/Translation
Cytoplasm
Eukaryotic Transcription/Translation
Nucleus
Cytoplasm
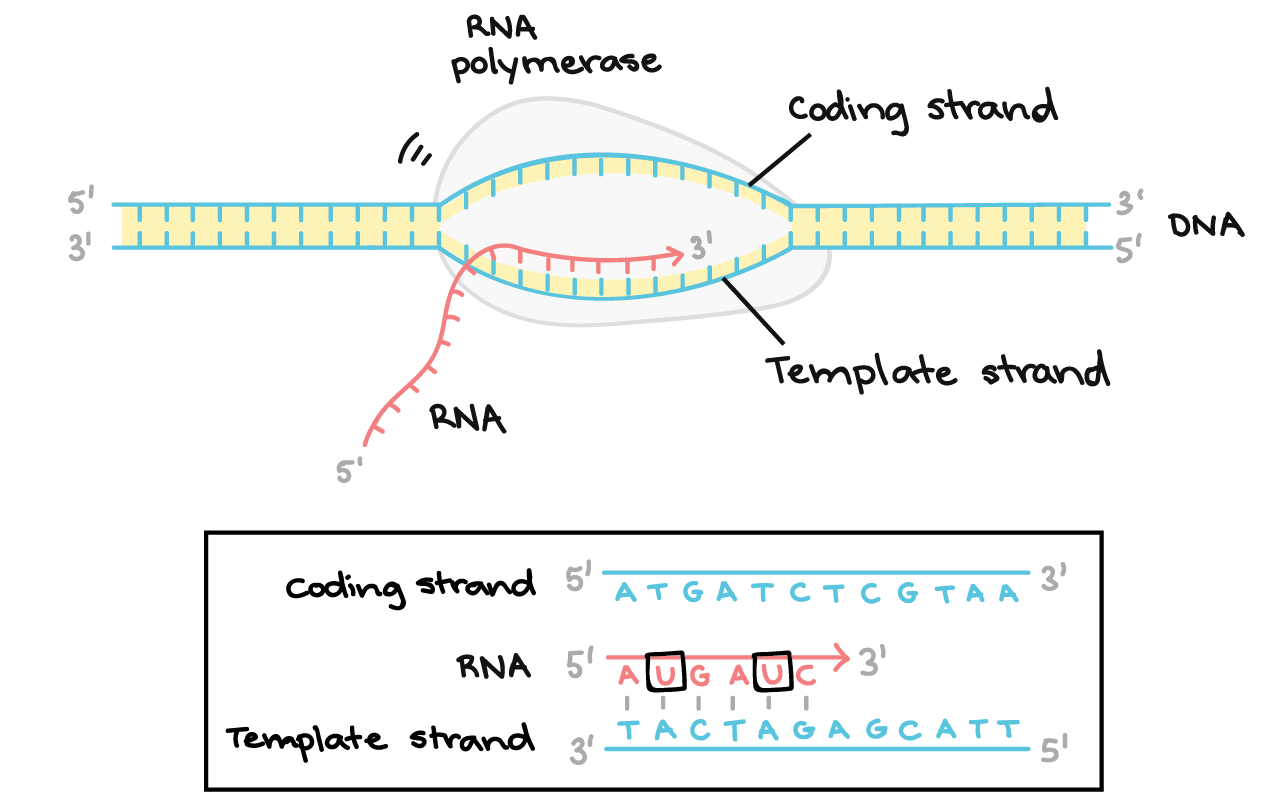
Transcription
a cell makes an RNA copy of a piece of DNA
Codon
3 nucleotides in mRNA that code for a specific amino acid or signal the end of protein synthesis. (Stop Signals)
Template strand
DNA Strand used as a guide to transcribe complementary RNA strand during transcription.
Coding Strand
Not used as template for transcription
Has same nucelutide sequence as transcribed RNA strand, but has T (Thymine) instead of U (Uracil)
TATA box
Found in Eukaryotic promoters
Transcription initiation complex
Transcription Factors
Proteins that bind to DNA, helping RNA polymerase attach to the template strand to initiate transcription.
Describe Eukaryotic pre-mRNA processing.
- Both exons and introns are transcribed into RNA.
Introns: interrupted, removed via splicing.
Exons: expressed.
5' cap is added.
Poly-A tail is added.
Location: Nucleus.
Alternative RNA splicing
Different mRNA versions can be produced from the original transcript
Depends on the exon included/excluded pattern.
Results of alternative RNA splicing
More than one kind of protein
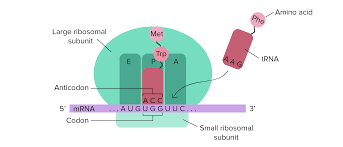
Translation
Happens in ribosomes in the cytoplasm
The ribosome reads the mRNA codons
tRNA has an anticodon that matches the mRNA codon.
tRNA (transfer RNA) brings specific amino acids to the ribosome.
They form a polypeptide chain, which becomes a protein
Wobble
Third base of codon.
Can pair with more than one base in tRNA anticodon
What happens to the polypeptide when released from ribosome?
Modifications can happen
Adding sugars, lipids, phosphates
Join or cut polypeptides
Remove amino acids
Signal proteins
Amino acid tags on a polypeptide
Tell cell where it should go
Nonsense mutation
Premature stop codon
Short protein
Often non-functional
Mutation
Change in nucleotides in DNA
Affects all new genes
Silent Mutation
Same amino acid
‘Silent’
Missense Mutation
Different amino acid.
Can be a slight or drastic effect on protein.
Nonsense Mutation
Introduces a premature stop codon
Short and often non-functional protein.
What is the difference between substitution, insertion, and deletion? What results in each case?
Substitution: One nucleotide is replaced by another. Can result in silent, missense, or nonsense mutations.
Insertion: Addition of one or more nucleotides. Can cause frameshift mutations, altering the entire protein sequence after the insertion point.
Deletion: Removal of one or more nucleotides. Similar to insertions, can cause frameshift mutations.