unit 6 biology
1/147
Earn XP
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
148 Terms
rosalind franklin
- performed x-ray crystallography of DNA
- revealed a pattern that was regular and repetitive
edwin chargaff
- analyzed DNA samples from different species
- chargaff rule
chargaff rule
- amount of adenine = amount of thymine
- amount of cytosine = amount of guanine
nucleotide structure
- purines: double ring (A, G)
- pyrimidines: single ring (C, U, T)
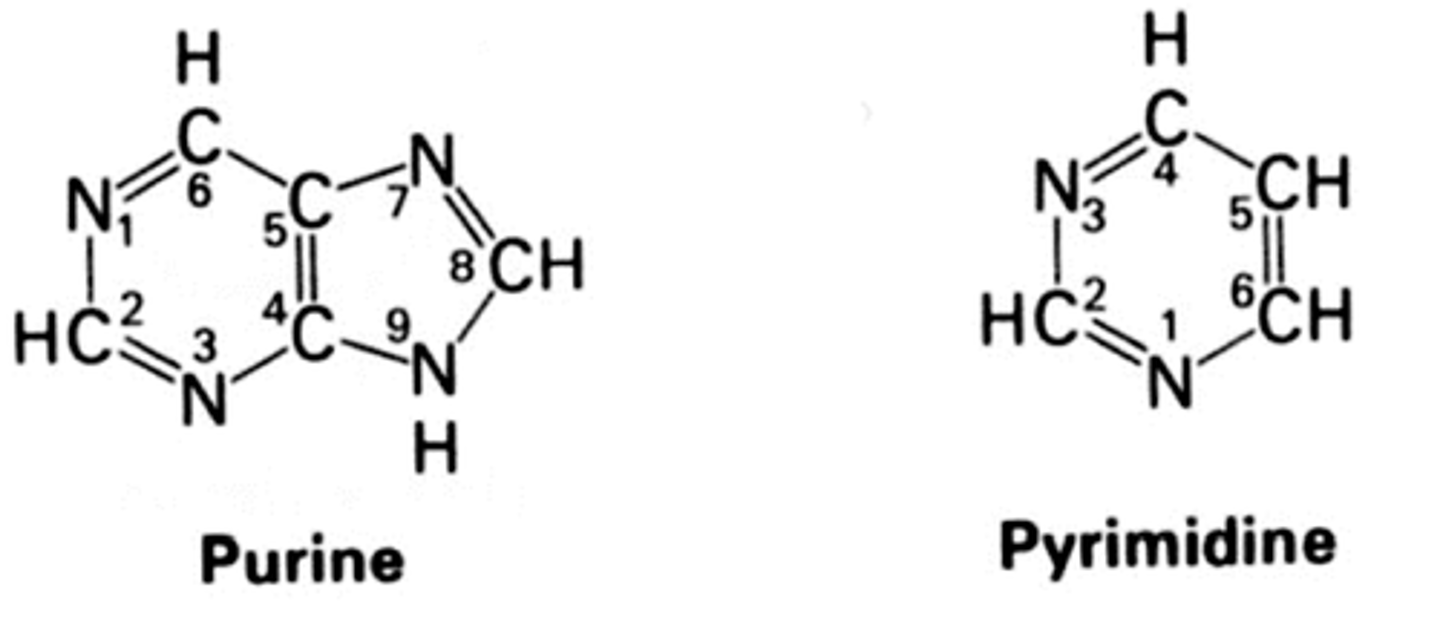
base pairs
held together by hydrogen bonds
two
number of hydrogen bonds for adenine and thymine
three
number of hydrogen bonds for cytosine and guanine
hydrogen bonds
easy to break and easy to put back together (weak)
watson and crick
combined the findings of franklin (helix shape) and chargaff (base pairing) to create the first 3D double helix model of DNA
DNA structure
- double stranded helix
- antiparallel strands: one strand runs 5' to 3' and the other runs in opposite, upside-down direction 3' to 5'
sugar phosphate
backbone of DNA
nucleotides pairing
center of DNA
5' end
free phosphate group
3' end
free hydroxyl group
DNA function
- primary source of heritable information
- genetic information is stored in and passed from one generation to the next through DNA (exception: RNA is the primary source of heritable information in some viruses)
eukaryotic cells DNA
- found in nucleus
- linear chromosomes
prokaryotic cells DNA
- in nucleoid region
- circular chromosomes
plasmids
- small, circular DNA molecules that are separate from the chromosomes
- replicate independently from the chromosomal DNA
- found in prokaryotes and some eukaryotes
- contain genes that may be useful to the prokaryote when it is in a particular environment, but may not be required for survival
manipulating plasmids
- can be removed from bacteria, then a gene of interest can be inserted into the plasmid to form recombinant plasmid DNA
- when the recombinant plasmid is inserted back into the bacteria the gene will be expressed
- bacteria can exchange genes found on plasmids with neighboring bacteria. once DNA is exchanged, the bacteria can express the genes acquired. helps with survival of prokaryotes
RNA
- ribonucleic acid
- single stranded
- AU, CG
DNA
- deoxyribonucleic acid
- double stranded
- AT, CG
DNA replication
- during S phase of cell cycle
- 3 alternative models: conservative, semi conservative, dispersive
conservative model of DNA replication
- parental strands direct synthesis of an entirely new double stranded molecule
- the parental strands are fully "conserved"
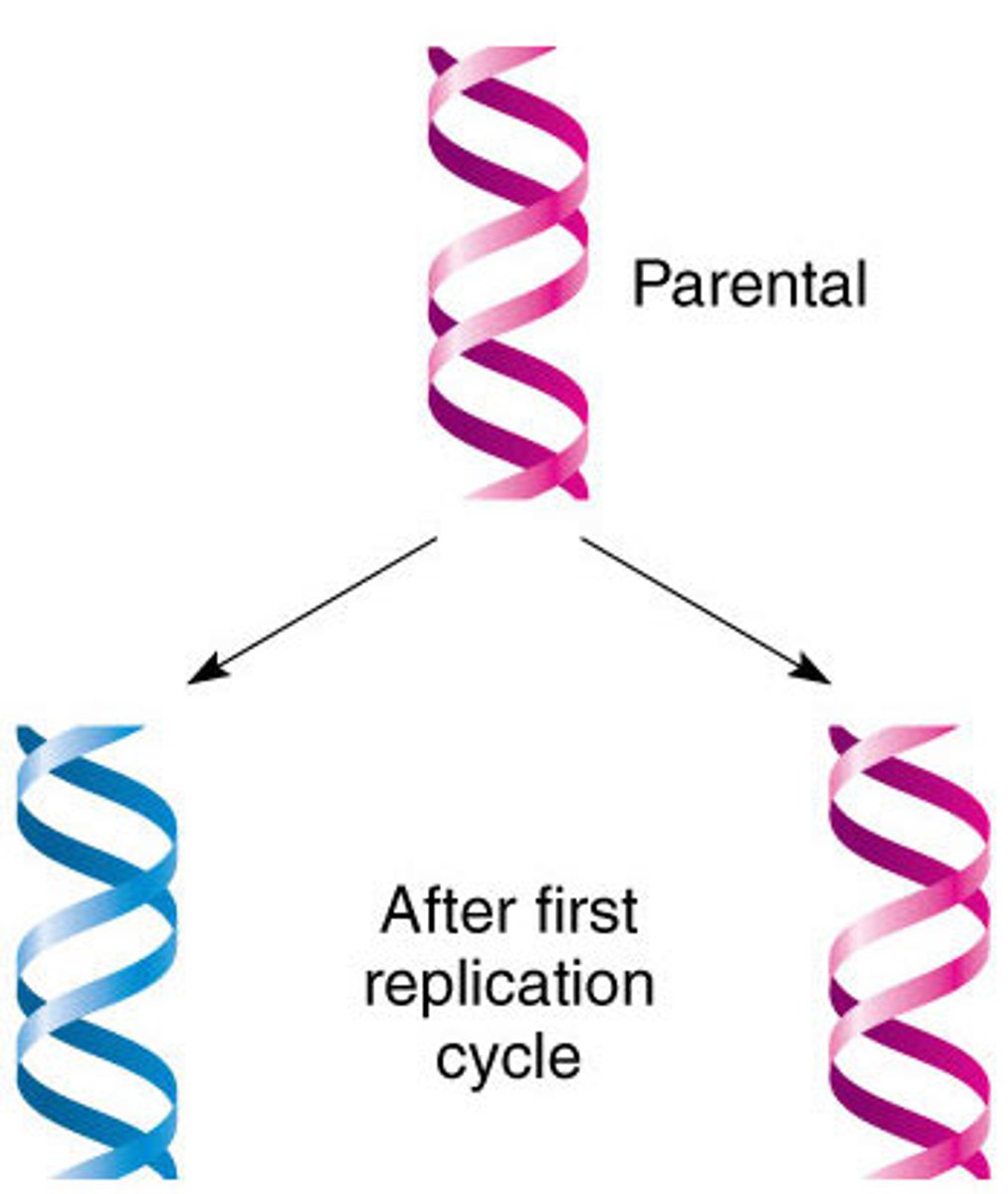
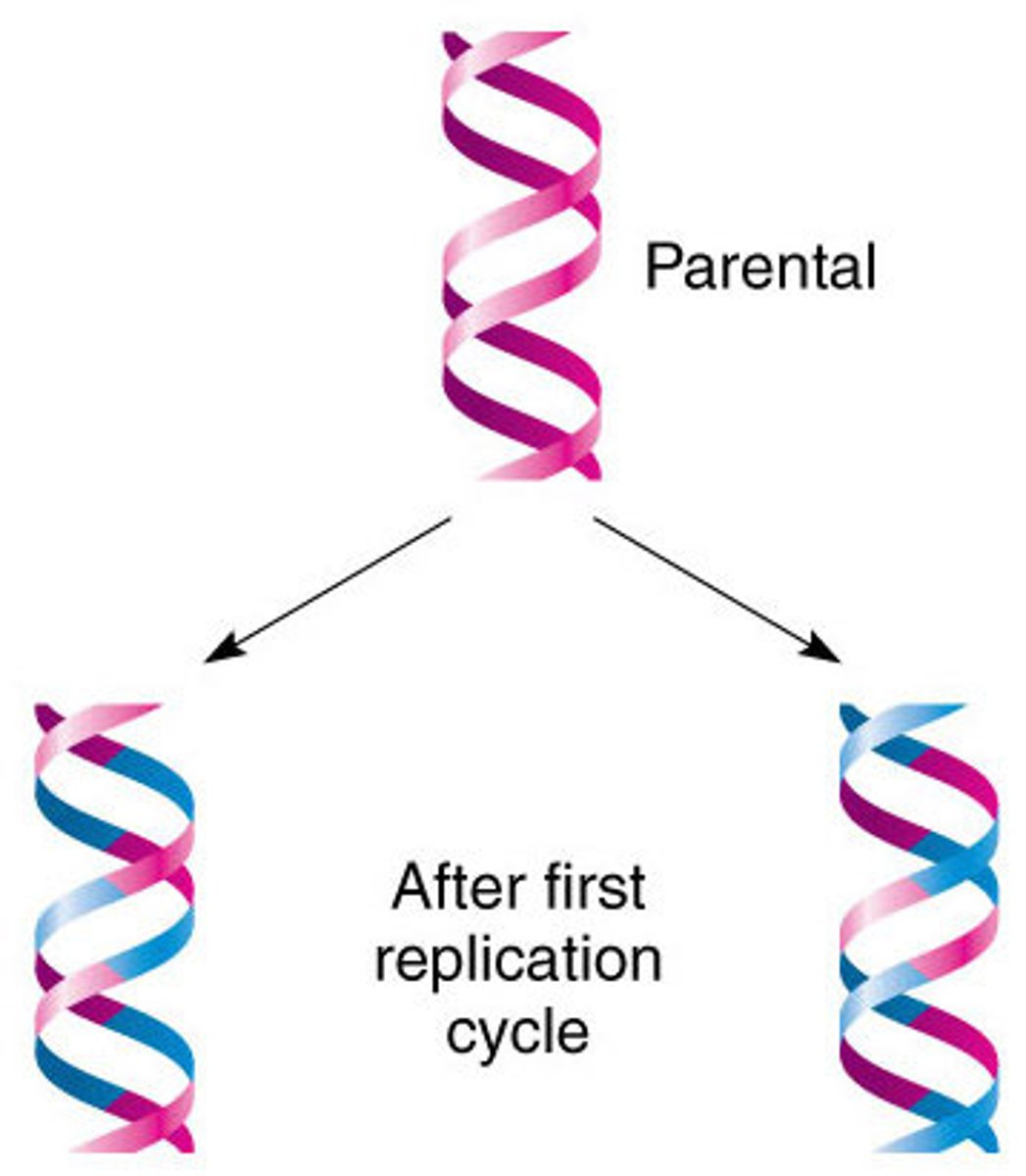
semi conservative model of DNA replication
- two parental strands each make a copy of itself
- after one round of replication, the two daughter molecules each have one parental and one new strand
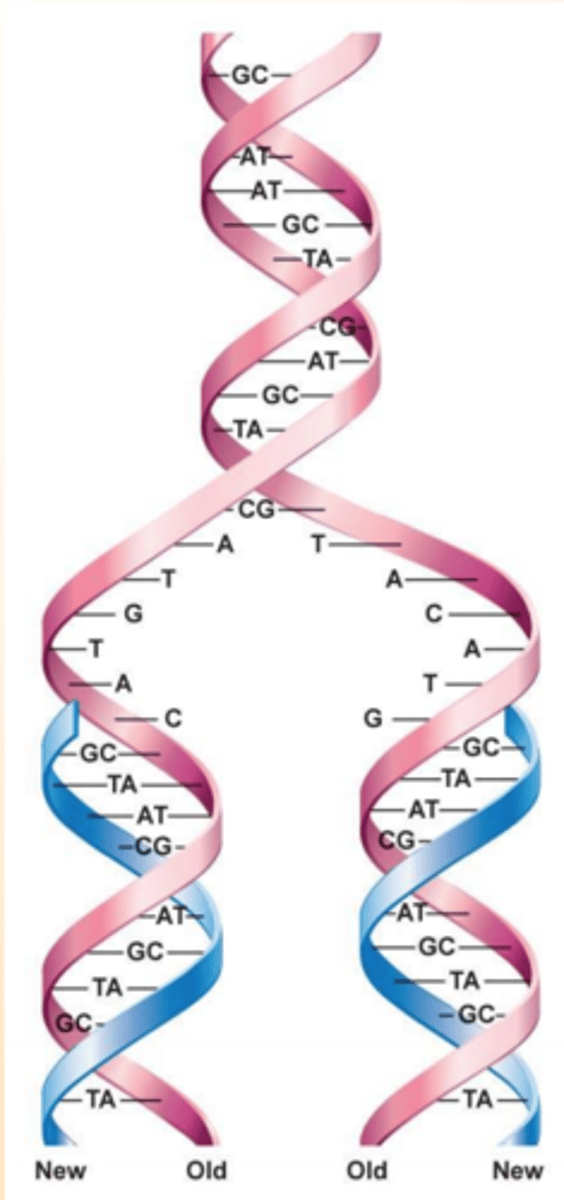
dispersive model of DNA replication
- the material in the two parental strands is dispersed randomly between the two daughter molecules
- after one round of replication, the daughter molecules contain a random mix of parental and new DNA
meselson and stahl experiment
by analyzing samples of DNA after each generation, it was found that the parental strands were following the semi-conservative model of DNA replication
DNA replication steps
1. origins of replication
2. helicase
3. primase
4. antiparallel elongation (DNA polymerase III)
5. okazaki fragments
6. ligase
origins of replication
- beginning site for DNA replication
- various proteins/enzymes attach to the origin of replication and open the DNA to form a replication fork
helicase
unwind DNA strands at each replication fork
single strand binding proteins (SSBPs)
to keep the DNA from rebonding with itself, these proteins bind to the DNA to keep it open
topoisomerase
help prevent strain ahead of the replication fork by relaxing supercoiling
primase
- initiates replication by adding short segments of RNA (primers) to the parental DNA strand
- the enzymes that synthesize DNA can only attach new DNA nucleotides to an existing strand of nucleotides
primers
foundation for DNA synthesis
DNA polymerase III (DNAP III)
- attaches to each primer on the parental strand and moves in the 3' to 5' direction
- can only move in one direction (3' to 5')
- adds nucleotides to the new strand in the 5' to 3' direction
leading strand
- DNAP III follows helicase and only requires one primer
- synthesized in one continuous segment
lagging strand
- DNAP III on the other parental strand that moves away from helicase and requires many primers
- synthesized in chunks bc it moves away from the replication fork
- since DNAP III can only add nucleotides to a 3' end, so there is no way to finish replication on the 5' end -> DNA becomes shorter and shorter over many replications
okazaki fragments
segments of the lagging strand
DNA nucleotides
after DNAP III forms an okazaki fragment, DNAP I replaces RNA nucleotides with ___
ligase
joins the okazaki fragments forming a continuous DNA strand
telomeres
- repeating units of short nucleotide sequences that do not code for genes
- forms a cap at the end of DNA to help postpone erosion
telomerase
adds telomeres to DNA
DNA polymerase
- proofreads the bases added while adding nucleotides to the new DNA strand
- if error still occur, mismatch repair will take place
mismatch repair
- enzymes remove and replace the incorrectly paired nucleotide
- if segments of DNA are damaged, nuclease can remove segments of nucleotides and DNA polymerase and ligase can replace the segments
proteins
polypeptides made up of amino acids
amino acids
linked by peptide bonds
gene expression
- the process by which DNA directs the synthesis of proteins
- two states: transcription and translation
- occur in all organisms
central dogma of biology
DNA --transcription--> RNA --translation--> protein
transcription
- the synthesis of RNA using information from DNA
- allows for the "message" of the DNA to be transcribed
- occurs in the nucleus: if no nucleus, then nucleoid region, cytosol, cytoplasm
translation
- the synthesis of a polypeptide using information from RNA
- occurs at the ribosome
- nucleotide sequence becomes an amino acid sequence
messenger RNA (mRNA)
-synthesized during transcription using a DNA template
- carries information from the DNA at the nucleus to the ribosomes in the cytoplasm
transfer RNA (tRNA)
- carries the amino acid that the mRNA codon code for
- can attach to mRNA via their anticodon region (complementary and antiparallel to mRNA)
- allow information to be translated into a peptide sequence
- carries an amino acid = "charged"
anticodon
complementary codon to mRNA
ribosomal RNA (rRNA)
- helps form ribosomes
- helps link amino acids together
DNA
contains the sequence of nucleotides that codes for proteins
triplet code
sequence of nucleotides is read in groups of threes
template/noncoding/minus/antisense strand
only one DNA strand is being transcribed during transcription
mRNA molecules
- antiparallel and complementary to the DNA nucleotides
- base pairing: AU, CG
codons
- mRNA nucleotide triplets
- codes for amino acids
redundancy
more than one codon code for each amino acid
codons
- 64 different combinations
- 61 code for amino acids
- 3 are stop codons
- universal to all life
reading frame
- the codons on the mRNA must be read in the correct groupings during translation to synthesize the correct proteins
- shift by even one letter will produce a completely different outcome
steps of transcription and translation
1. initiation
2. elongation
3. termination
transcription initiation step
- begins when RNA polymerase molecules attach to a promoter region of DNA
- do not need a primer to attach
- eukaryotes: promoter region = TATA box, transcription factors help RNA polymerase bind
- prokaryotes: RNA polymerase can bind directly to promoter
promoter region
upstream of the desired gene to transcribe
transcription elongation step
- RNA polymerase opens the DNA and reads the triplet code of the template strand
- moves in 3' to 5' direction
- mRNA transcript elongates 5' to 3'
- RNA polymerase moves downstream: opens small sections of DNA at a time, pairs complementary RNA nucleotides, growing mRNA strand peels away from the DNA template strand, and DNA double helix reforms
- single gene can be transcribed simultaneously by several RNA polymerase molecules, helping increase the amount of mRNA synthesized and protein production
transcription termination step
- prokaryotes: transcription proceeds through a termination sequence (stop codon is reached), causes a termination signal, RNA polymerase detaches, mRNA is released and proceeds to translation, mRNA does not need modifications
- eukaryotes: RNA polymerase transcribes a sequence of DNA called polyadenylation signal sequence, releases the pre-mRNA from the DNA, must undergo modifications before translation
pre-mRNA modifications
- 5' cap (GTP)
- poly-a tail
- RNA splicing
- 5' cap and poly-a tail function to help the mature mRNA leave the nucleus, protect the mRNA from degradation, and help ribosomes attach to the 5' end of the mRNA when it reaches the cytoplasm
- once all modifications have occurred, the pre-mRNA is now considered mature mRNA and can leave the nucleus and proceed to the cytoplasm for translation at the ribosomes
5' cap (GTP)
the 5' end of the pre-mRNA receives a modified guanine nucleotide "cap"
poly-a tail
the 3' end of the pre-mRNA receives adenine nucleotides
RNA splicing
sections of the pre-mRNA, called introns, are removed and exons are joined together
introns
- intervening sequence
- do not code for amino acids
- gets spliced out
exons
- expressed sections
- code for amino acids
alternative splicing
a single gene can code for more than one kind of polypeptide
aminoacyl-tRNA synthease
responsible for attaching amino acids to tRNA
ribosomes
- two subunits (small and large)
- large subunit has three sites: A, P, E
A site (amino acid site)
holds the next tRNA carrying an amino acid
P site (polypeptide site)
holds the tRNA carrying the growing polypeptide chain
E site
exit site
translation initiation step
- beings when the small ribosomal subunit binds to the mRNA and a charged tRNA binds to the start codon on the mRNA
- tRNA carries methionine
- large subunit binds
- the first tRNA carrying Met will go to the P site, every other tRNA will go to the A site first
translation elongation step
- starts when the next tRNA comes into the A site
- mRNA is moved through the ribosome and its codons are read
- each mRNA codon codes for a specific amino acid
- occurs in steps: codon recognition, peptide bond formation, translation
codon charts
used to determine the amino acid
common ancestry
since all organisms use the same genetic code, it supports the idea of ___
codon recognition
the appropriate anticodon of the next tRNA goes to the A site
peptide bond formation
peptide bonds are formed that transfer the polypeptide to the A site tRNA
translocation
- tRNA in the A site moves to the P site
- tRNA in the P site goes to the E site
- A site is open for the next tRNA
translation termination step
- occurs when a stop codon in the mRNA reaches the A site of the ribosome
- stop codons signals for a release factor: hydrolyzes the bond that holds the polypeptide to the P site, polypeptide releases, all translational units disassemble
stop codons
do not code for amino acids
genes
determine primary structure
primary structure
determines the final shape
growing polypeptide chain
begins to coil and fold as translation takes lace
chaperone proteins
some polypeptides require this to fold correctly and some require modification before it can be functional in the cell
retroviruses
- exception to the standard flow of genetic information
- information flows from RNA to DNA
- uses an enzyme known as reverse transcriptase
reverse transcriptase
- couples viral RNA to DNA
- DNA then becomes part of the RNA
gene expression
- prokaryotes and eukaryotes must be able to regulate which genes are expressed at any given time
- genes can be turned "on" or "off" based on environmental and internal cues; on/off refers to whether or not transcription will take place
- allows for cell specialization
operons
- group of genes that cab be turned on or off
- three parts: promotor, operator, genes
- repressible or inducible: both regulate transcription of genes
promotor (operon)
where RNA polymerase can attach
operator (operon)
the on/off switch
genes (operon)
code for related enzymes in pathway
repressible operon
transcription is usually on but can be repressed/stopped (on to off)
inducible operon
transcription is usually off but can be induced/started (off to on)