Biochemistry I Module of the MCAT Self Prep eCourse: Lesson 5: Transcription, Translation, and DNA Repair (Pro)
1/98
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
99 Terms
Lesson 5: Transcription, Translation, and DNA Repair
Lesson 5: Transcription, Translation, and DNA Repair
In eukaryotic cells, transcription takes place in the _____________. In bacteria, transcription takes place in the _____________.
(A) Cytosol, Nucleus
(B) Nucleus, Cytosol
(C) Endoplasmic Reticulum, Nucleus
(D) Nucleus, Endoplasmic Reticulum
(B) Nucleus, Cytosol
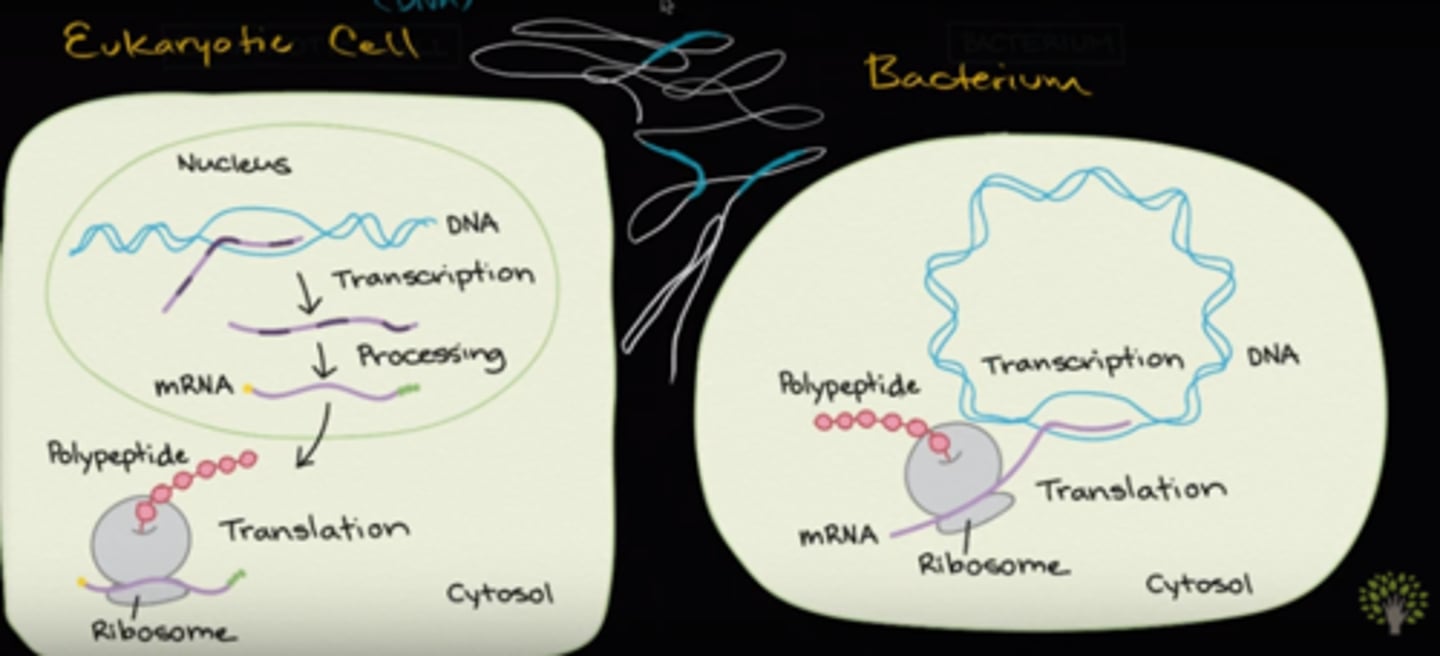
In eukaryotic cells, transcription takes place in the Nucleus. In bacteria, transcription takes place in the Cytosol.
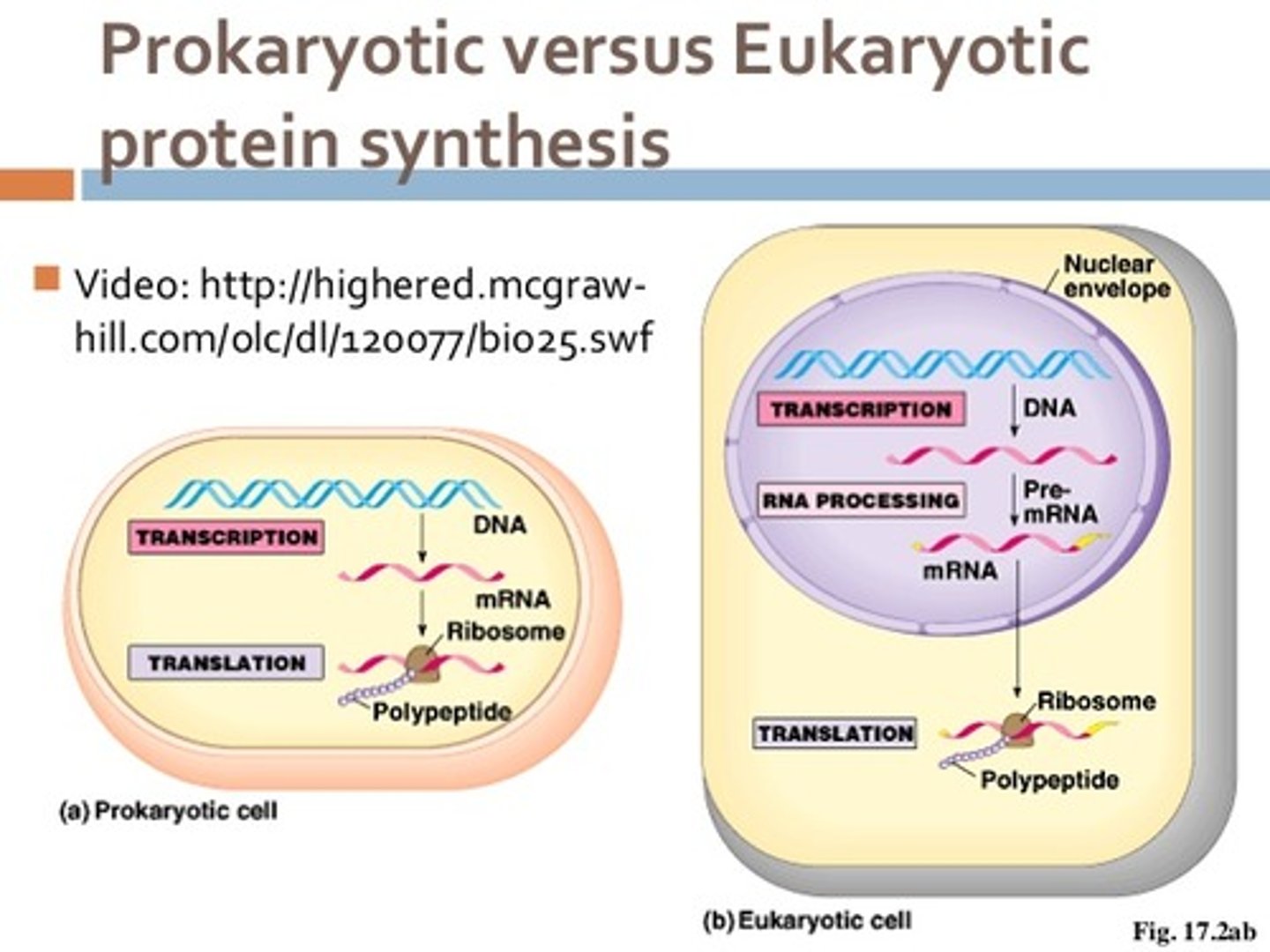
During the process of transcription, the RNA goes through some extra steps such as adding a cap, adding a tail, and the splicing of introns. In which type of cell does this extra processing occur?
(A) Prokaryote
(B) Eukaryote
(C) Both Prokaryote and Eukaryote
(D) Neither
(B) Eukaryote
Extra processing of the RNA such as adding a cap, adding a tail, and the splicing of introns take place only in eukaryotic cells.
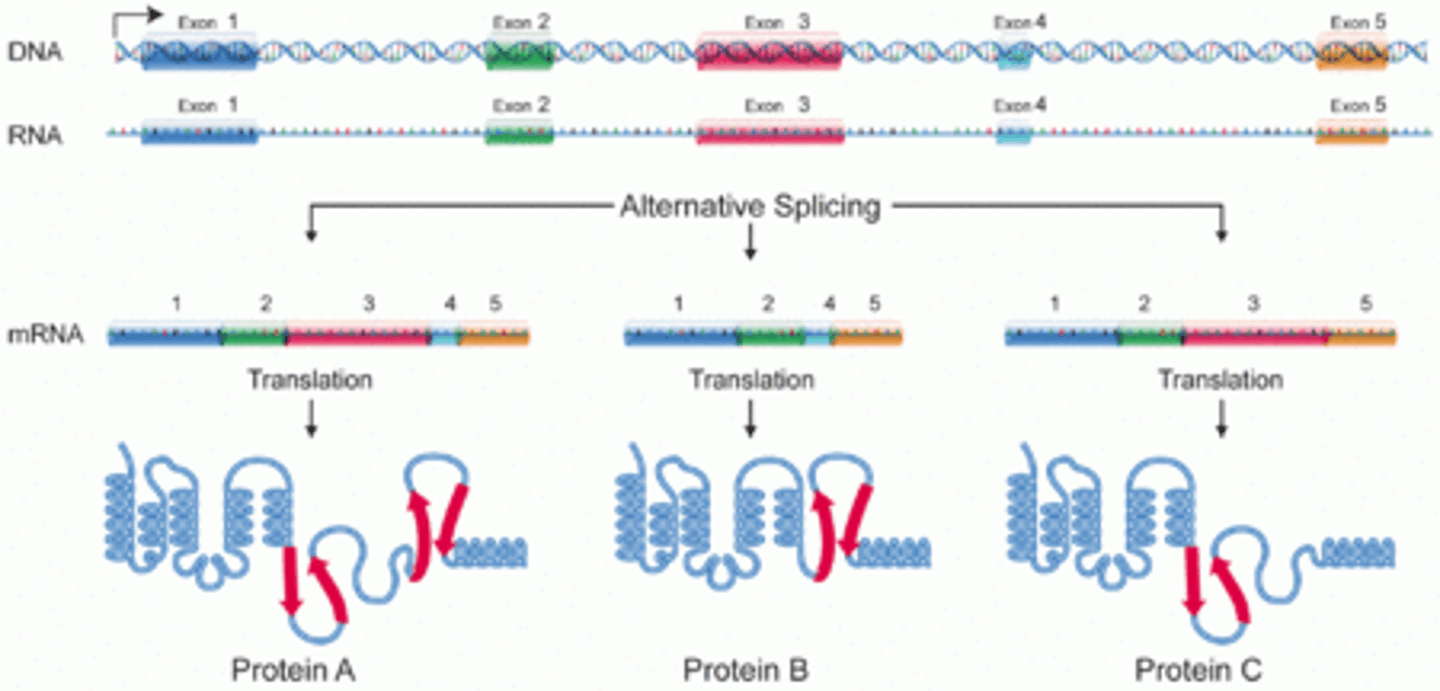
CRB Describe the process of Alternative Splicing. Does it happen in Prokaryotes, Eukaryotes, or Both?
Alternative Splicing occurs in Eukaryotes, and is when exons (as well as introns) could be removed from the RNA during processing to form mRNA. This allows multiple proteins to be formed from the same sequence of DNA.
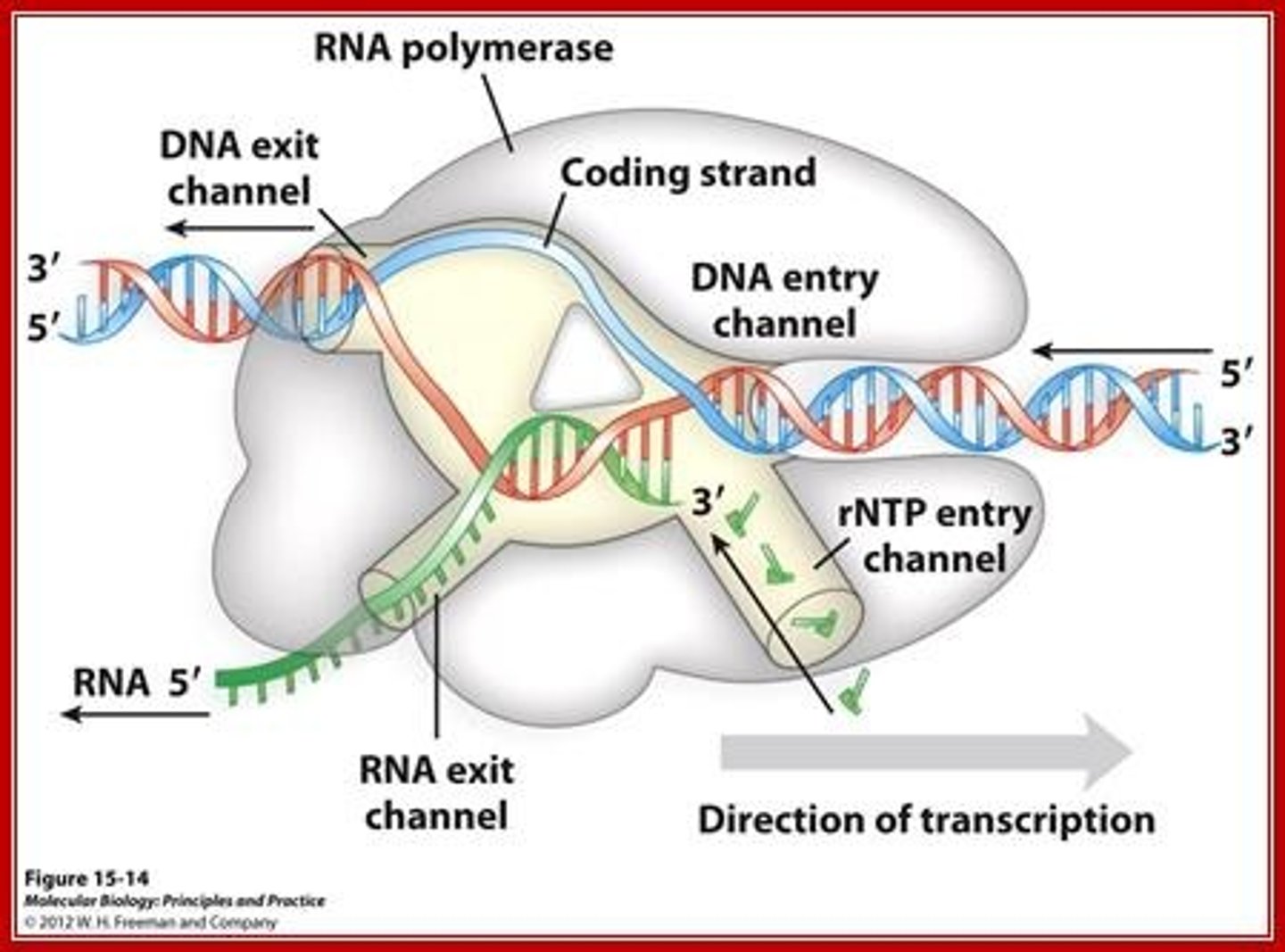
What enzyme is responsible for constructing mRNA using DNA as a template?
(A) DNA Polymerase
(B) RNA Polymerase
(C) DNA Transcriptase
(D) RNA Transcriptase
(B) RNA Polymerase
RNA polymerase is responsible for constructing mRNA using DNA as a template.
What is the primary difference between the promoters found in the DNA of eukaryotic versus prokaryotic cells?
Promoters in eukaryotic DNA are longer and more abundant than those in prokaryotic cells.
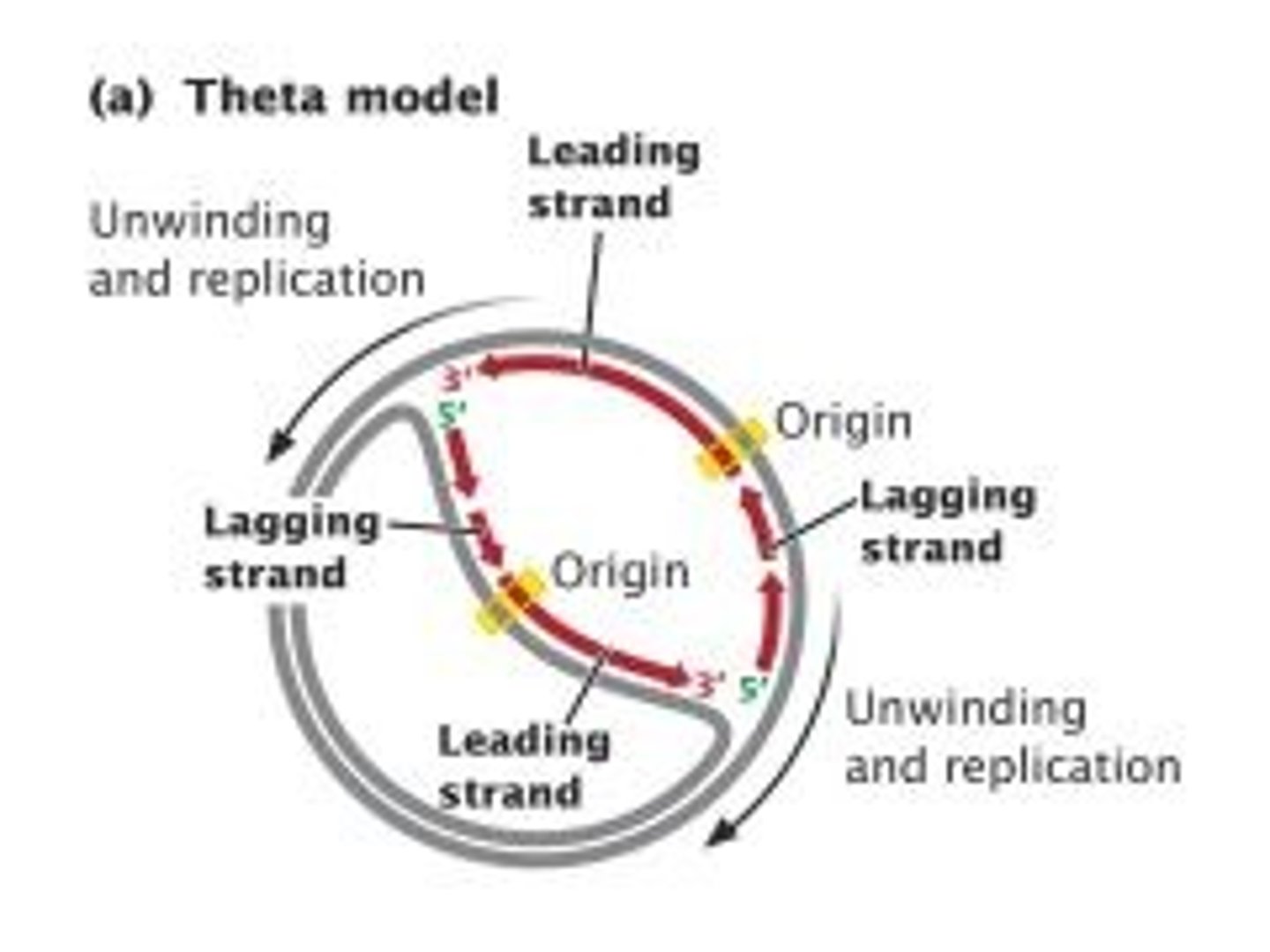
CRB True or false? Because Prokaryotes have circular chromosomes, they look like the Greek letter ϴ when being transcribed, and their replication is sometimes called Theta Replication.
True. Because Prokarytoes have circular chormosomes, they look like the Greek letter ϴ when being transcribed, and their replication is sometimes called Theta Replication.
CRB Which of the following is the common name for the Eukaryotic Promoter?
(A) TATA Box
(B) TATAAT Pribnow Box
(C) GCCG Box
(D) GCGCCG Pribnow Box
(A) TATA Box
The TATA Box is the common name for the Eukaryotic Promoter, based off of its sequence.
Prokaryotes have the TATAAT Pribnow Box.
What is the difference between the template strand and coding strand in the process of transcription?
The template strand is the strand of DNA that the RNA Polymerase reads as it generates mRNA.
The coding strand is not read by RNA Polymerase. Its sequence resembles the newly synthesized strand of mRNA except that the coding strand contains Thymine instead of Uracil.
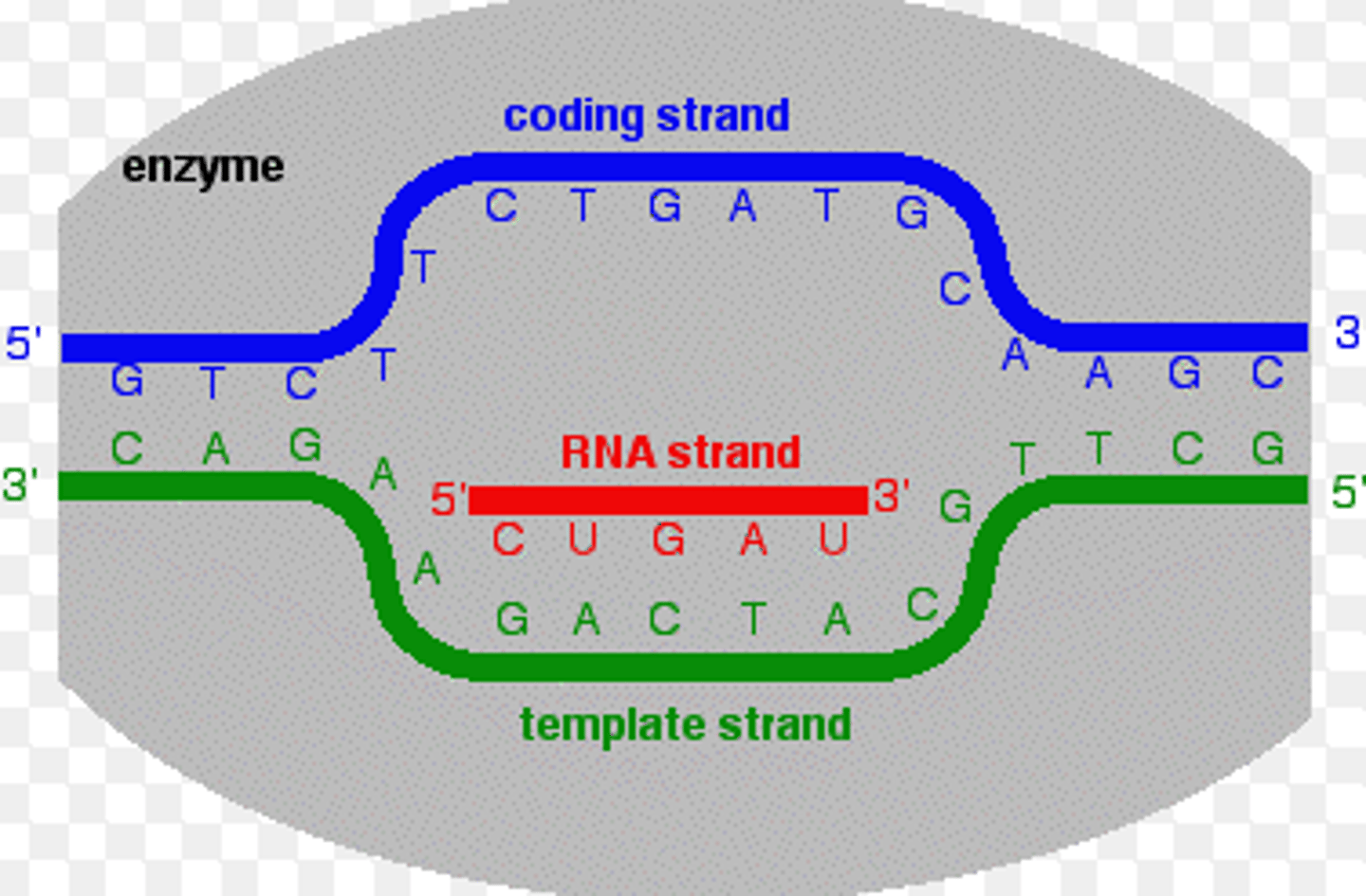
CRB True or false? The Coding Strand is also called the Sense Strand.
True. The Coding Strand is also called the Sense Strand.
RNA Polymerase reads the template strand in the _______ direction, and RNA is generated in the ________ direction.
(A) 5' to 3', 5' to 3'
(B) 5' to 3', 3' to 5'
(C) 3' to 5', 3' to 5'
(D) 3' to 5', 5' to 3'
(D) 3' to 5', 5' to 3'
RNA Polymerase reads the template strand in the 3' to 5' direction, and RNA is generated in the 5' to 3' direction.
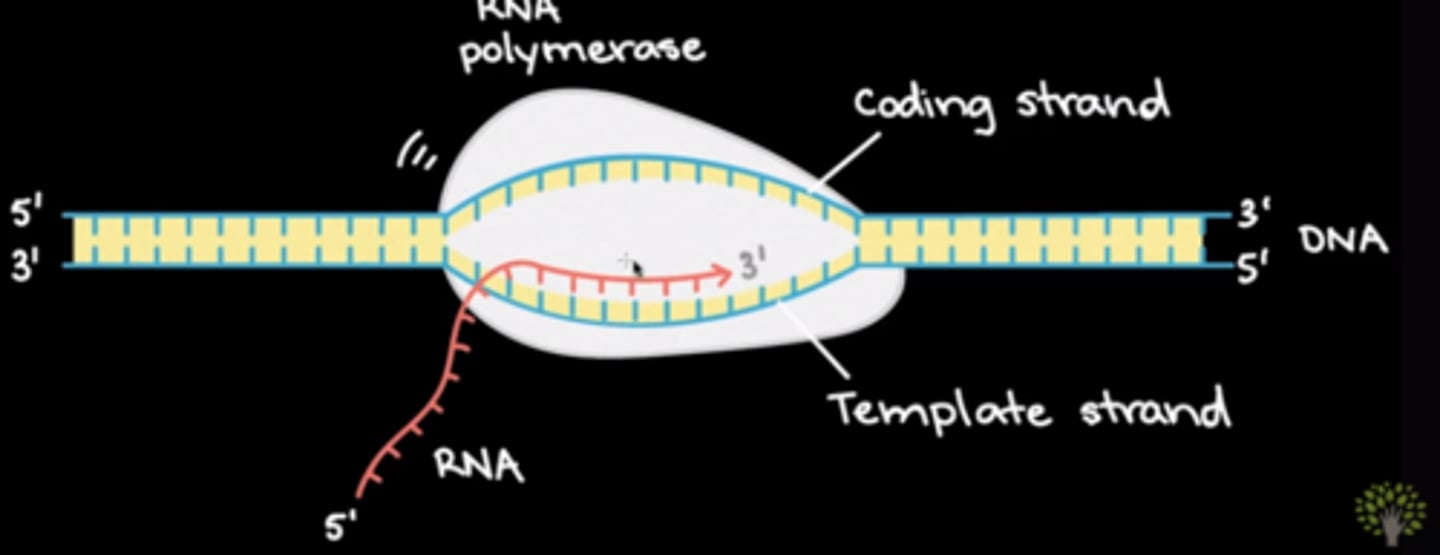
Which of the following nitrogenous bases is NOT found in mRNA?
(A) A
(B) T
(C) C
(D) G
(B) T
You will not find the base Thymine in RNA. Instead, Adenine will pair with Uracil (A-U). The base pair Cytosine-Guanine (C-G) remains the same.
What would be the mRNA strand from the template strand:
3'-TACTAG-5'?
(A) 5'-AUGAUC-3'
(B) 5'-ATGATC-3'
(C) 5'-GAUCAU-3'
(D) 5'-GACTAT-3'
(A) 5'-AUGAUC-3'
The template strand 3'-TACTAG-5' will have an mRNA strand of 5'-AUGAUC-3'.
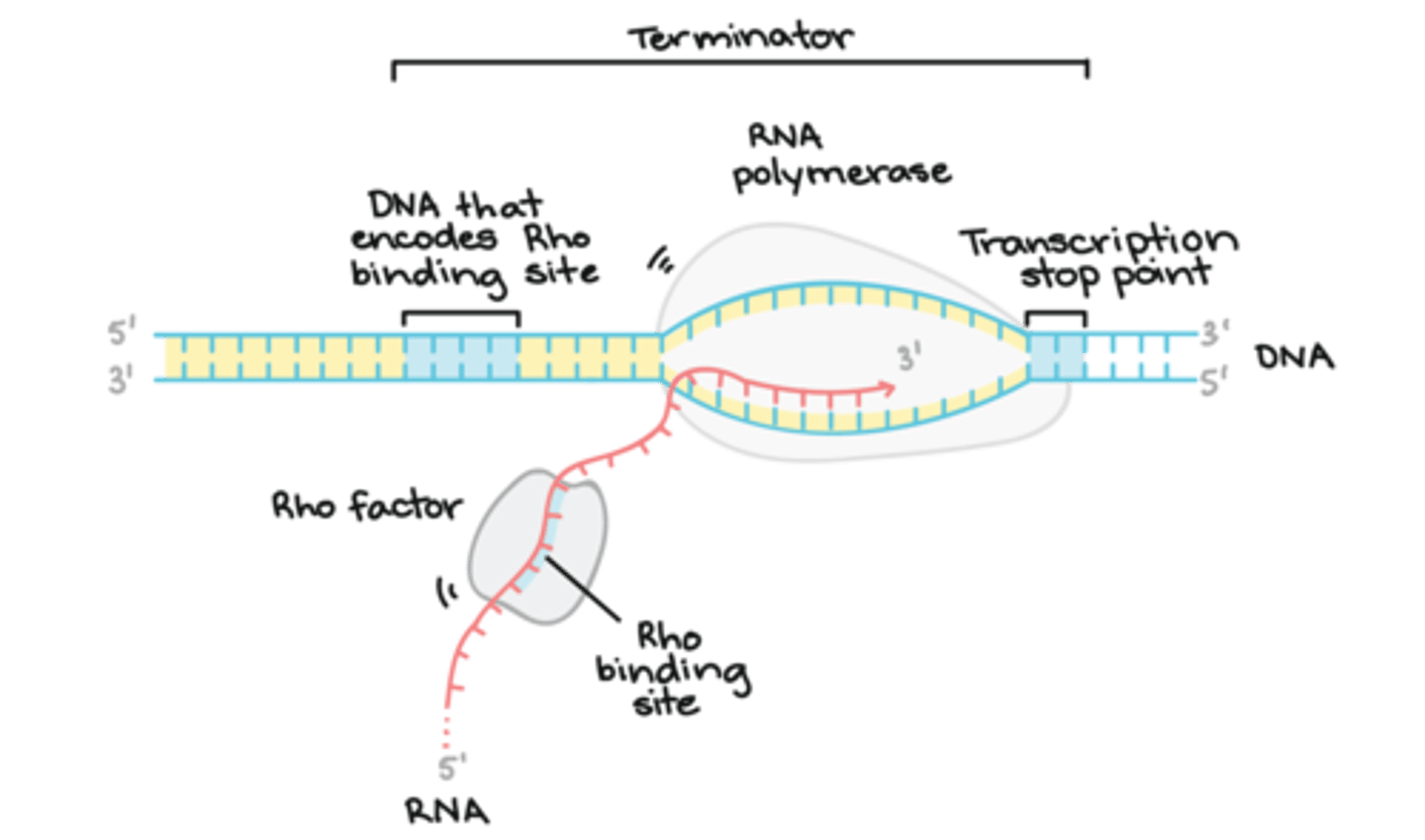
Which of the following are mechanisms that may allow an RNA polymerase to know when to stop transcription?
I. RNA polymerase recognizes a known sequence of DNA called a terminator that signals the RNA polymerase to stop transcription.
II. mRNA forms a hairpin on itself that impairs the polymerase from moving forward.
III. RNA Polymerase receives a signal from telomerase that the DNA template is ending, causing transcription to end.
(A) I Only
(B) I and II Only
(C) II and III Only
(D) I and III Only
(B) I and II Only
The following are mechanisms that allows an RNA polymerase to know when to stop transcription
1. RNA polymerase recognizes a known sequence of DNA called a terminator that signals the RNA polymerase to stop transcription.
2. mRNA forms a hairpin on itself that impairs the polymerase from moving forward.
Which of the following is a difference between the mRNA from Prokaryotic cells versus Eukaryotic cells?
I. In Prokaryotic cells, the RNA polymerase directly makes the mRNA without any extra processing.
II. In Eukaryotic cells, mRNA contains exons, which are later removed with further processing.
III. In Prokaryotic cells, mRNA undergoes translation and transcription simultaneously.
(A) I Only
(B) I and II Only
(C) I and III Only
(D) I, II, and III
(C) I and III Only
The following are differences between the mRNA from Prokaryotic cells versus Eukaryotic cells:
- In Prokaryotic cells, the RNA polymerase directly makes the mRNA without any extra processing.
- In Eukaryotic cells, mRNA contains INTRONS, which are later removed with further processing.
- In Prokaryotic cells, mRNA undergoes translation and transcription simultaneously.
CRB Fill in the blanks: In Eukaryotes, mRNA is ______________, so each mRNA molecule can only be translated into one protein. In _____________, each mRNA molecule can be translated into different proteins, making this mRNA _________________.
(A) Monoprotomic, Archaea, Polyprotomic
(B) Monoprotomic, Prokaryotes, Polyprotomic
(C) Monocistronic, Prokaryotes, Polycistronic
(D) None of the above.
(C) Monocistronic, Prokaryotes, Polycistronic
In Eukaryotes, mRNA is Monocistronic, so each mRNA molecule can only be translated into one protein. In Prokaryotes, each mRNA molecule can be translated into different proteins, making this mRNA Polycistronic.
Introns are removed via a process known as __________________.
(A) Dicing
(B) Splicing
(C) Cutting
(D) Crossing
(B) Splicing
Introns are removed via a process known as splicing.
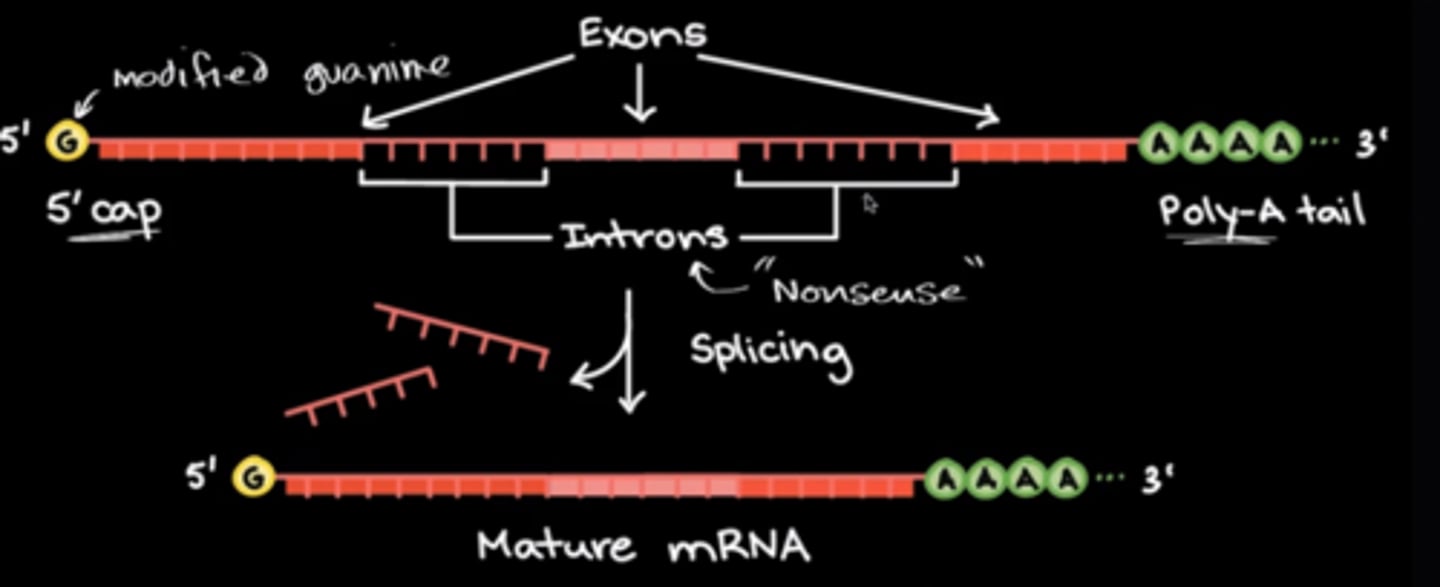
What is the difference between introns vs. exons in terms of expression?
Introns are noncoding regions while exons are coding regions. This means that introns will not be expressed and exons will encode all of the expressed proteins.
Which of the following is not a modification made to pre-mRNA before it becomes mature mRNA?
(A) The addition of 5' cap
(B) The addition of the poly-A tail
(C) The splicing of introns
(D) The removal of telomeric mRNA
(D) The removal of telomeric mRNA
After RNA polymerase makes pre-mRNA, the pre-mRNA goes through some modifications such as:
- the addition of a 5' cap
- the addition of the poly-A tail
- the splicing of introns
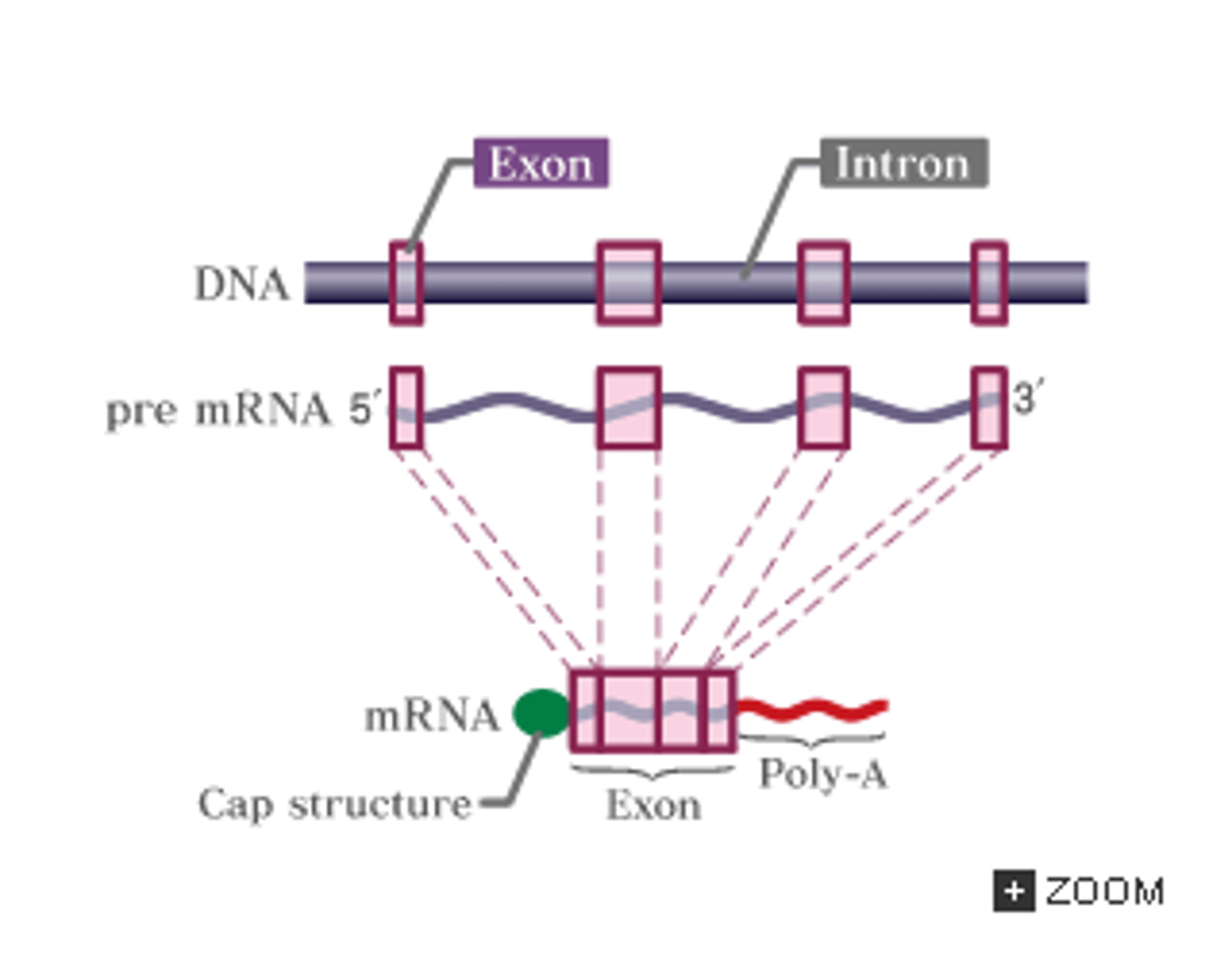
Technically, mRNA has two 3' ends. How is this possible?
An mRNA technically ends in both 3' and 3' because the addition of the 5' cap converts the 5' end into a 3' via a 5'-5' linkage which protects the mRNA from exonucleases.
Please note that we still refer to the mRNA as having a 3' and 5' end.
What is the role of polyadenylate polymerase?
Polyadenylate polymerase is the enzyme responsible for catalyzing the construction of the poly-A tail.
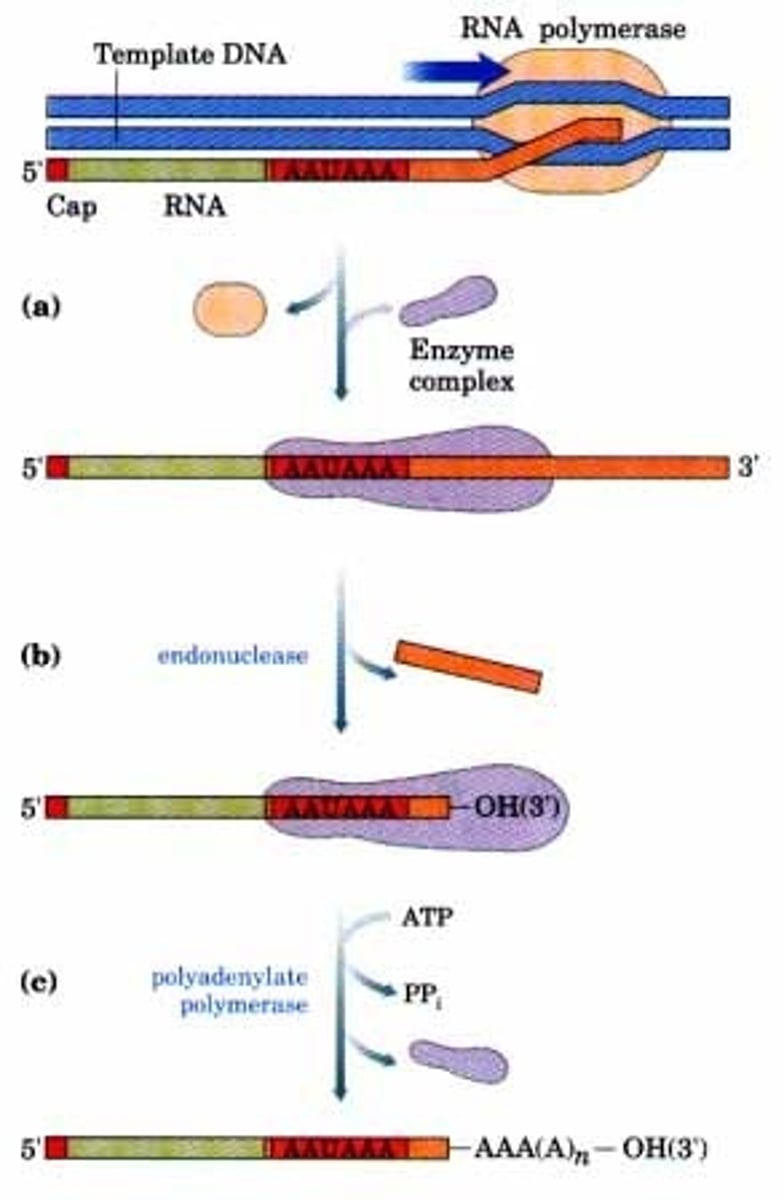
True or False? The poly-A tail may assist in the termination of transcription.
True. The poly-A tail may assist in the termination of transcription.
What is RNA editing?
RNA editing is a post-transcriptional process that may entail the insertion, deletion, and base substitution of nucleotides. This process is quite rare!
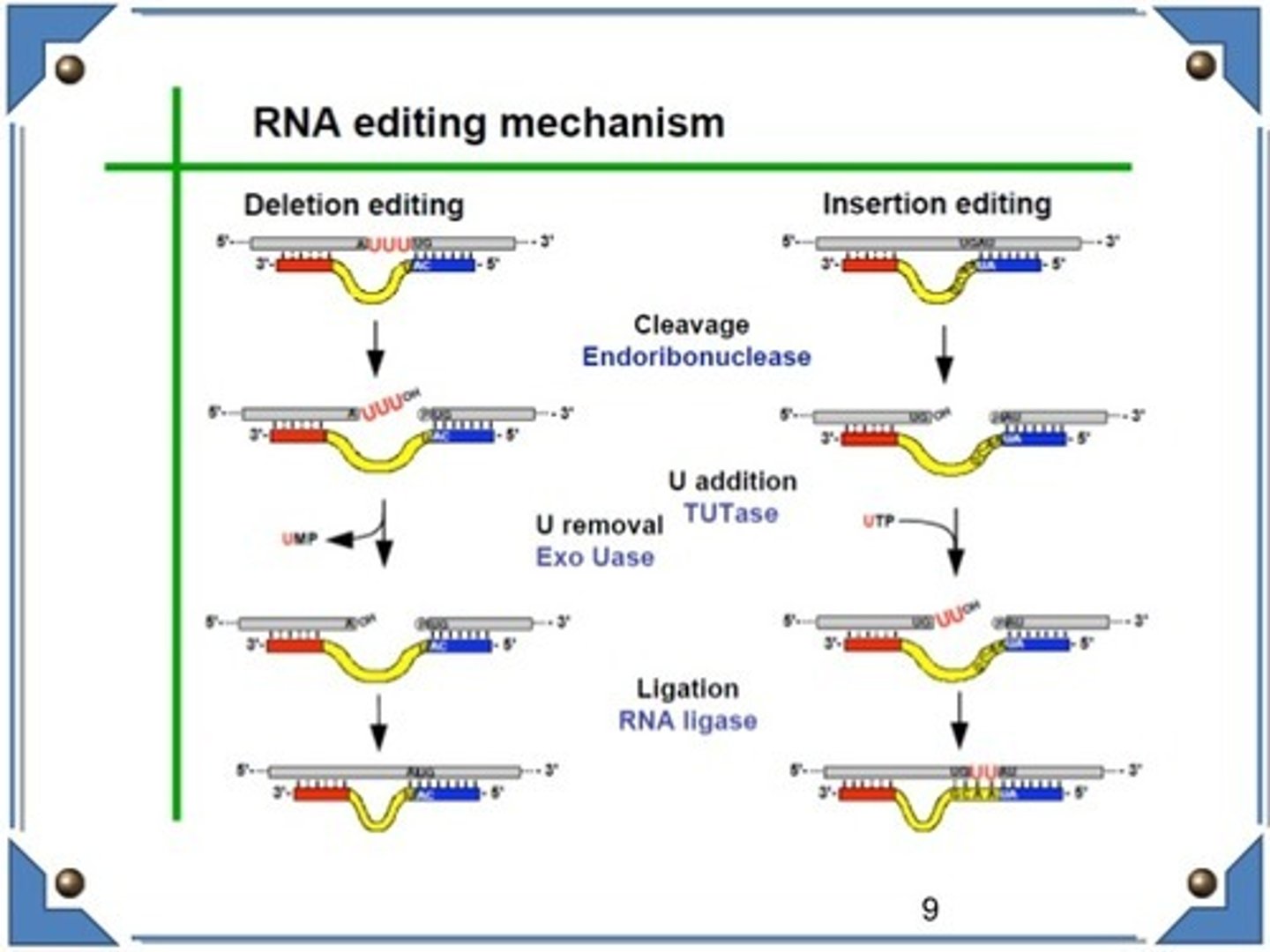
What does Adenosine Deaminase Acting on RNA (ADAR) do?
Adenosine Deaminase Acting on RNA (ADAR) is an enzyme involved in RNA editing that converts adenosine residues into inosine via hydrolytic deamination.
What does Cytosine Deaminase Acting on RNA (CDAR) do?
An RNA editing enzyme called Cytosine Deaminase Acting on RNA (CDAR) converts cytosine residues into uridine via deamination.

Which of the following is the umbrella term for "functional RNA that does not get translated into a protein"?
(A) miRNA
(B) snoRNA
(C) ncRNA
(D) siRNA
(C) ncRNA
non-coding RNA (ncRNA) is functional RNA that does not get translated into a protein. Examples of ncRNA include micro RNA (miRNA), small nucleolar RNA (snoRNA), and silencing RNA (siRNA).
Typically ncRNA plays a role in which two processes?
(A) Synthesis and Replication
(B) Translation and Transcription
(C) Translation and Replication
(D) Replication and Transcription
(B) Translation and Transcription
Typically ncRNA plays a role in translation and transcription.
What is the function of an micro RNA (miRNA)?
The function of miRNA is to silence mRNA via complementary base pairing.
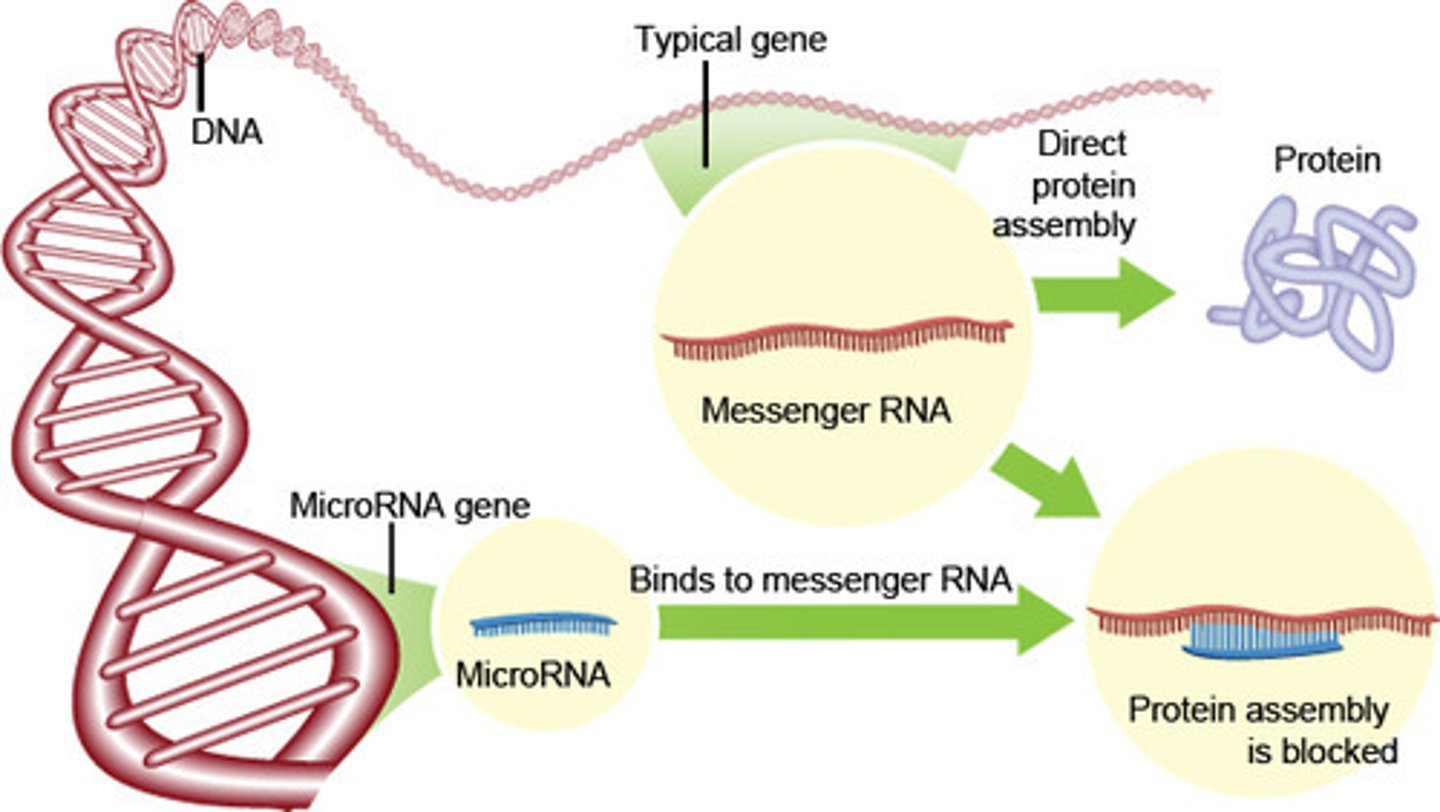
miRNA may silence gene expression through which of the following methods?
I. Blocking translation
II. mRNA modification
III. mRNA degradation
(A) I Only
(B) I and II Only
(C) I and III Only
(D) I, II, and III
(C) I and III Only
miRNA may silence gene expression through blocking translation or mRNA degradation. miRNA will not modify mRNA sequences.
What is the function of small nucleolar RNA (snoRNA) versus small nuclear RNA (snRNA)?
Small nucleolar RNA (snoRNA) guide covalent modifications of rRNA, tRNA, and small nuclear RNA primarily through methylation and pseudouridylation.
Small nuclear RNA (snRNA) process pre-mRNA in the nucleus, aide in the regulation of RNA polymerase and transcription factors, and help maintain telomeres.
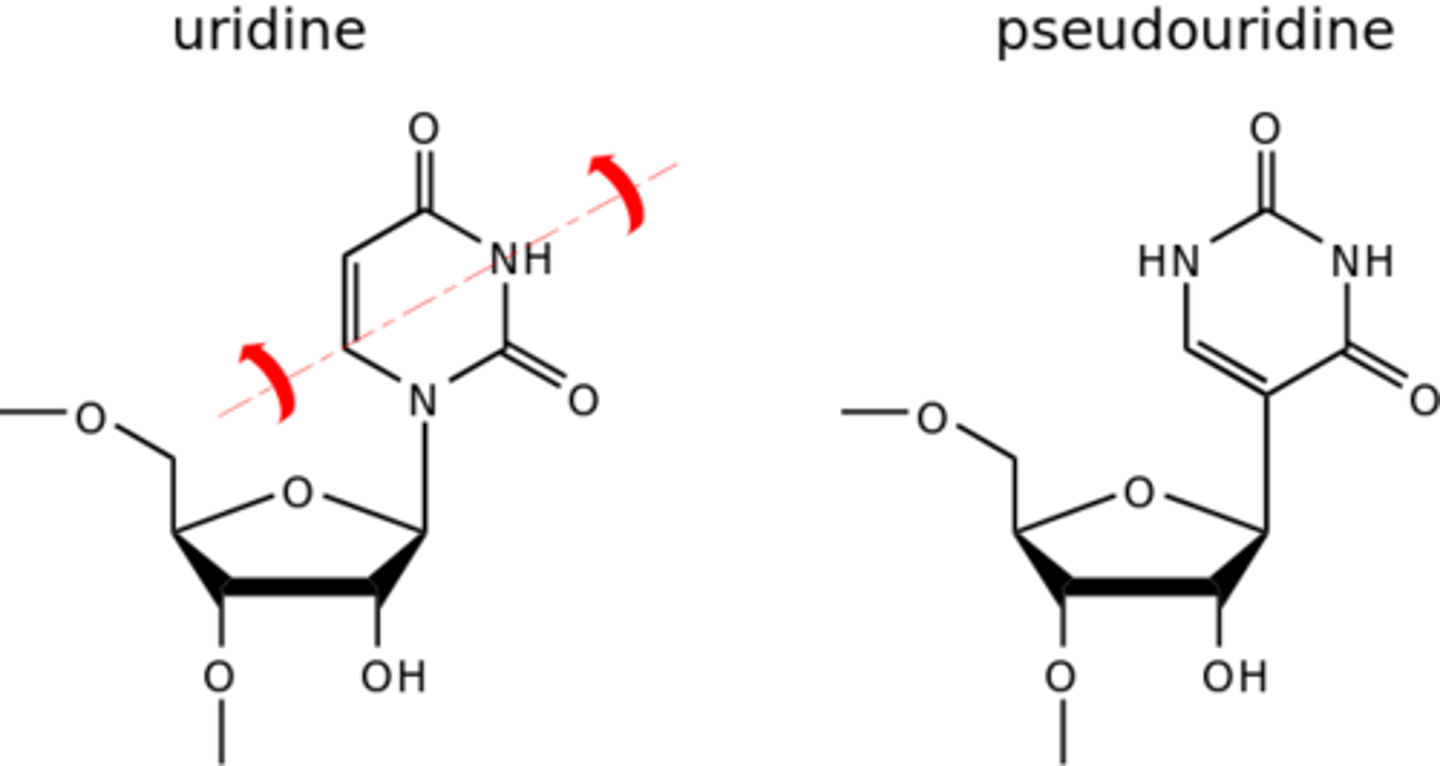
What is pseudouridylation?
Pseudouridylation is the addition of an isomer of the nucleoside uridine by small nucleolar RNA (snoRNA).
What is the function of the spliceosome?
The spliceosome is responsible for splicing introns out and ligating exons together in making mature mRNA.
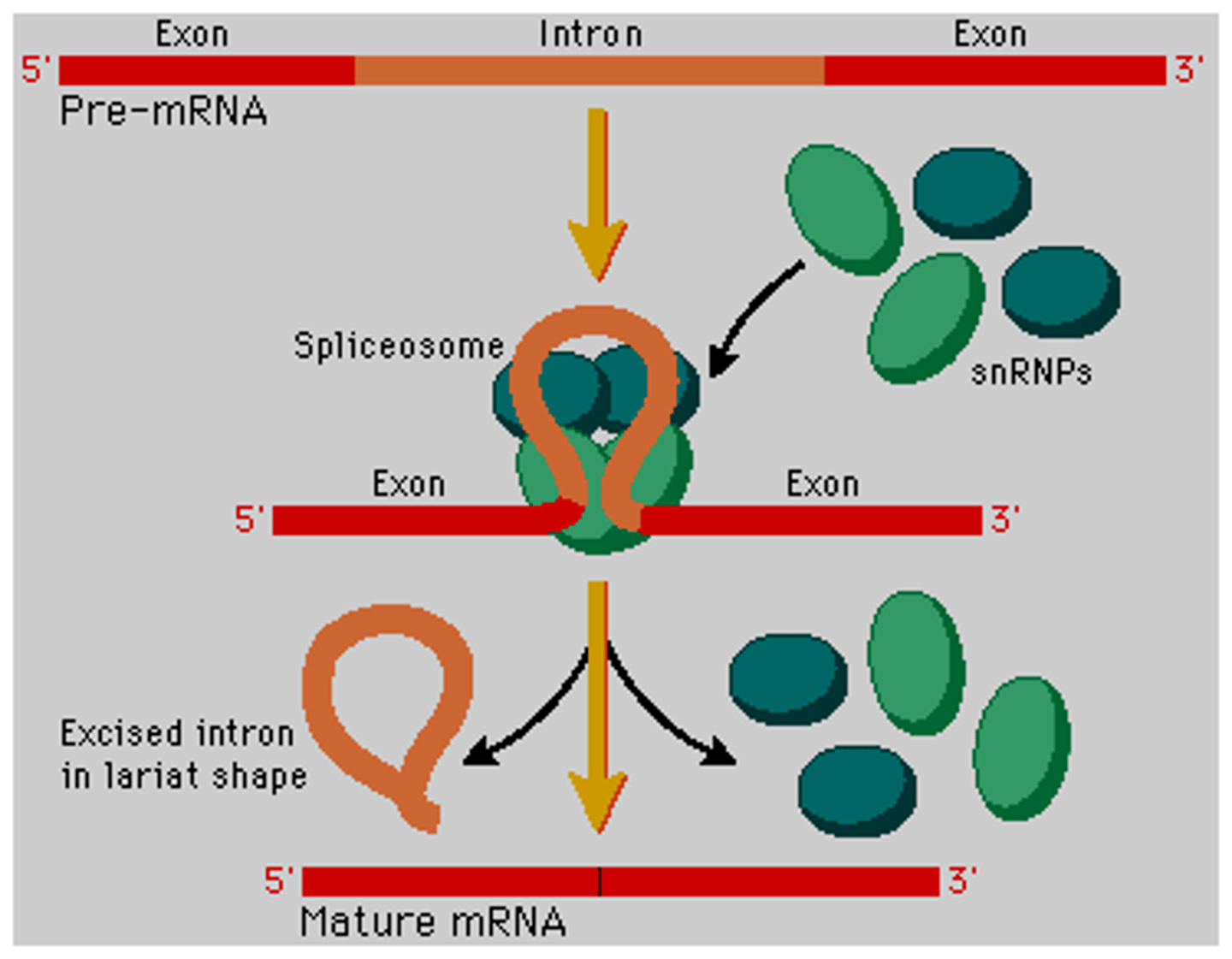
What is the spliceosome composed of?
(A) snRNA and snoRNA
(B) snRNA and snRNP
(C) snoRNA and snRNP
(D) snRNA and miRNA
(B) snRNA and snRNP
A spliceosome is a complex protein composed of small nuclear RNA (snRNA) and small nuclear ribonucleic proteins (snRNP) and it is responsible for post-transcriptional splicing of introns in mRNA.
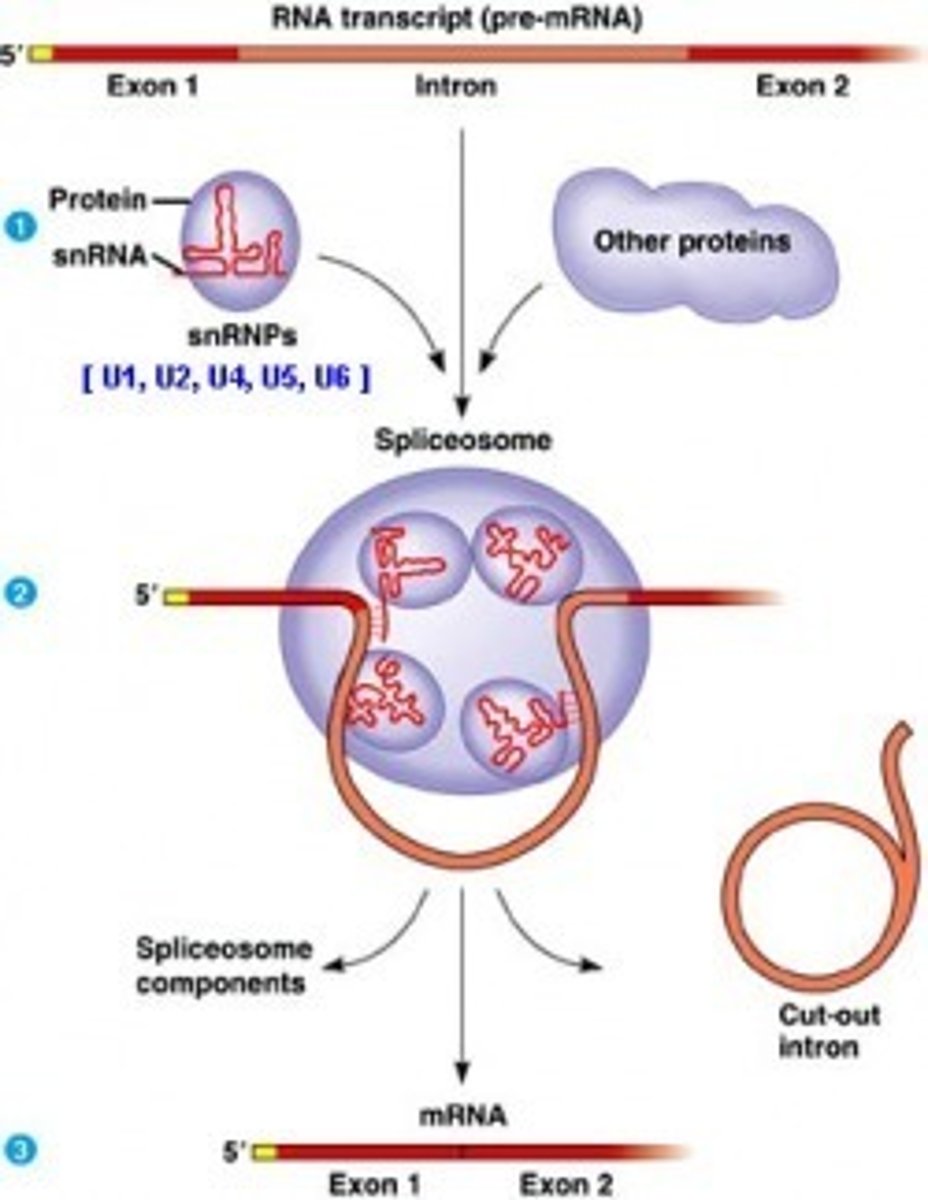
snRNA can combine with what to form snRNP (aka snRP)?
snRNA can combine with proteins to form small nuclear ribonucleic proteins (snRNP or snRP).
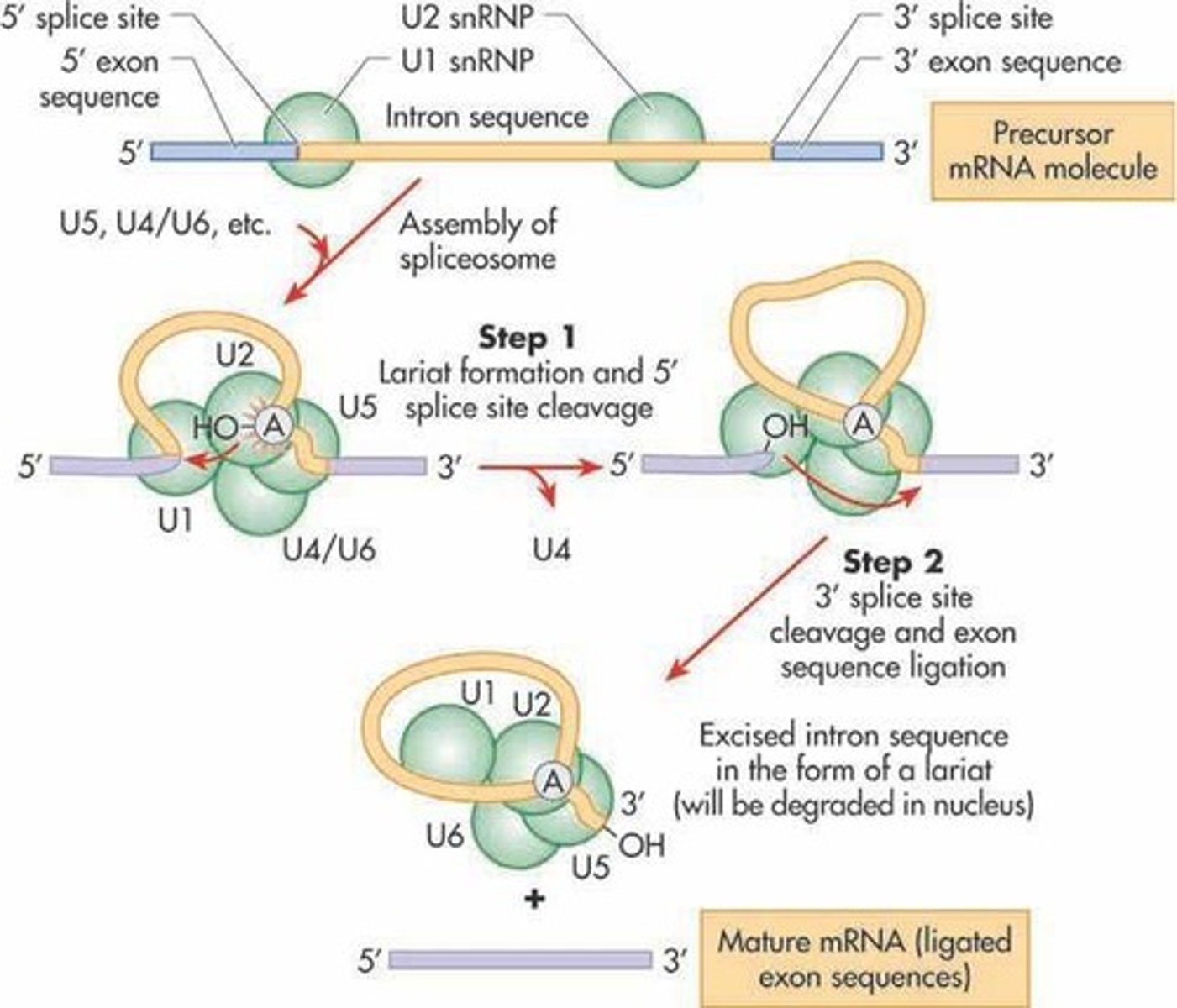
The spliceosome splices introns through:
(A) Two sequential transesterification reactions followed by a ligation reaction.
(B) Two sequential ligation reactions followed by a transesterification reaction.
(C) One transesterification reaction followed by two sequential ligation reactions.
(D) One ligation reaction followed by a two sequential transesterification reactions.
(A) Two sequential transesterification reactions followed by a ligation reaction.
The spliceosome splices introns through two sequential transesterification reactions and a ligation.
True or False? A single strand of mRNA is always transcribed and translated simultaneously.
False. A single strand of mRNA is transcribed and translated simultaneously in prokaryotic cells since both processes occur in the cytoplasm. In eukaryotic cells, however, mRNA is transcribed in the nucleus, processed, and then transported to the cytoplasm to be translated by the ribosome.
After post-transcriptional modifications are complete, where will the mature mRNA go next?
(A) Cytosol
(B) Golgi
(C) Nucleolus
(D) Lysosome
(A) Cytosol
After a eukaryotic cell makes a mature mRNA, the mRNA would exit the nucleus and go to the cytosol where the ribosomes are located. It will then undergo translation.
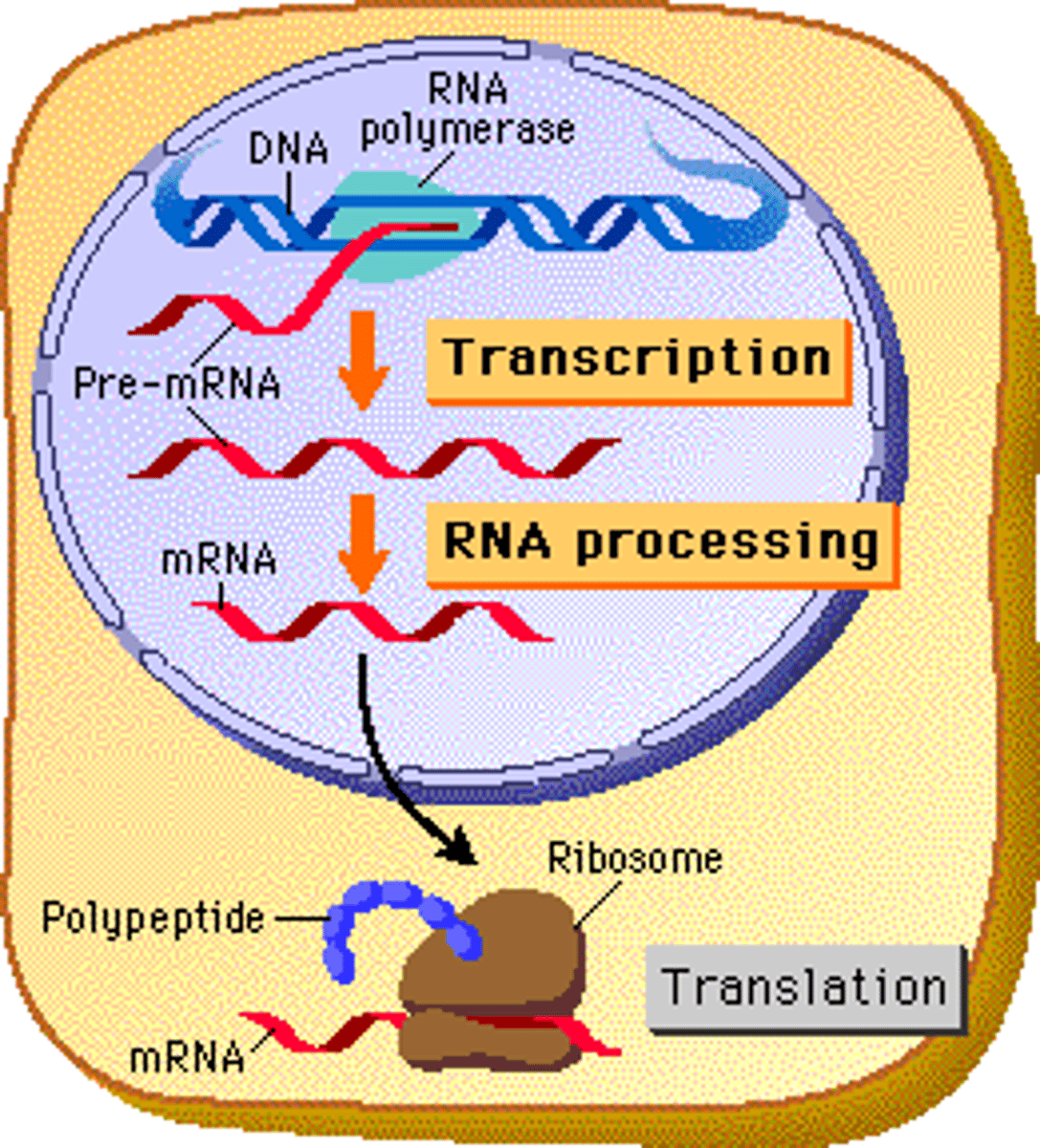
What are the primary components of a ribosome?
I. tRNA
II. rRNA
III. Proteins
(A) I and II Only
(B) I and III Only
(C) II and III Only
(D) I, II, and III
(C) II and III Only
A ribosome is composed of proteins and ribosomal RNA (rRNA).
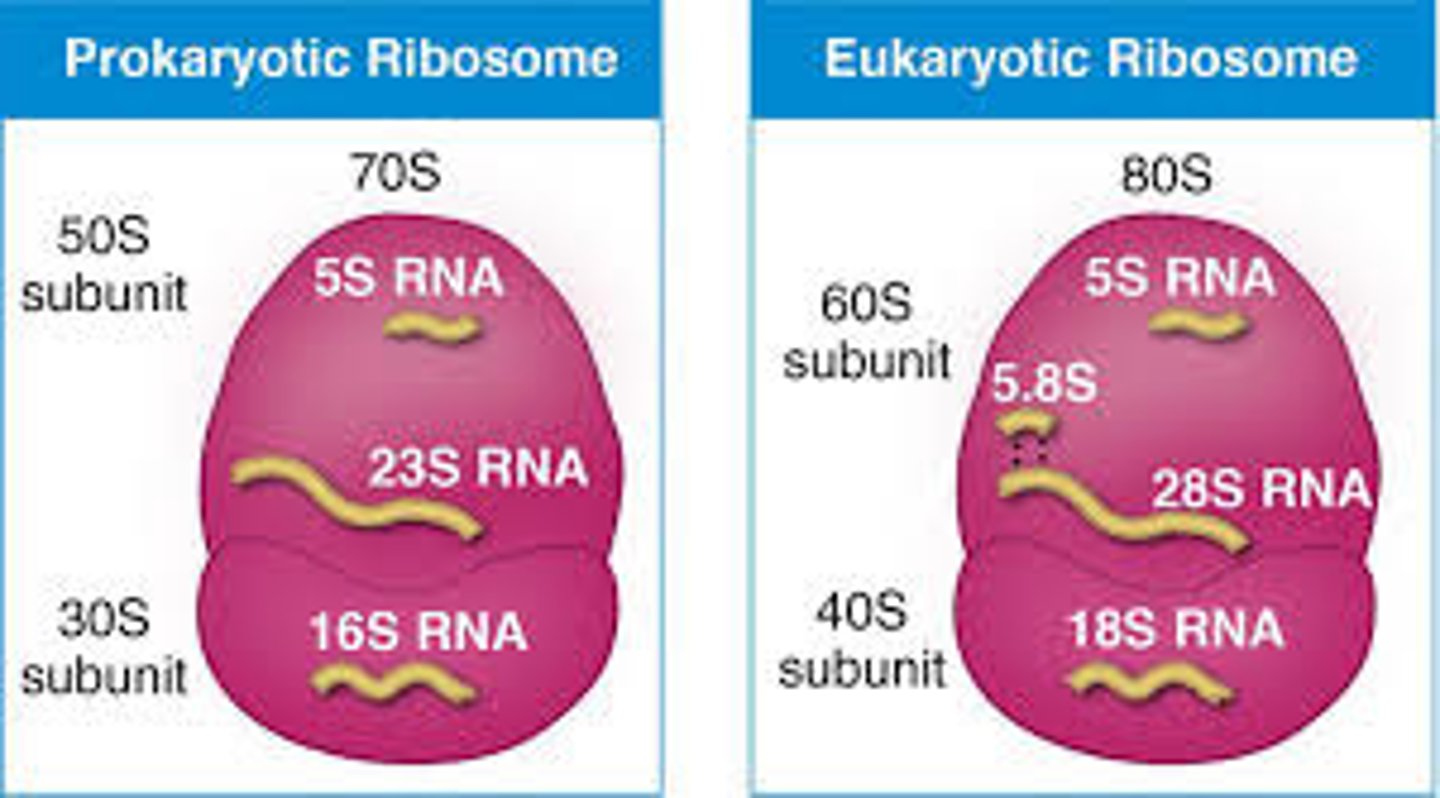
Compare the functions of ribosomal RNA (rRNA) and messenger RNA (mRNA).
Ribosomal RNA (rRNA) provides both a functional and structural role within the ribosome, a key participant in protein translation.
This is quite different than mRNA's information-storage role.
CRB True or false? Many rRNA actually form Ribozymes, which are like enzymes that are not made out of proteins.
True. Many rRNA actually form Ribozymes, which are like enzymes that are not made out of proteins.
What is the function of the ribosome?
Ribosomes read an mRNA template to produce a polypeptide.
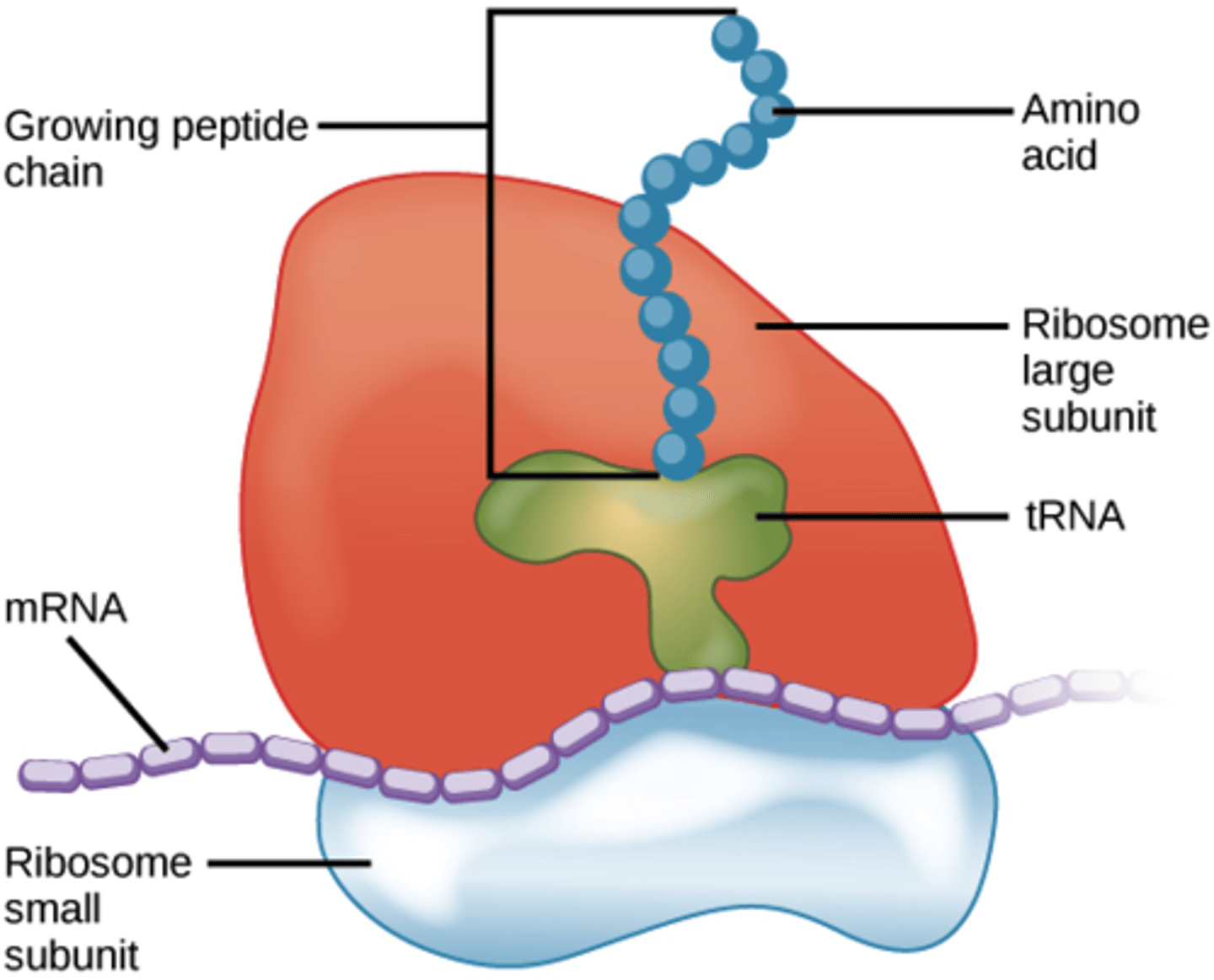
Ribosomes read the mRNA template from the ___ end to the ___ end.
(A) 3', 5'
(B) 3', 3'
(C) 5', 3'
(D) 5', 5'
(C) 5', 3'
Ribosomes read the mRNA template from the 5' end to the 3' end.
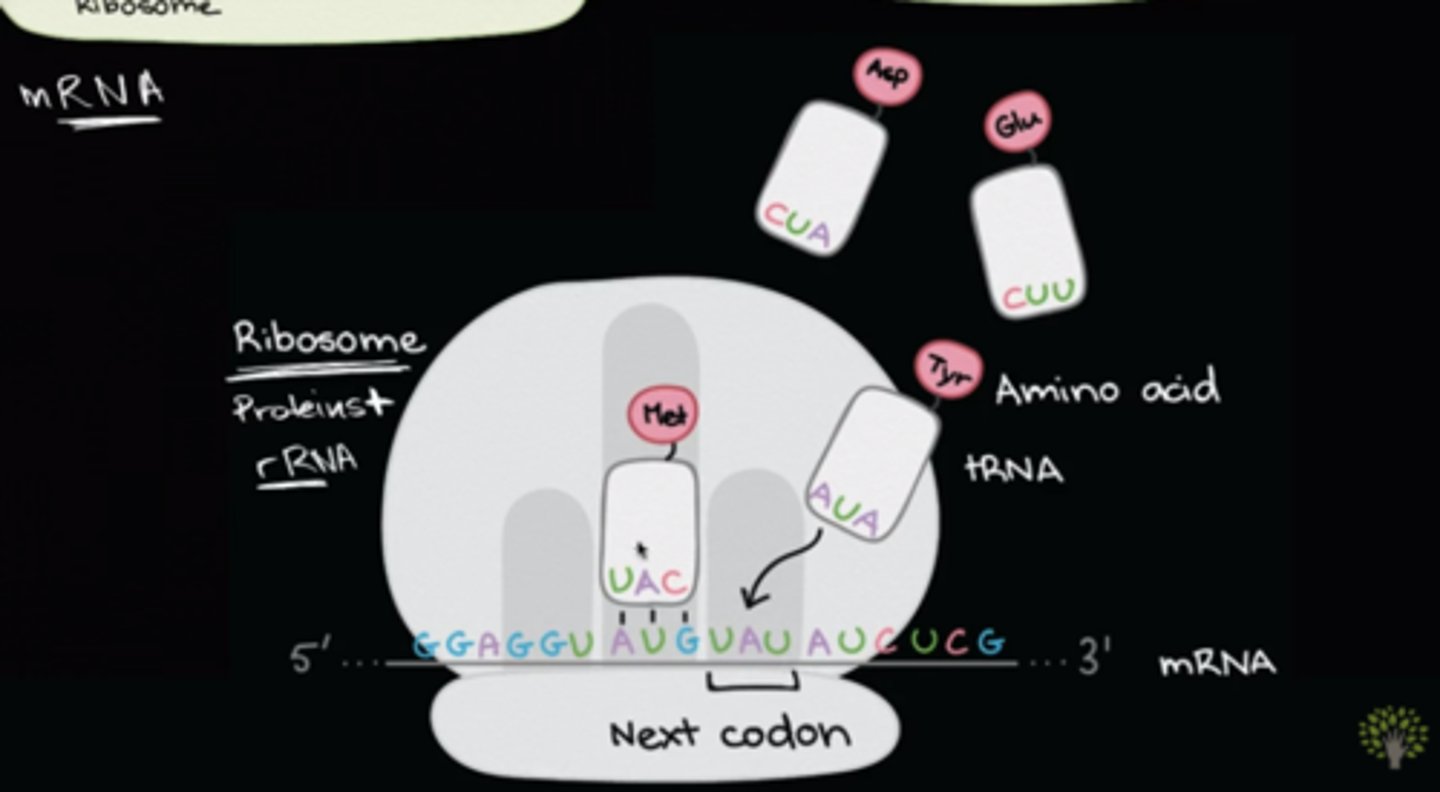
What is the difference between a codon vs. an anti-codon?
A codon is composed of three nucleotides found on the mRNA that code for an amino acid.
An anticodon is part of transfer RNA (tRNA) that complements the mRNA, positioning the tRNA and its attached amino acid for addition to the polypeptide. tRNA are specific for each amino acid.
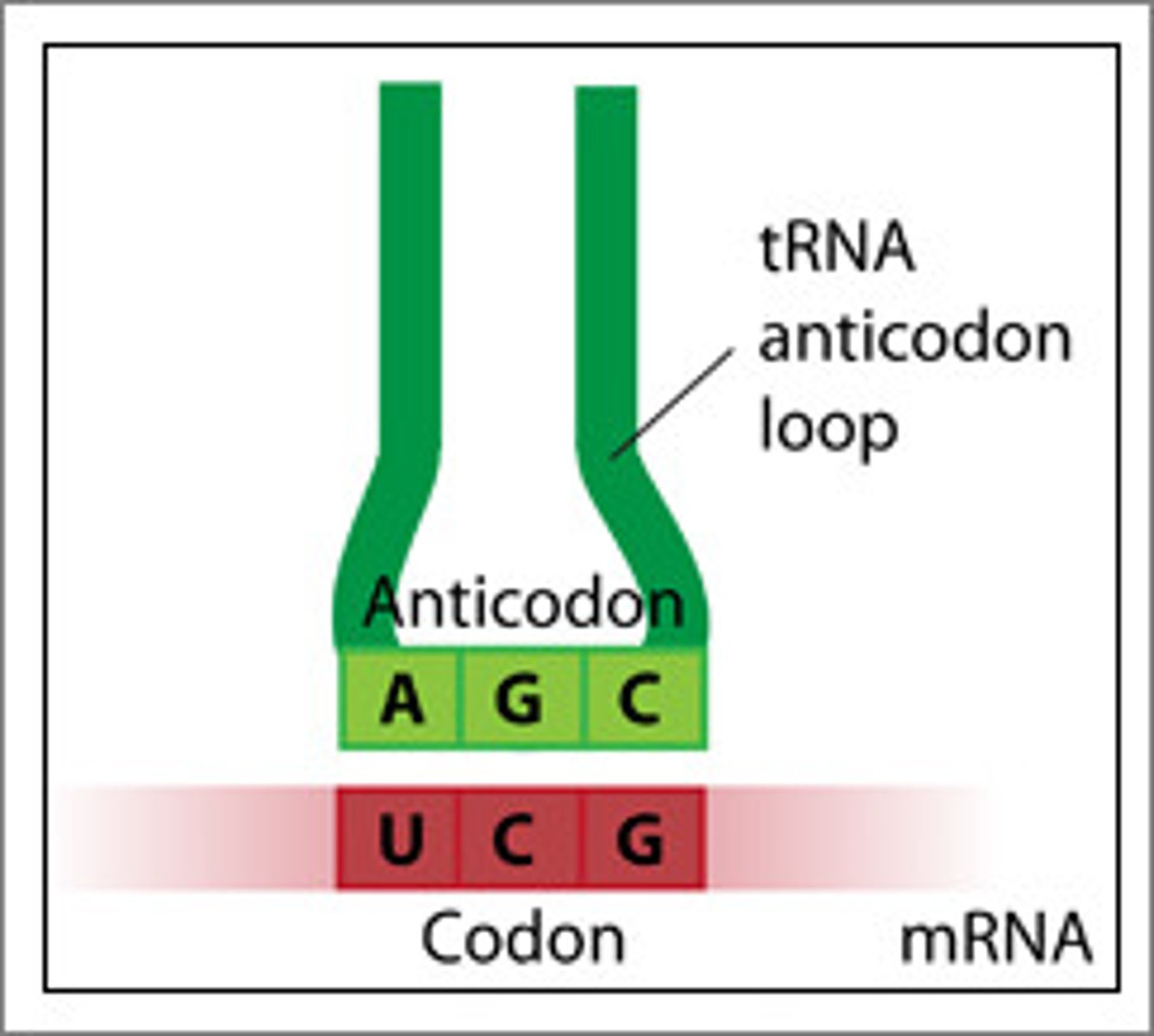
What is the start codon's function and structure?
The start codon (AUG) is where the ribosome initially attaches to start translating the messenger RNA. This AUG sequence encodes for the amino acid Methionine.
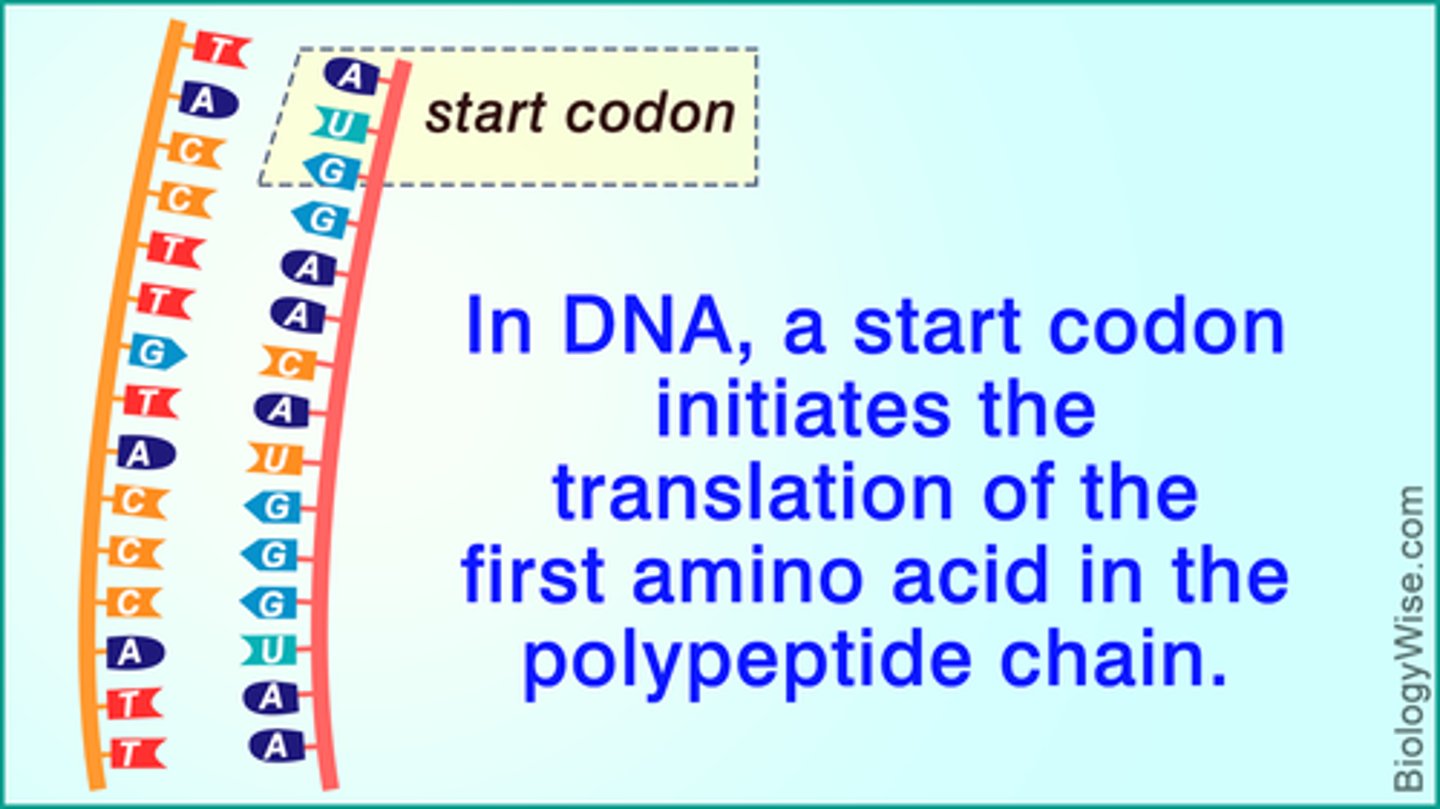
What are the stop codons' function and structures?
The stop codons (UAA, UGA, UAG) signal the ribosome to stop translating. They do not encode for any amino acids.
Think "U Are Annoying", "U Go Away", and "U Are Gone".

The t-RNA with the anticodon of UAC will initially bind at which site within the ribosome?
(A) A-site
(B) E-site
(C) P-site
(D) S-site
(C) P-site
UAC is the anticodon for AUG, which is the start codon. This tRNA molecule carries methionine into the P site to start the translation process.
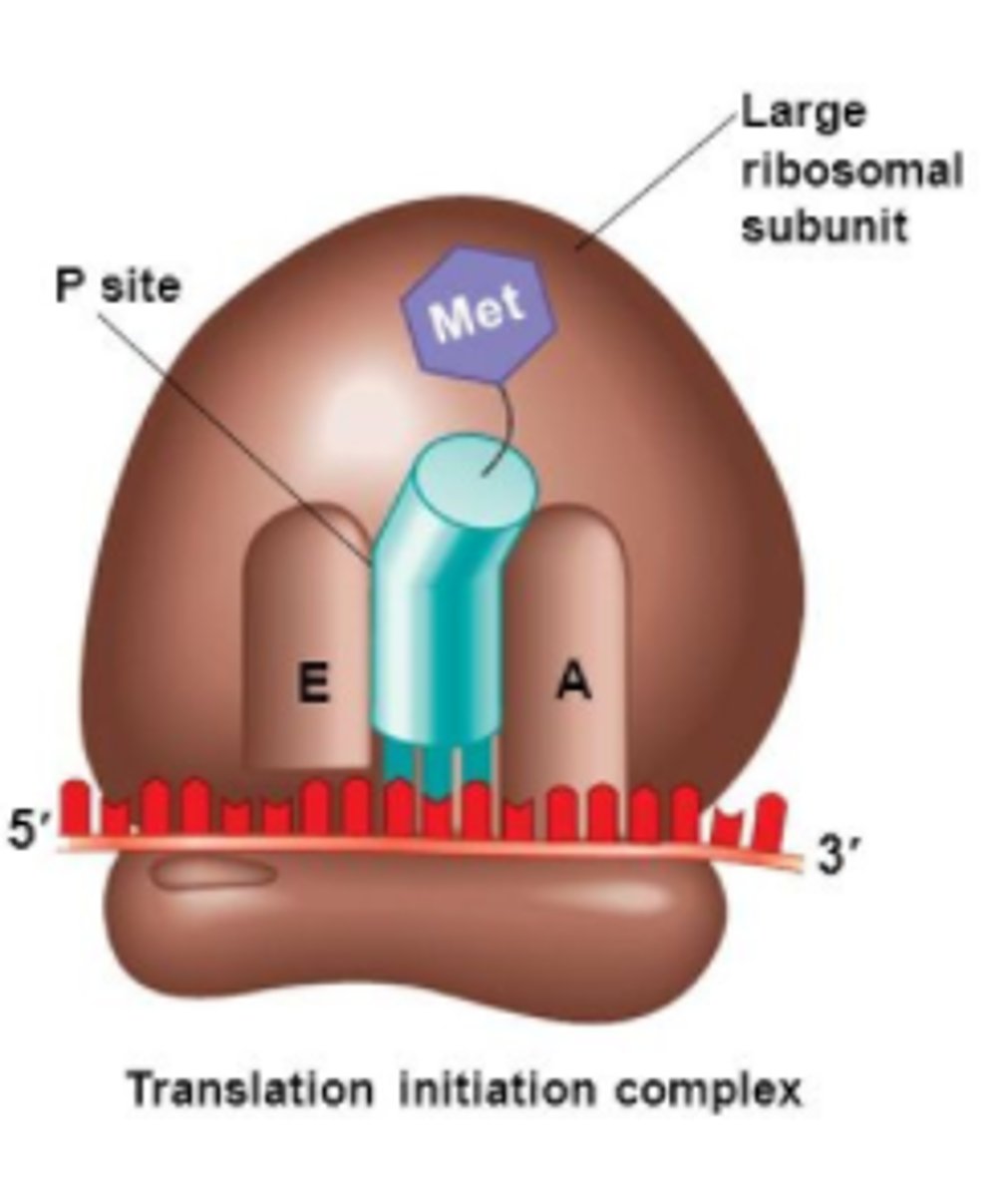
CRB In Prokaryotes, where does the small Ribosomal Subunit first bind for translation?
(A) Shine-Dalgarno Sequence
(B) TATA box
(C) TATAAT
(D) Inititator sequence
(A) Shine-Dalgarno Sequence
The Shine Dalgarno-sequence is in the 5' untranslated region of the RNA, and is where the small Ribosomal Subunit first binds.
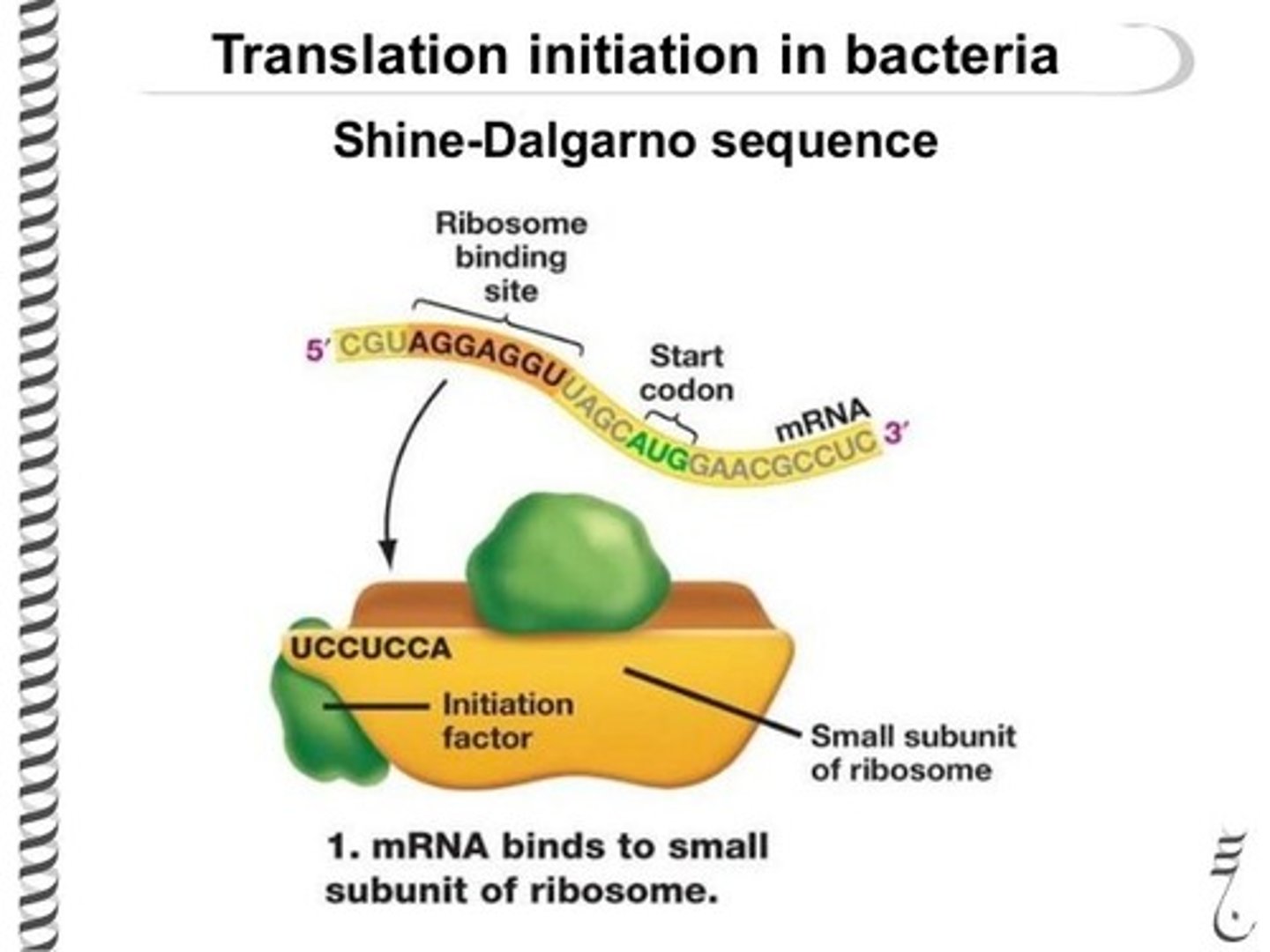
At which site on the ribosome will an incoming tRNA (other than the tRNA with the anticodon of UAC) bind?
(A) A-site
(B) E-site
(C) P-site
(D) S-site
(A) A-site
The A-site on the ribosome is where the tRNA initially enters carrying an amino acid. The name "A-site" comes from the word "amino acid."
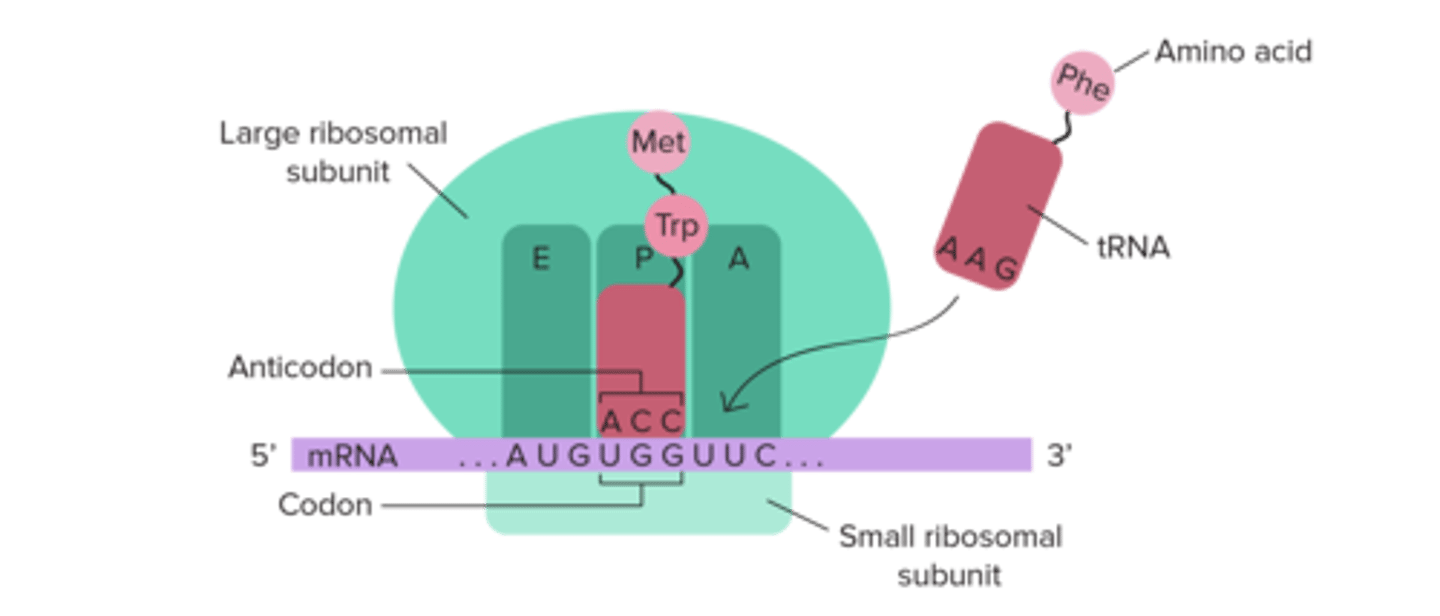
Which site on the ribosome holds the growing polypeptide chain?
(A) A-site
(B) E-site
(C) P-site
(D) S-site
(C) P-site
The P-site on the ribosome is where the polypeptide chain is forming. The name "P-site" comes from the word "polypeptide."
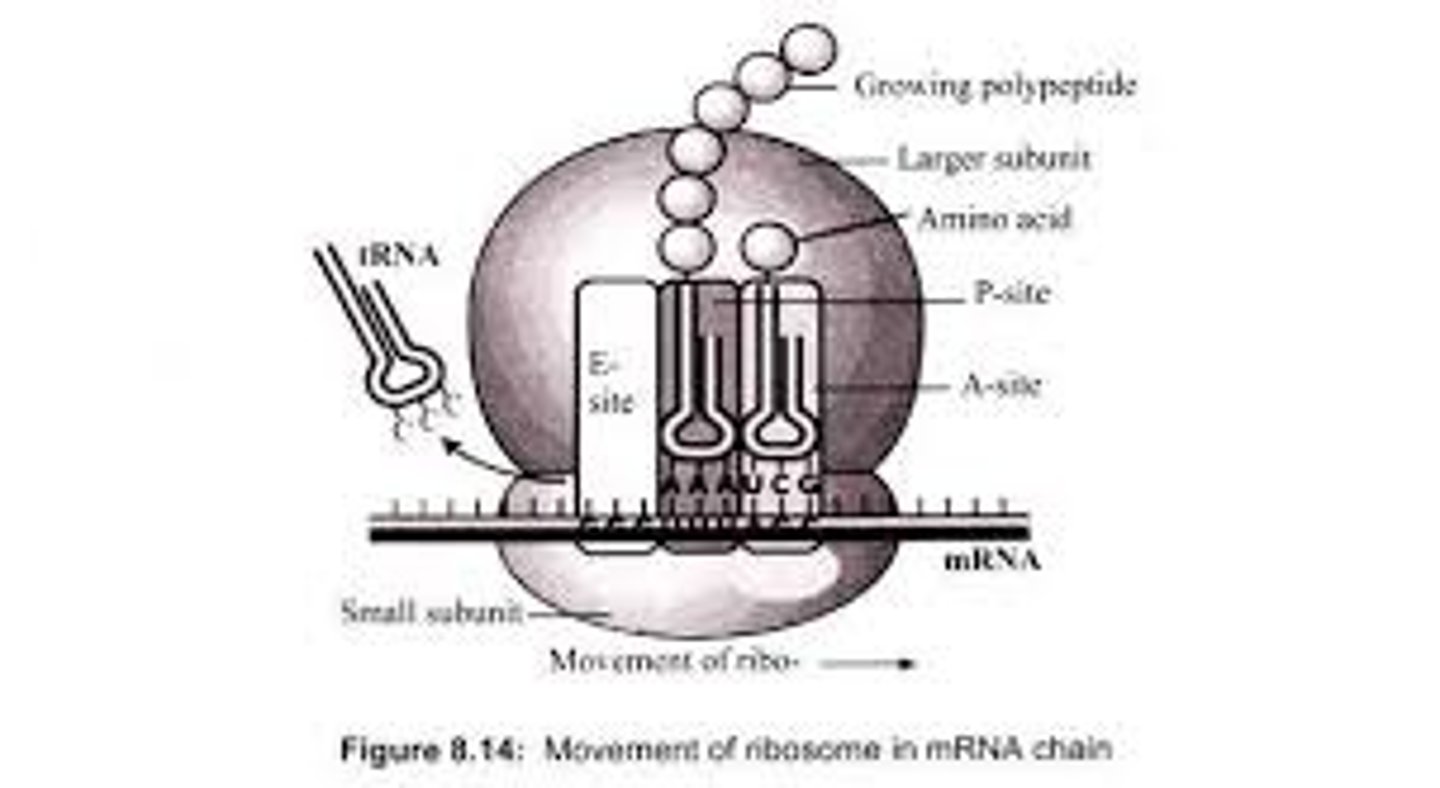
From which site on the ribosome does the empty tRNA exit?
(A) A-site
(B) E-site
(C) P-site
(D) S-site
(B) E-site
The E-site on the ribosome is where the empty tRNA exits. That name "E-site" comes from the word "exit."
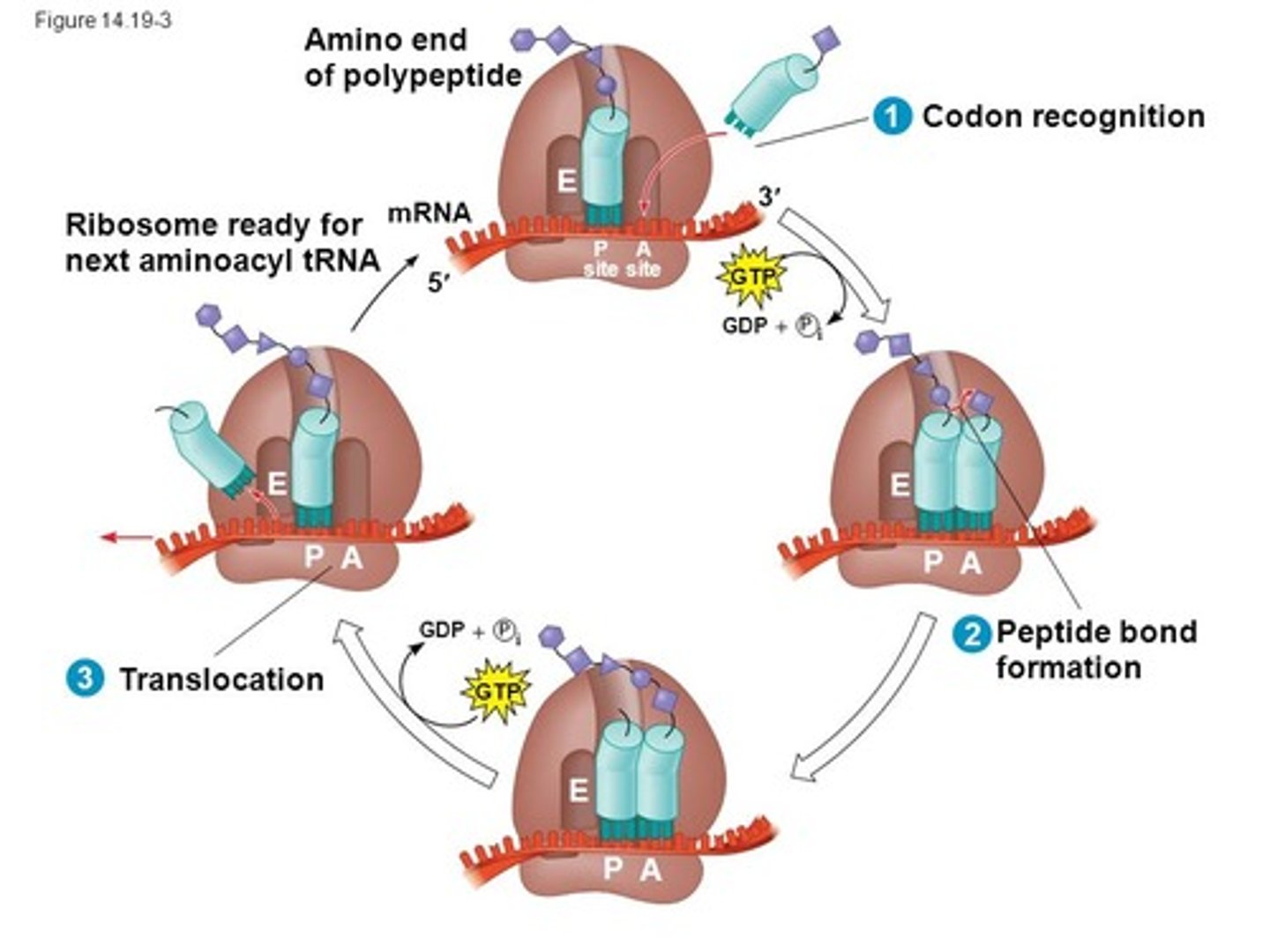
CRB True or false? After the ribosome releases the newly-formed proteins, another class of proteins called Chaperones help this protein be transported.
False. After the ribosome releases the newly-formed proteins, another class of proteins called Chaperones help this protein fold properly.
As the tRNA in the P-site shifts to the E-site, which of the following is true?
I. The ribosome shifts towards the 5' end of the mRNA.
II. The tRNA in the A-site shifts to the P-site.
III. The amino acid from the A-site moves to bind to the polypeptide chain in the P-site.
(A) I Only
(B) II Only
(C) I and II Only
(D) I, II, and III
(B) II Only
As the tRNA in the P-site shifts to the E-site...
- The ribosome shifts towards the 3' (NOT 5') end of the mRNA.
- The tRNA in the A-site shifts to the P-site.
- The polypeptide chain in the P-site moves to bind to the amino acid in the A-site.
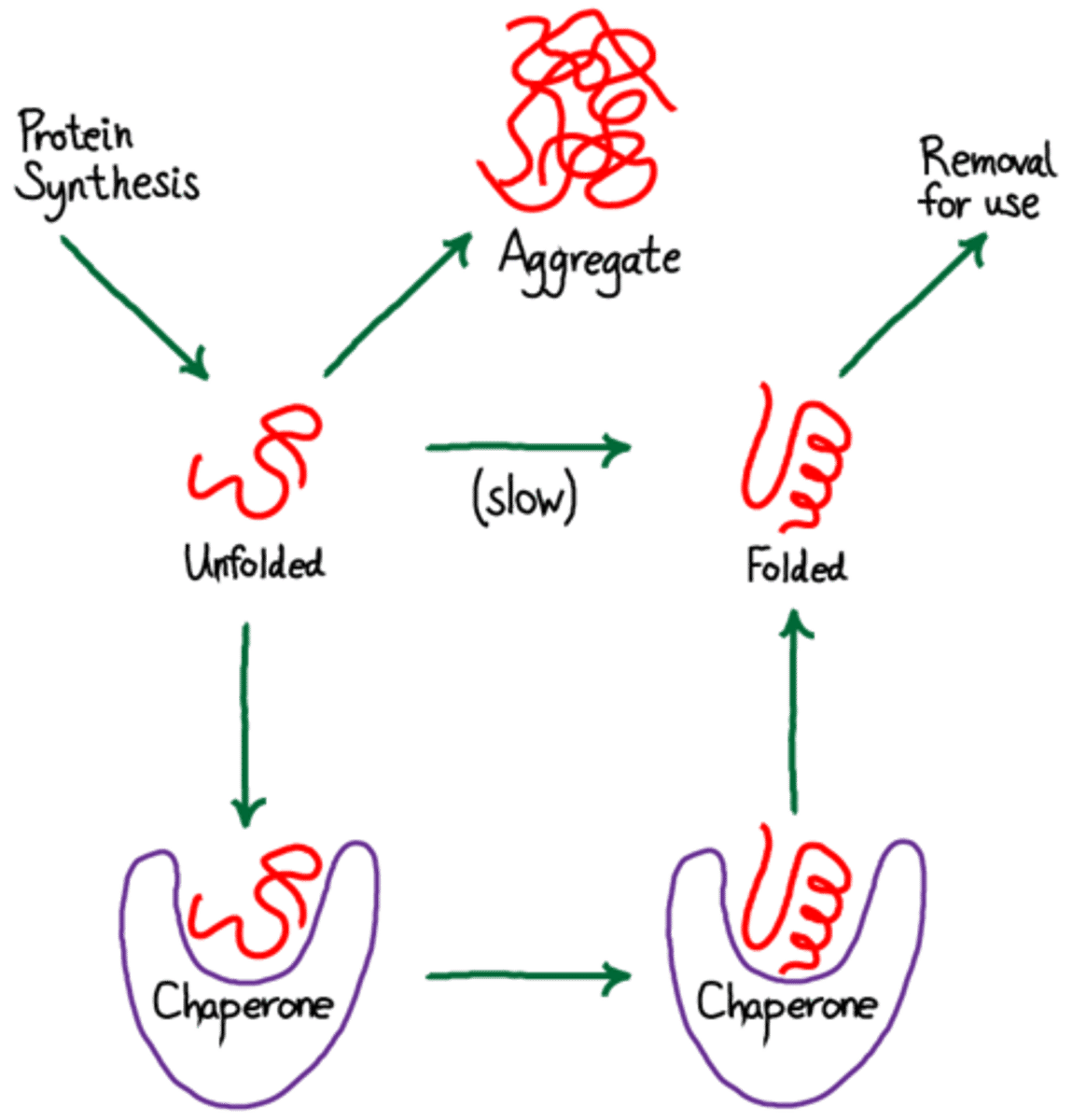
CRB True or false? There are multiple Hairpin Loops typically seen in tRNA.
There are multiple Hairpin Loops typically seen in tRNA.
CRB Which of the following terms can be used to describe a tRNA molecule that is bound to an amino acid?
I. Charged
II. Paired
III. Activated
(A) II only
(B) I and III only
(C) II and III only
(D) I, II and III
(B) I and III only
A tRNA molecule that is bound to an amino acid is typically described as Charged or Activated.
What site does the ribosome recognize and initially bind to in eukaryotic vs. prokaryotic cells?
The ribosome recognizes and binds to the 5' cap of eukaryotic mRNA.
The ribosome recognizes and binds to the Shine-Delgarno Sequence of prokaryotic mRNA.
True or False? The Shine-Delgarno Sequence acts as a both a 5' cap and a start codon.
False. The Ribosome recognizes and binds to the Shine-Delgarno Sequence and then moves down the mRNA strand until it reaches the start codon (AUG) where it will start translation. Furthermore, the Shine-Delgarno-Sequence doesn't provide protection to the mRNA strand like the 5' cap does.
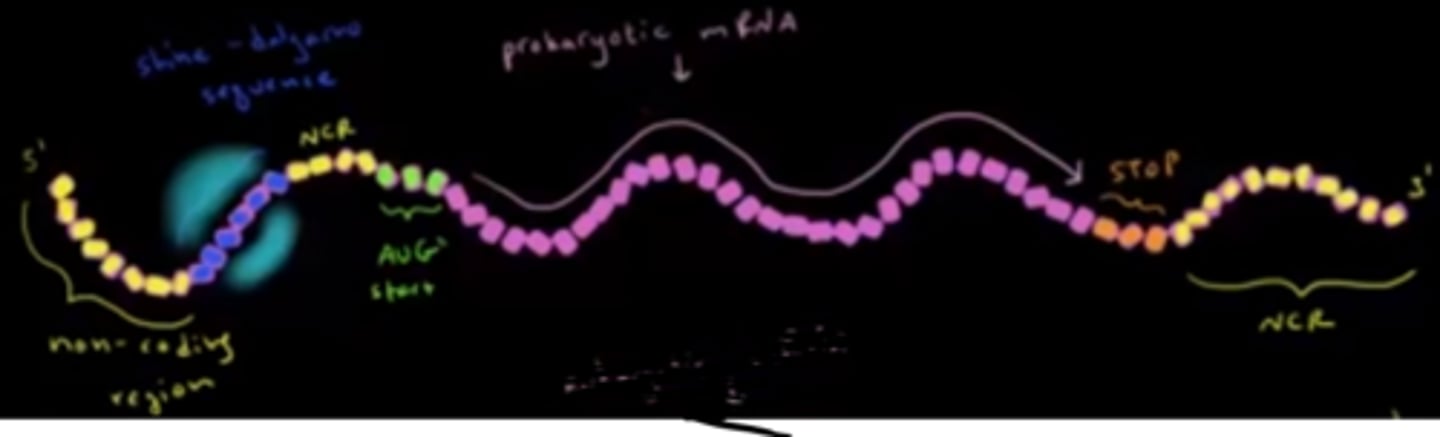
True or False? Prokaryotic mRNA does not contain any non-coding regions.
False. From the 5' to 3' end, prokaryotic mRNA contains non-coding mRNA, the Shine-Delgarno Sequence, non-coding mRNA, the start codon, coding mRNA, the stop codon, and then more non-coding mRNA.
Eukaryotes contain a 5' cap and a poly-A tail to protect their mRNA. Compare the composition of these two structures.
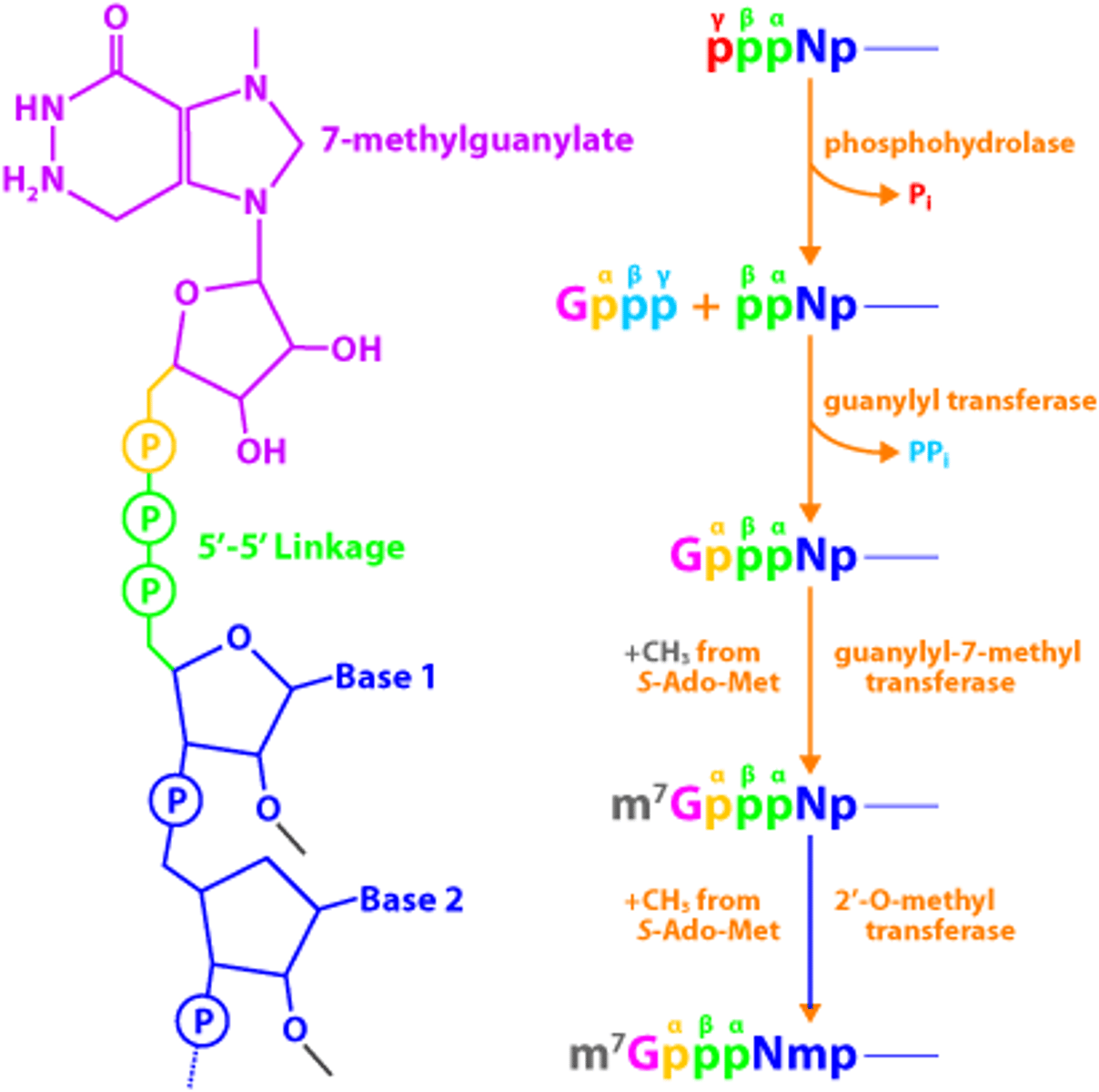
The 5' cap is a methylated guanosine triphosphate while the poly-A tail is a string of hundreds of adenine nucleotides.
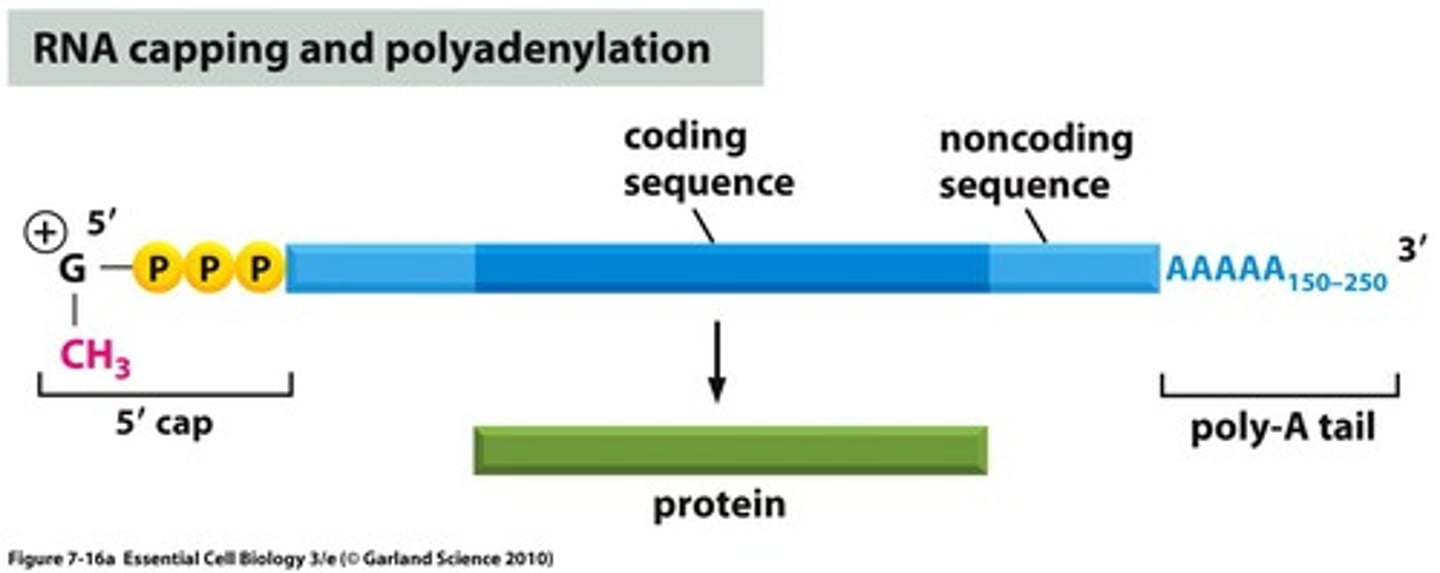
Why doesn't prokaryotic mRNA need the 5' cap and poly-A tail?
The prokaryotic mRNA does not have the 5' cap and poly-A tail because transcription and translation both happen simultaneously in the cytosol; thus, the mRNA does not need to "travel."
For eukaryotic mRNA, on the other hand, it needs to exit the nucleus and travel to the ribosome, and the 5' cap and poly-A tail are added on as extra protection.
During translation in _____________ cells, the first amino acid in the polypeptide chain is formylmethionine. During translation in _________________ cells, the first amino acid in the polypeptide chain is methionine.
(A) Eukaryotic, Eukaryotic
(B) Eukaryotic, Prokaryotic
(C) Prokaryotic, Prokaryotic
(D) Prokaryotic, Eukaryotic
(D) Prokaryotic, Eukaryotic
During translation in Prokaryotic cells, the first amino acid in the polypeptide chain is formylmethionine. During translation in Eukaryotic cells, the first amino acid in the polypeptide chain is methionine.
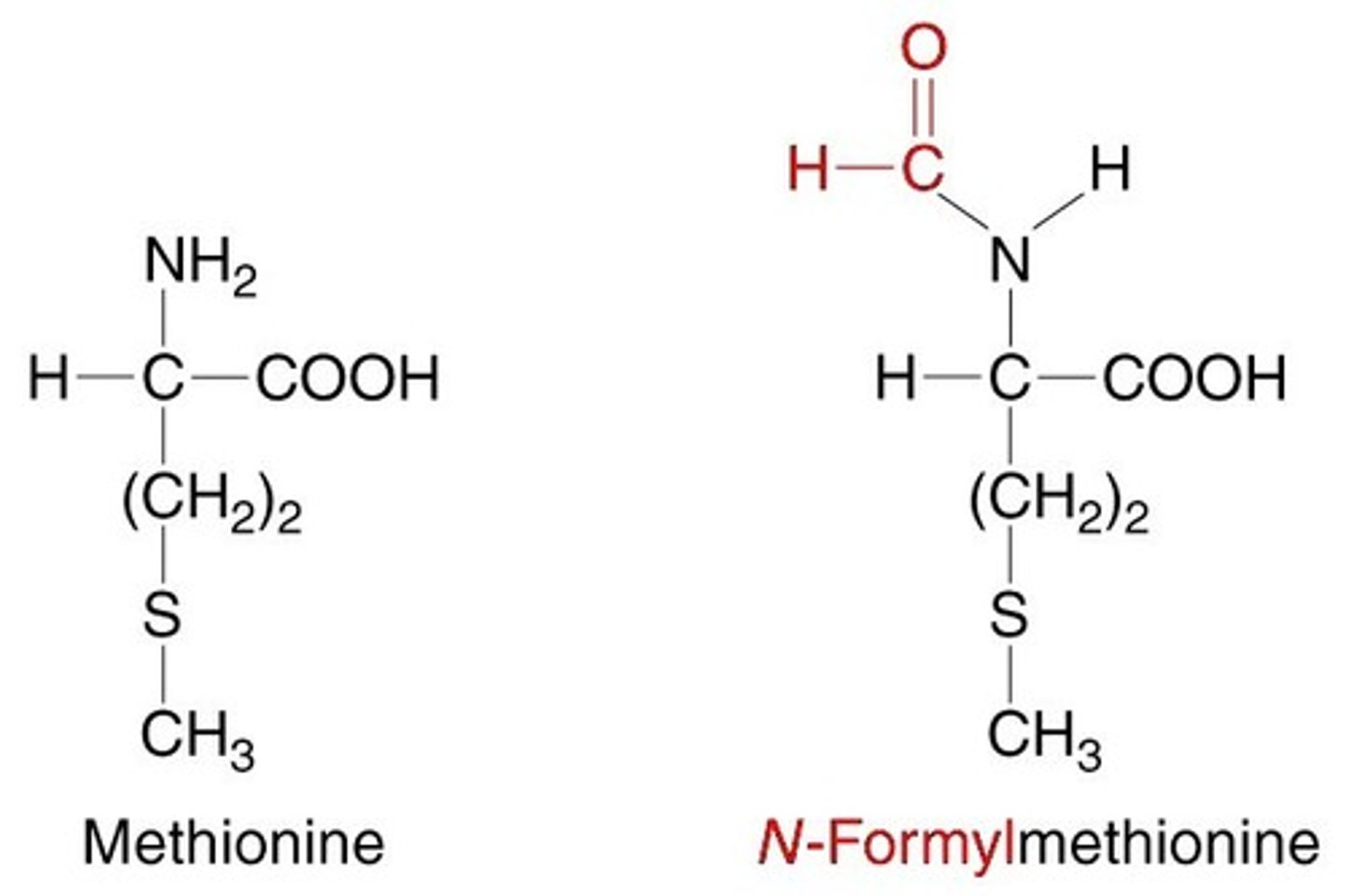
What role does formylmethionine play in the human body?
Formylmethionine acts as an alarm system in the human body and can trigger an immune response. It basically tells your body that there is a foreign bacteria invader.
DNA replication is:
(A) Fast and precise
(B) Fast but not precise
(C) Slow and precise
(D) Slow but not precise
(A) Fast and precise
DNA replication is very fast and very precise. For example, DNA polymerase can add an average of 700 base pairs per second and makes only 1 mistake every 10 million nucleotides.
Which enzyme(s) is/are responsible for proofreading?
I. DNA Polymerase I
II. DNA Polymerase II
III. DNA Polymerase III
(A) I Only
(B) II Only
(C) I and II Only
(D) I and III Only
(D) I and III Only
The enzyme DNA polymerase III and DNA polymerase I can both proofread.
CRB True or false? DNA Ligase, which is also involved in linking Okazaki Fragments, can also proofread the DNA.
DNA Ligase, which is also involved in linking Okazaki Fragments, cannot proofread the DNA.
CRB Since DNA Ligase does not have Proofreading capabilities, which of the following is most likely to have mutations?
(A) Beginning of a strand
(B) Leading Strand
(C) Lagging Strand
(D) End of a strand
(C) Lagging Strand
Since DNA Ligase does not have Proofreading capabilities, the lagging strand is most likely to have mutations.
CRB Which of the following is the proper term for any compound that can cause mutations?
(A) Cancerous
(B) Mutagen
(C) Carcinogen
(D) None of the above
(B) Mutagen
A Mutagen is any compound that can cause mutation. This differs from a Carcinogen, which would aid in the process of tumor development in general.
True or False? Proofreading takes place during replication.
True. Proofreading (the correction of misplaced nucleotides) takes place during replication.
What is the difference between exonuclease activity and endonuclease activity?
Enzymes engage in exonuclease activity when they remove an incorrect nucleotide from the end of a DNA strand.
Enzymes engage in endonuclease activity when they remove an incorrect nucleotide from the middle of a DNA strand.
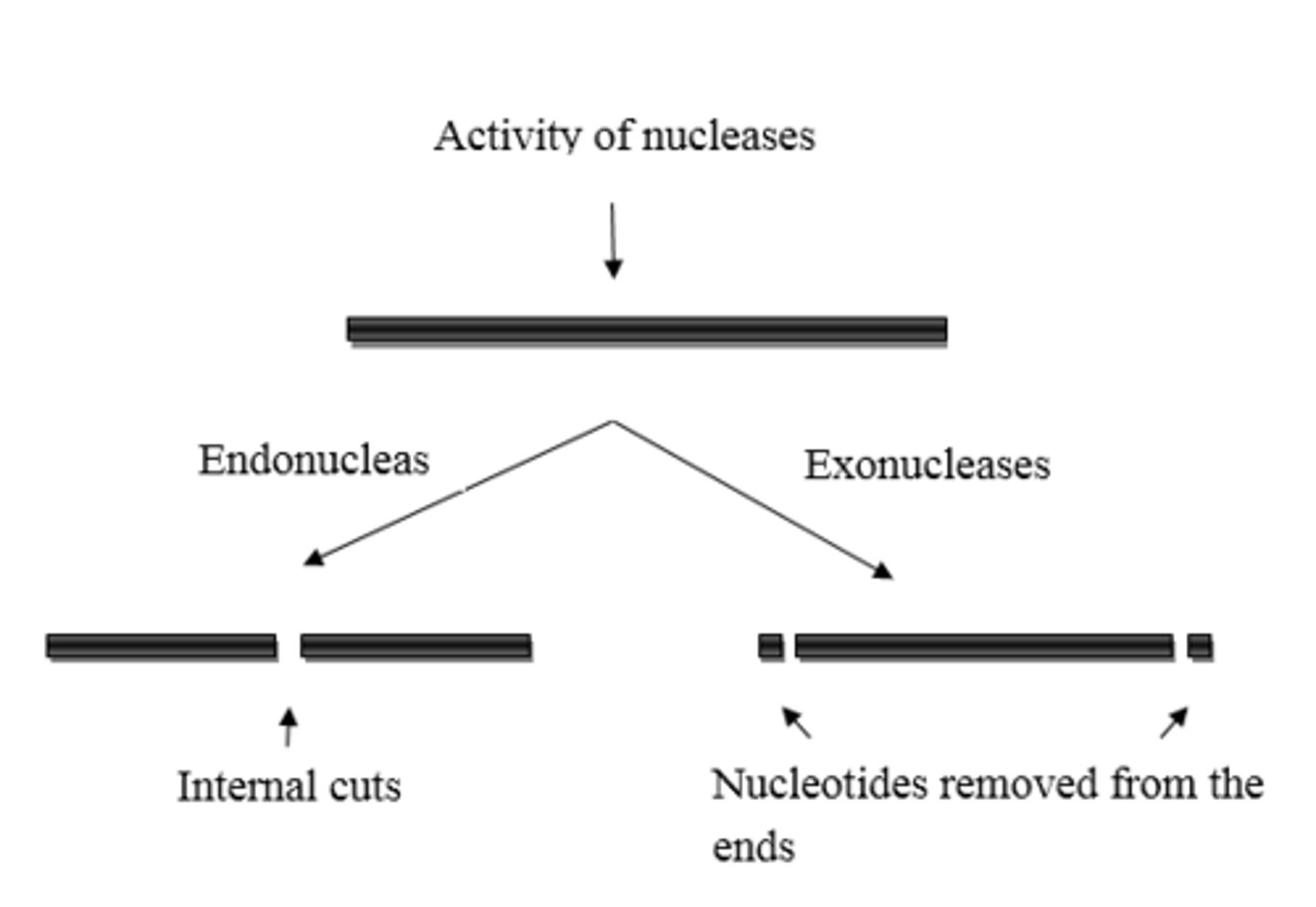
DNA Polymerase I is an ____________________, and DNA Polymerase III is an ____________________.
(A) Endonuclease, Endonuclease
(B) Endonuclease, Exonuclease
(C) Exonuclease, Exonuclease
(D) Exonuclease, Endonuclease
(C) Exonuclease, Exonuclease
DNA Polymerase I is an Exonuclease, and DNA Polymerase III is an Exonuclease.
Which DNA Polymerase is responsible for removing the RNA Primer during replication?
(A) DNA Polymerase I
(B) DNA Polymerase II
(C) DNA Polymerase III
(D) DNA Polymerase IV
(A) DNA Polymerase I
DNA Polymerase I is responsible for removing the RNA Primer during replication.
CRB Which of the following statements about Base Excision Repair are true?
I. Base Excision Repair can detect non-helix distorting mutations
II. Base Excision repair will remove the affected base using a Glycosylase Enzyme.
III. An AP Endonuclease will then remove the damaged DNA sequence, so DNA polymerase and ligase can finish the repairs.
(A) III only
(B) I and II only
(C) I and III only
(D) I, II and III
(D) I, II and III
Each of the following statements about Base Excision Repair are true:
I. Base Excision Repair can detect non-helix distorting mutations
II. Base Excision repair will remove the affected base using a Glycosylase Enzyme.
III. An AP Endonuclease will then remove the damaged DNA sequence, so DNA polymerase and ligase can finish the repairs.
CRB True or false? Base Excision repair would notice if a Cytosine is Deaminated (forming a Uracil) and trigger that repair.
True. Base Excision repair would notice if a Cytosine is Deaminated (forming a Uracil) and trigger that repair.
CRB Which of the following is NOT a possible name for the site where the Glycosylase Enzyme has removed the affected base?
(A) Anucleotidic site
(B) Apurinic site
(C) Apyrimidinic site
(D) Abasic site
(A) Anucleotidic site
Once the base has been removed, that site lacking a base can be called an Apurinic/Apyrimidic Site (AP Site) or an Abasic Site.
After replication is completed, there is a thymine base-paired with a guanine. What will fix this issue?
(A) The DNA Polymerase III Proofreading Mechanism
(B) The Base Excision Repair Mechanism
(C) The Mismatch Repair Mechanism
(D) The Nucleotide Excision Repair Mechanism
(C) The Mismatch Repair Mechanism
The mismatch repair mechanism happens after DNA replication. Through this mechanism, mismatches in the DNA sequence missed by the DNA polymerases during replication will be fixed.
CRB In which of the following stages of the Cell Cycle would you expect Mismatch Repair to be active?
(A) G1
(B) G2
(C) S
(D) M
(B) G2
In the G2 phase (right after S phase, where transcription occurs) is where the Mismatch Repair Mechanism is active.
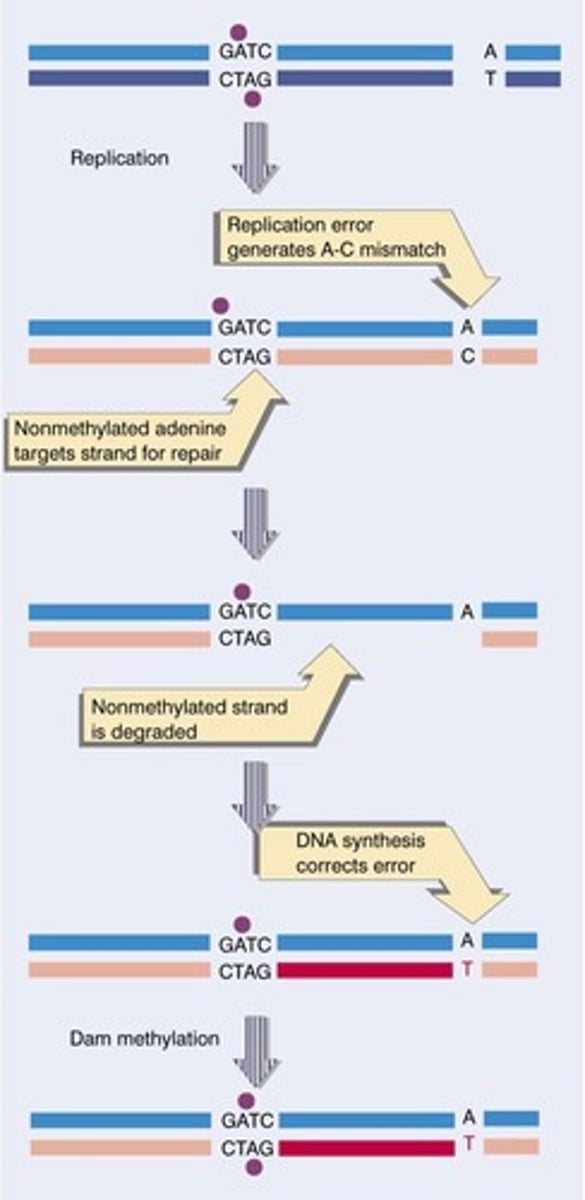
How does the mismatch repair mechanism distinguish between the parent strand and the newly synthesized strand that has the mistake on it in bacteria?
The mismatch repair mechanism can distinguish between the parent strand and the newly synthesized strand that has the mistake on it because the parent strand will have more adenine nucleotides that are methylated.
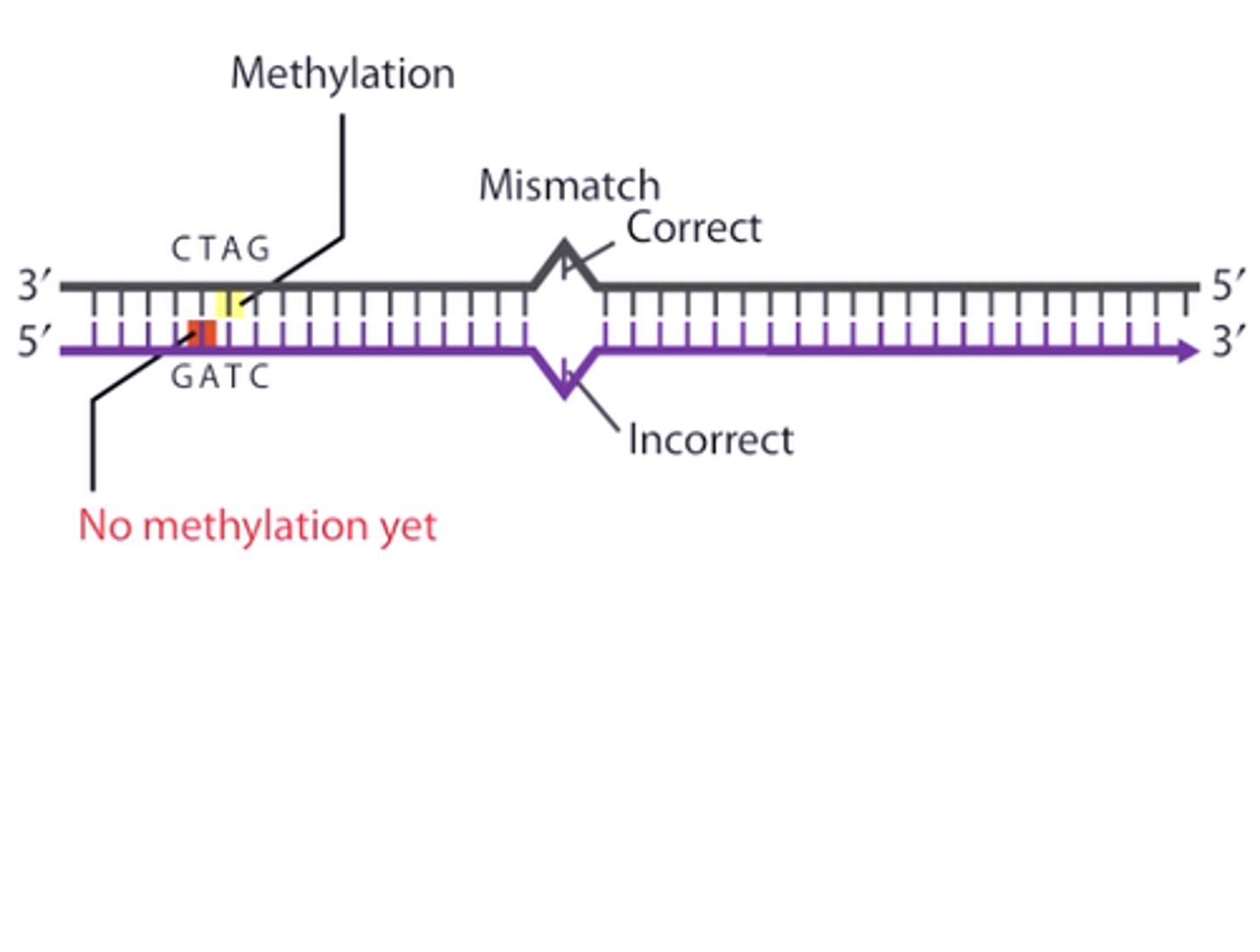
CRB True or false? These levels of methylation can also impact the transcriptional activity of the DNA.
True. These levels of methylation can also impact the transcriptional activity of the DNA.
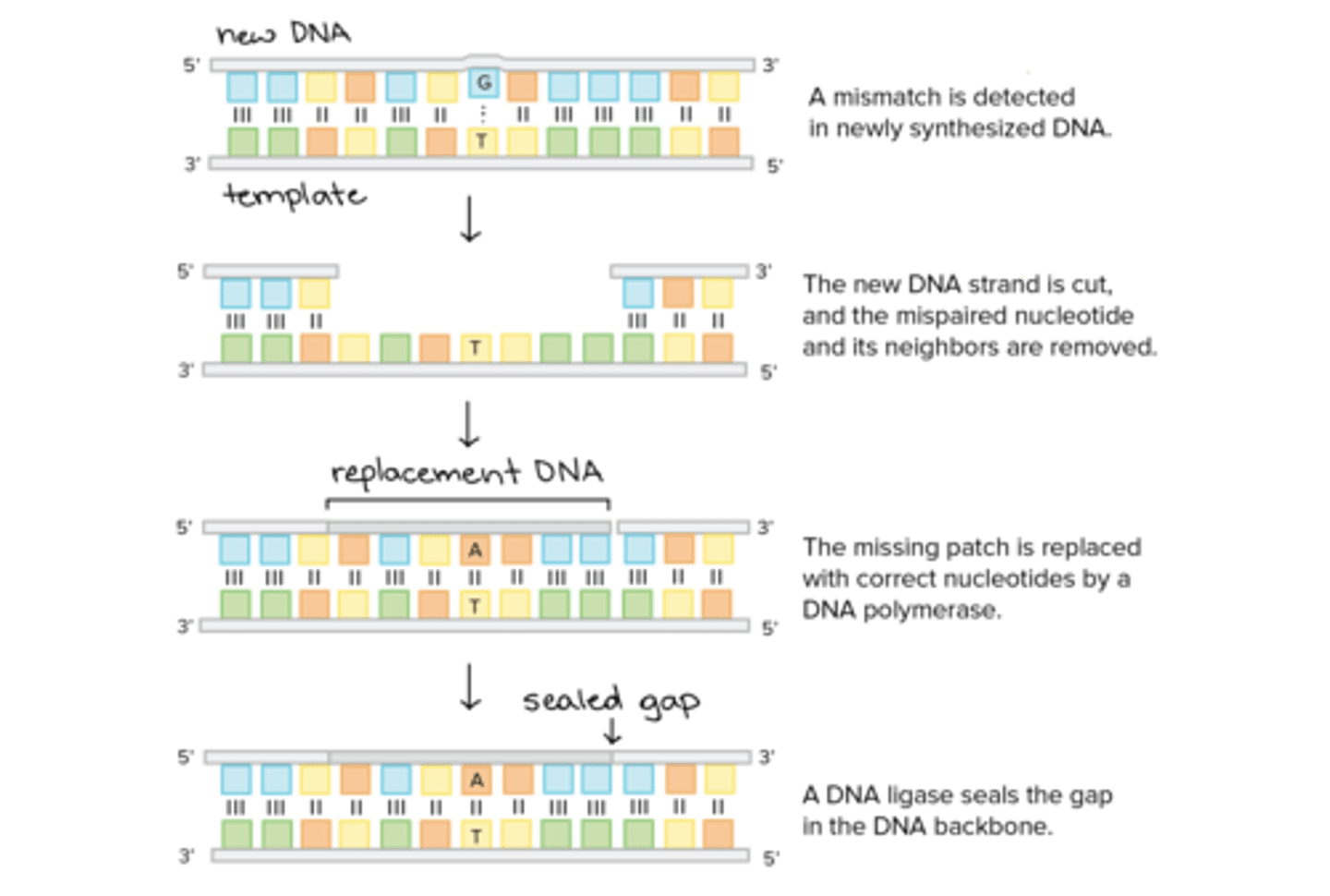
Put the following steps of the Mismatch Repair Mechanism in order:
I. DNA Ligase repairs the DNA backbone.
II. Exonuclease removes the misplaced nucleotide.
III. DNA Polymerase adds the correct nucleotide.
IV. The mismatched DNA is marked with a cut.
(A) IV, I, II, III
(B) I, IV, II, III
(C) IV, II, III, I
(D) I, II, III, IV
(C) IV, II, III, I
Mismatch Repair Mechanism entails the following steps in order:
1. The mismatched DNA is marked with a cut.
2. Exonuclease removes the misplaced nucleotide (and perhaps some of its neighbors) from the newly synthesized strand.
3. DNA Polymerase adds the correct nucleotide.
4. DNA Ligase repairs the DNA backbone by binding the newly placed nucleotide to the neighboring nucleotides.
What is a pyrimidine dimer?
A pyrimidine dimer is a type of DNA damage in which two pyrimidine nucleotides (typically thymine) are stuck together. The structural damage of pyrimidine dimers includes the protrusion of phosphate backbone and the breaking of hydrogen bonds between adjacent nucleotide base pairs.
You look into a microscope and see a pyrimidine dimer in DNA. What has the DNA likely been exposed to?
(A) X-rays
(B) Visible Light
(C) Infrared Light
(D) Ultraviolet Light
(D) Ultraviolet Light
You can conclude that the DNA has been exposed to UV rays because UV rays cause the formation of pyrimidine dimers.
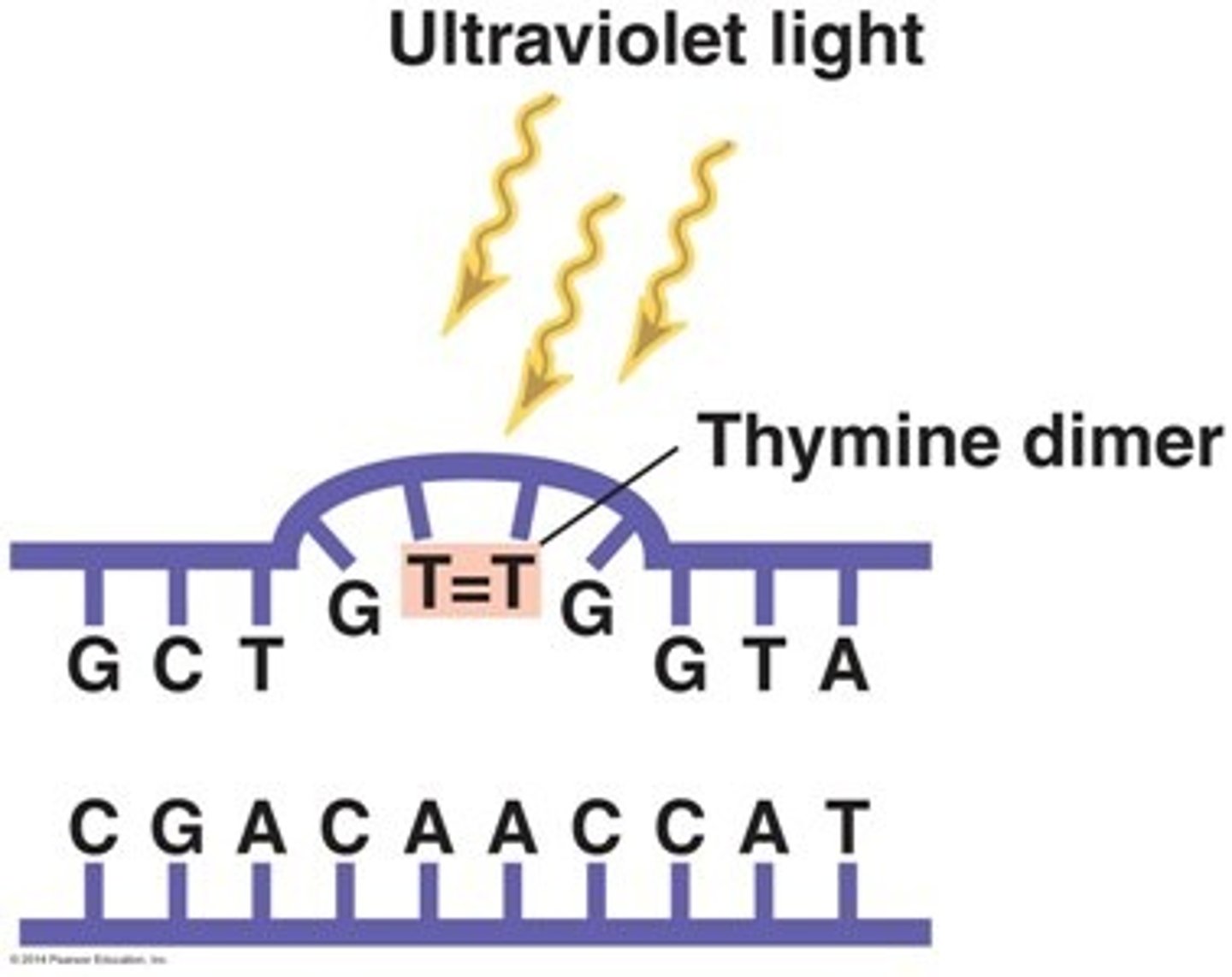
What is the difference between DNA mutations and DNA damage? Think of an example of each.
DNA mutations change the order of the DNA sequence. An example would be a mutation from 5'-ATCG-3' to 5'-AACG-3'.
DNA damage is the structural damage to DNA that leaves nucleotides in the correct order. An example of this is a pyrimidine dimer.
What is the difference between endogenous agents vs exogenous agents when talking about DNA damage?
Endogenous agents are internal factors, such as reactive oxygen species found within our own cells, that can cause DNA damage.
Exogenous agents are external factors, such as UV rays, gamma rays and x-rays, that can cause DNA damage.
Superoxide anions (O2) and peroxides are examples of what kind of DNA damage? Exogeneous or endogeneous agents?
Superoxide anions and peroxides are examples of Reactive Oxygen Species (ROS), which are endogenous agents that can cause DNA damage.
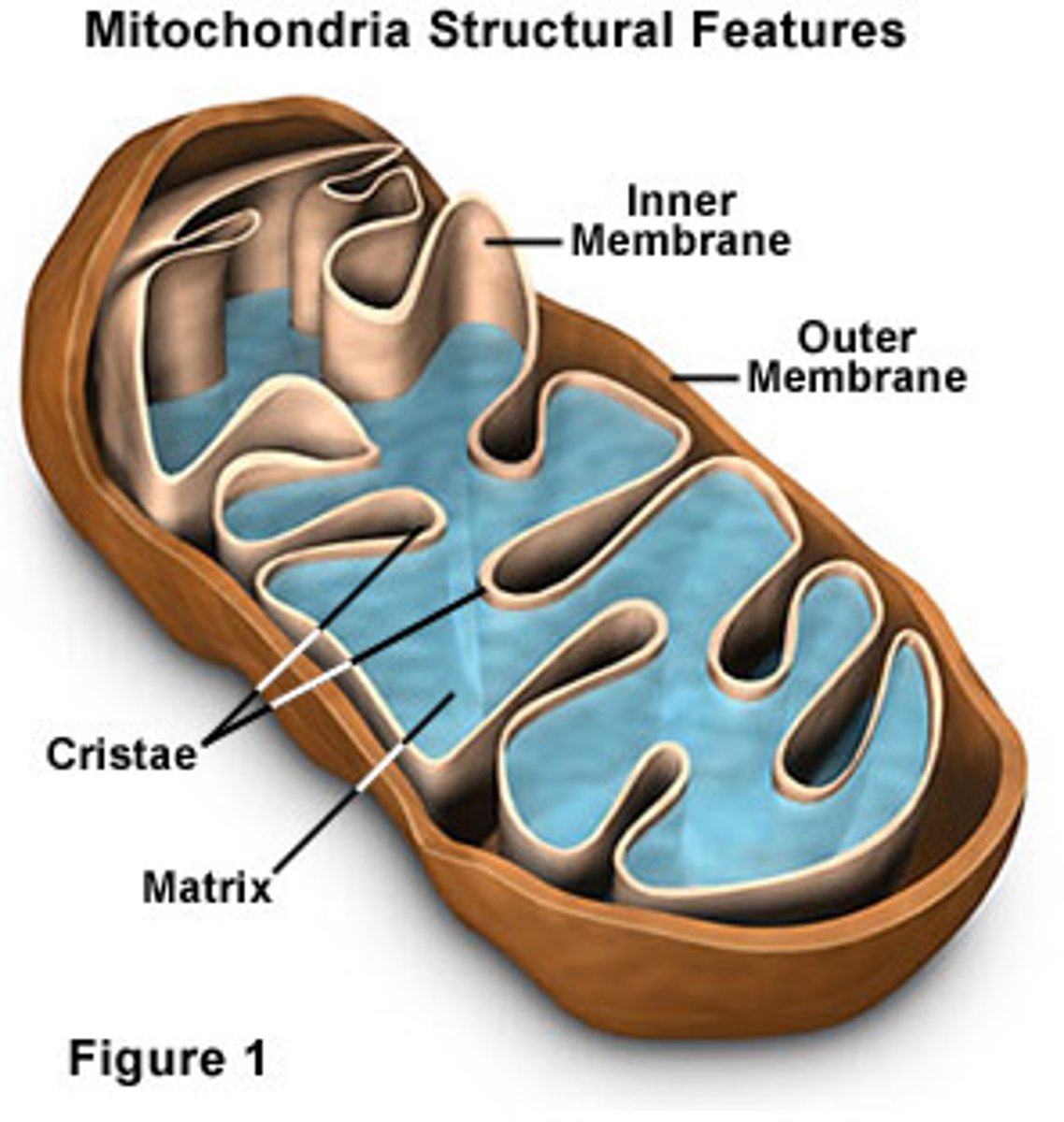
Which organelle might be considered a potential producer of endogeneous agents?
(A) Golgi Apparatus
(B) Cytosol
(C) Cell Membrane
(D) Mitochondria
(D) Mitochondria
The mitochondria can be considered a potential threat to the nucleus because the electron transport chain in mitochondria naturally produces reactive oxygen species, which could damage DNA that is stored in the nucleus.
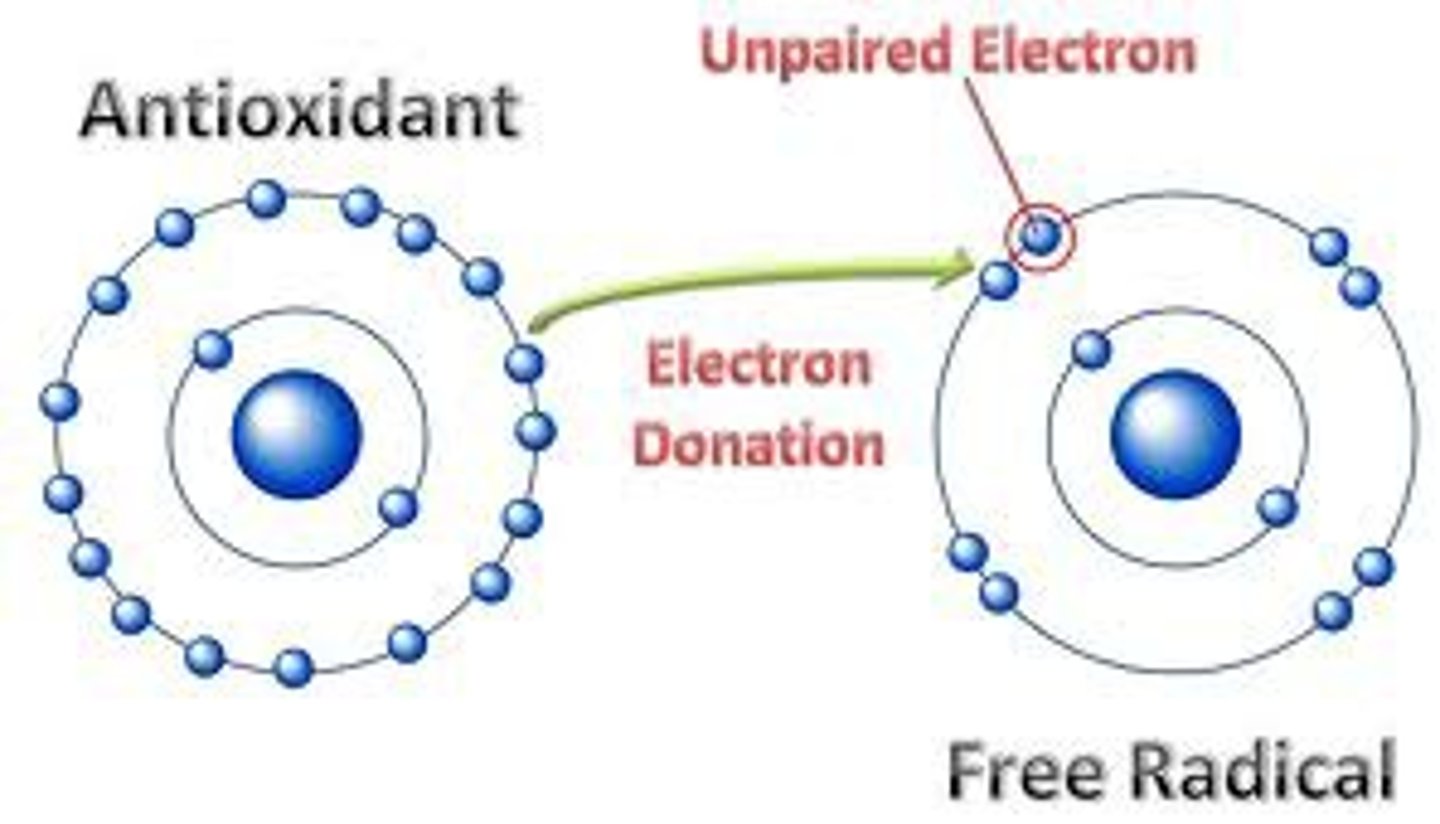
What is the role of an antioxidant molecule in preventing DNA damage/mutation?
An antioxidant molecule helps protect DNA aganist the damaging effect of reactive oxygen species.
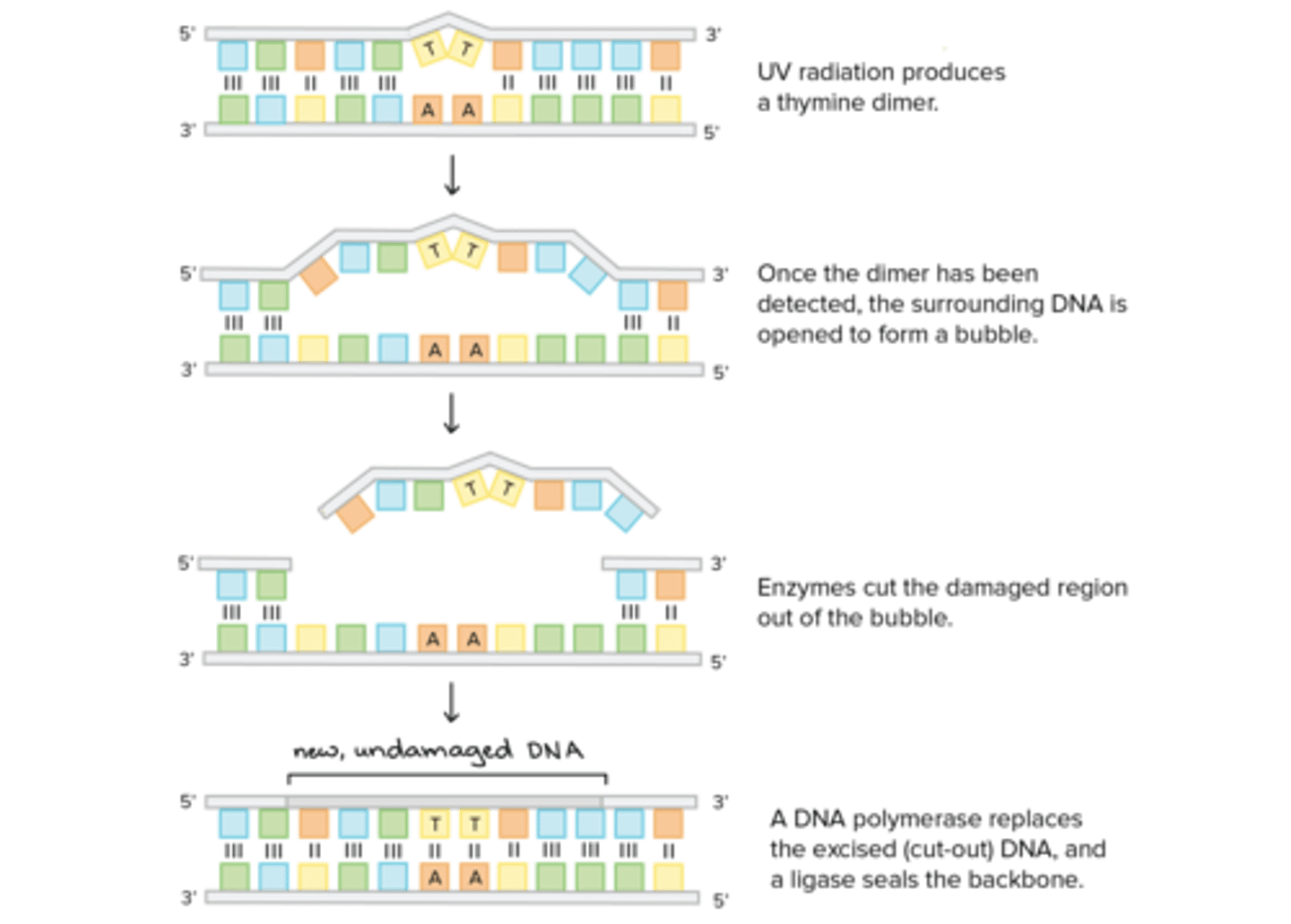
Which repair mechanism is responsible for fixing Pyrimidine Dimers?
(A) The DNA Polymerase III Proofreading Mechanism
(B) The Base Excision Repair Mechanism
(C) The Mismatch Repair Mechanism
(D) The Nucleotide Excision Repair Mechanism
(D) The Nucleotide Excision Repair Mechanism
Nucleotide excision repair is used to remove and repair pyrimidine dimers.
Which enzyme is involved in a nucleotide excision repair? An endonuclease or exonuclease?
An endonuclease is involved in a nucleotide excision repair because nucleotides need to be removed from the middle of a DNA strand.
There are only three steps to Nucleotide Excision Repair. What are they?
Nucleotide Excision Repair entails the following three steps:
(1) Endonuclease removes pyramidine dimer and surrounding nucleotides.
(2) DNA Polymerase will fill in the gap with the proper nucleotides.
(3) DNA Ligase will repair the backbone by connecting the new nucleotides to the old nucleotides.
Which enzyme(s) is/are required in proofreading, mismatch repair, and nucleotide excision repair?
I. Endonuclease
II. DNA Polymerase
III. DNA Ligase
(A) I Only
(B) II Only
(C) I and III Only
(D) II and III Only
(D) II and III Only
Proofreading, mismatch repair, and nucleotide excision repair all require the support of DNA polymerase to fill in the gap and DNA ligase to seal the strand.
The mismatch repair process can occur in an endonuclease-independent pathway, though, so it is not required for all of the listed processes.
CRB True or false? Sometimes the Nucleotide Excision Repair mechanism needs to function even when the DNA had been properly replicated.
True. Sometimes the Nucleotide Excision Repair mechanism needs to function even when the DNA had been properly replicated.
CRB Describe how UV Light and multiple Thymines on the same strand could require the Nucleotide Excision Repair mechanism to function.
UV light can excite the electrons of Thymines and cause neighboring Thymines to bond, forming a Thymine Dimer, distorting the double-helix and proper functions. The Nucleotide Excision Repair mechanism will recognize this error and replace the affected DNA section.
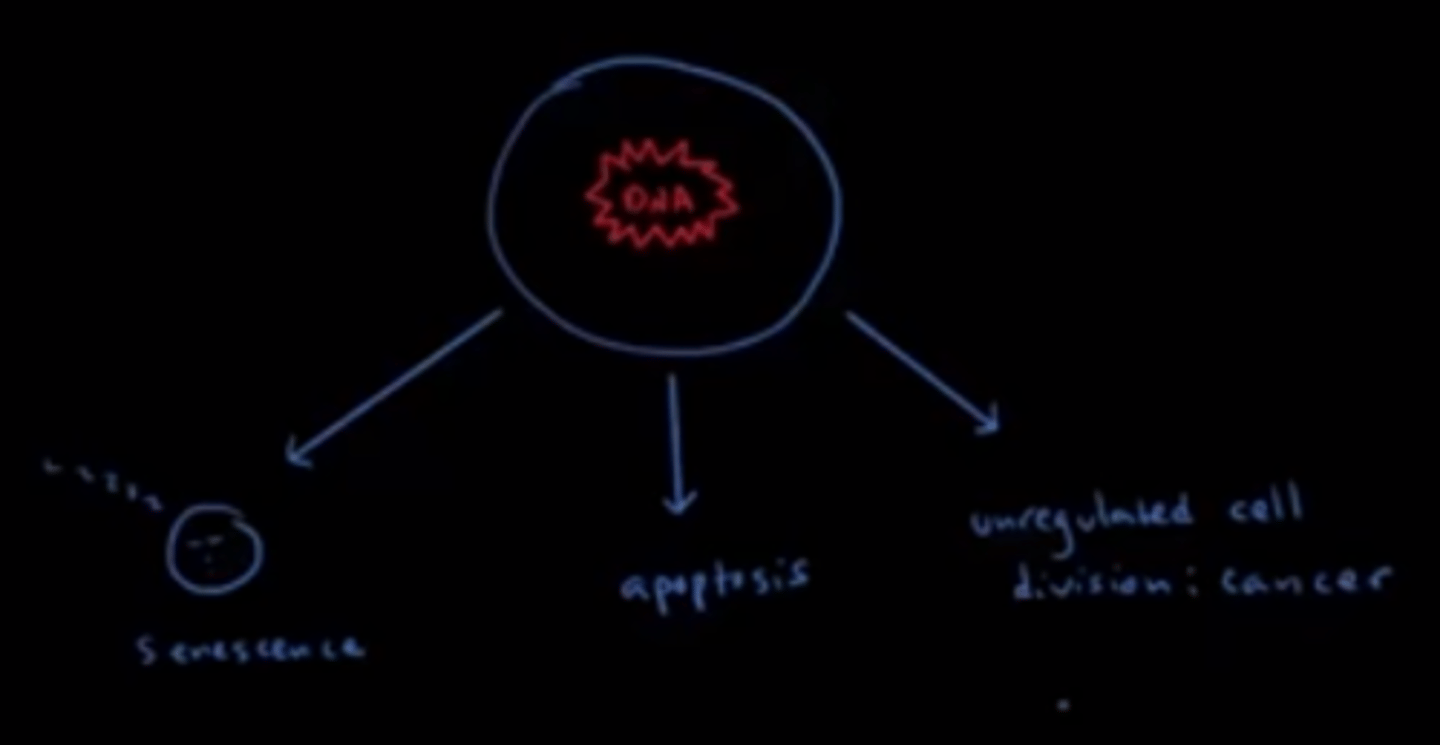
Which of the following may happen to a cell that has accumulated a lot of DNA damage and/or mutations?
I. The cell enters a dormant state and cell does not divide anymore.
II. The cell dies.
III. The cell becomes a cancer cell due to unregulated cell division.
(A) I Only
(B) I and III Only
(C) II and III Only
(D) I, II, and III
(D) I, II, and III
A cell that has accumulated a lot of DNA damage and/or mutations may engage in any of the following:
I. The cell enters a dormant state and cell does not divide anymore.
II. The cell dies.
III. The cell becomes a cancer cell due to unregulated cell division.
CRB Compare Missense, Nonsense and Frameshift Mutations.
Missense mutations replace one amino acid for another.
Nonsense mutations will add a premature stop codon, and can also be called a Truncation Mutation.
Frameshift mutations alter the number of nucleotides in the sequence, and will affect the amino acid sequence for all following codons.
True or false? Because the genetic code is Degenerate and the third nucleotide could be changed and not alter the amino acid coded for, there are also Silent or Degenerate Mutations that do not affect the protein.
True. Because the genetic code is Degenerate and the third nucleotide could be changed and not alter the amino acid coded for, there are also Silent or Degenerate Mutations that do not affect the protein.
Compare senescence versus apoptosis.
To say a cell is senescent means that the cell no longer divides.
Apoptosis is programmed cell death in which a cell digests/destroys itself.
CRB When DNA Repair processes fail, cancer is likely to follow. Which of the following would likely cause an increased risk of cancer?
I. Breaking the DNA backbone
II. Altering bases and base pairs
III. Incorporating incorrect bases during replication.
(A) II only
(B) I and III only
(C) II and III only
(D) I, II and III
(D) I, II and III
Each of the following could increase the risk of cancer:
I. Breaking the DNA backbone
II. Altering bases and base pairs
III. Incorporating incorrect bases during replication.
CRB Fill in the blanks: __________________ are genes that, if one copy is mutated, then they can cause cancer. ____________ are genes that would need both copies to be mutated to promote cancer.
(A) Oncogenes, Proto-oncogenes
(B) Oncogenes, Antioncogenes
(C) Antioncogenes, Proto-oncogenes
(D) Proto-oncogenes, antioncogenes.
(D) Proto-oncogenes, antioncogenes.
Proto-oncogenes are genes that, if one copy is mutated, then they can cause cancer. Antioncogenes are genes that would need both copies to be mutated to promote cancer.
CRB Which of the following statements about Proto-oncogenes and Antioncogenes is NOT true?
(A) Antioncogenes are often called Tumor Suppressor Genes
(B) Proto-oncogenes typically code for cell-cycle related proteins.
(C) Even one functional copy of an antioncogene is typically enough to inhibit tumor formation.
(D) Oncogenes could be described as speeding up the cell cycle, and antioncogenes are like cutting the brakes.
(D) Oncogenes could be described as speeding up the cell cycle, and antioncogenes are like cutting the brakes.
Having non-functional (mutated) antioncogenes are like cutting the brakes.
CRB True or false? Tumor Suppression genes are often described as being a "Two-Hit Model", since the two alleles must be mutated in the same tissue.
False. Tumor Suppression genes are often described as being a "Two-Hit Model", since the two alleles must be mutated in the same Cell.