D1.2 - PROTEIN SYNTHESIS
1/29
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
30 Terms
list and describe a few examples of proteins
collagen: structural protein found in skin, muscles, bones, tendons, ligaments, blood vessels etc. it is the most abundant protein by mass in the body
insulin: signalling protein (hormone) that makes cells absorb sugar from the blood
hemoglobin: transport protein found in RBC that carries oxygen throughout the body
trypsin: an enzyme that catalyses a chemical reaction
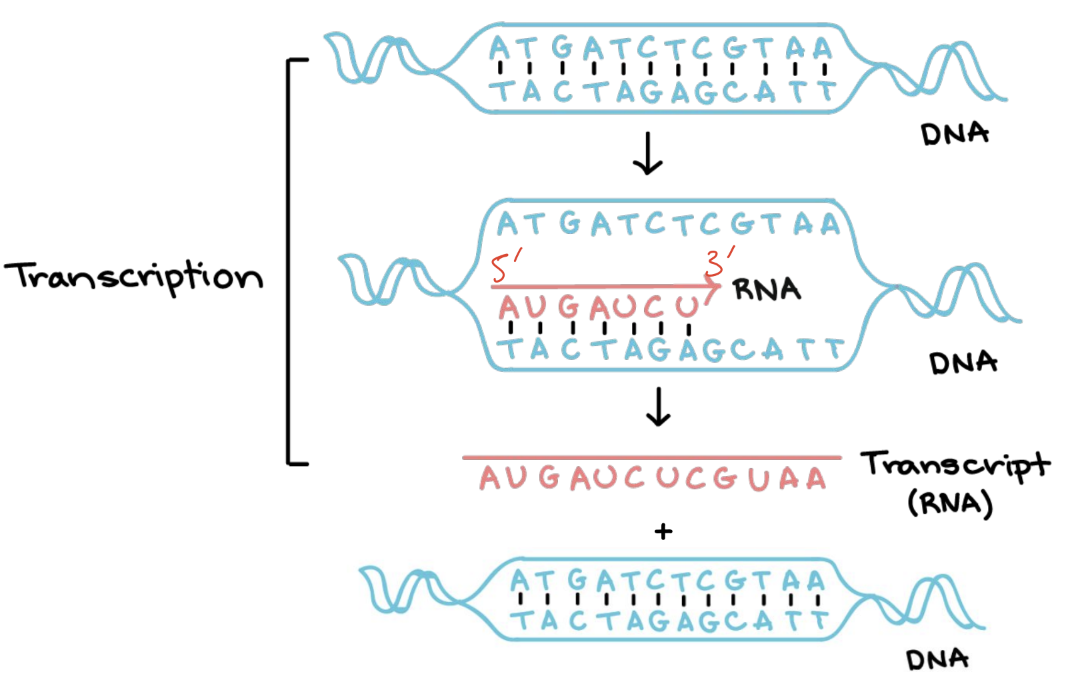
describe the process of transcription
occurs in nucleus for eukaryotic cells / nucleoid region (cytoplasm) in prokaryotic cells
RNA polymerase is an enzyme seperates DNA strands along a gene (only occurs on one of the DNA strands)
RNA nucleotides form complementary base pairs with one DNA strand (template) by hydrogen bonding:
A-U, T-A, G-C, C-G
RNA polymerase moves along the DNA in a 5’ → 3’ direction antiparallel to the DNA template and covalently bonds w the RNA nucleotides into a single strand
at the end of the gene, RNA polymerase releases the RNA, and the DNA returns to its original double helix structure
DNA is very stable (base sequence does not change) and is unaffected by transcription
RNA produced has a base sequence based on the DNA sequence
mRNA IS BEING SYNTHESISED IN A 5’ TO 3’ DIRECTION - ANTIPARALLEL TO DNA TEMPLATE
describe gene expression
refers to how active a gene is and how often it is transcribed and translated
genes can be “on” or “off”
genes can have a range of expression
frequent, sometimes, seldom, as needed, never
✓ transcription → gene expressed
✕ transcription → gene not expressed
gene - sequence of DNA that codes for a protein
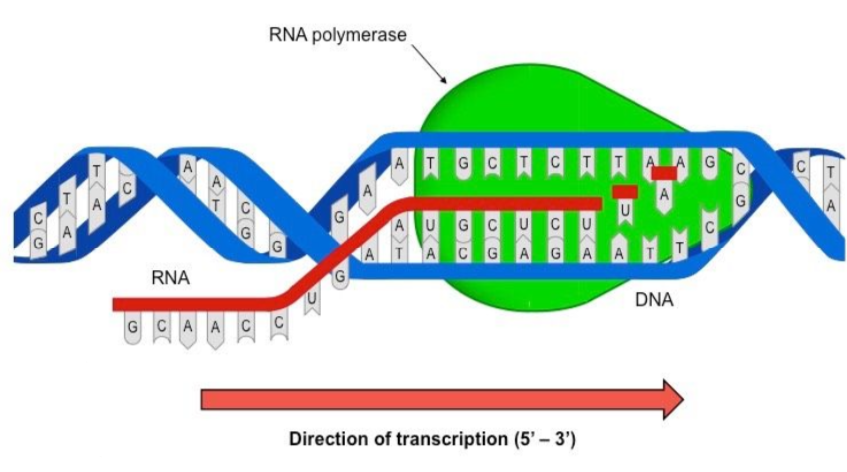
explain transcription using the diagram provided
occurs in the nucleus (euk) or cytoplasm (prok)
DNA double helix strands are separated by RNA polymerase
a gene is transcribed
RNA polymerase enzyme responsible for transcription
enzyme covalently bonds RNA nucleotides together
DNA is unchanged and returns back to double helix structure
complementary base pairing between newly synthesised RNA strand and DNA template strand
describe the structure of a protein
proteins are polypeptides and are composed of amino acids (polymers)
monomer: amino acid
polymer: polypeptide
“polypeptide” used during translation bc it hasn’t folded up into a fully functioning protein yet
amino acids are held together by peptide bonds
20 kinds of amino acids are used
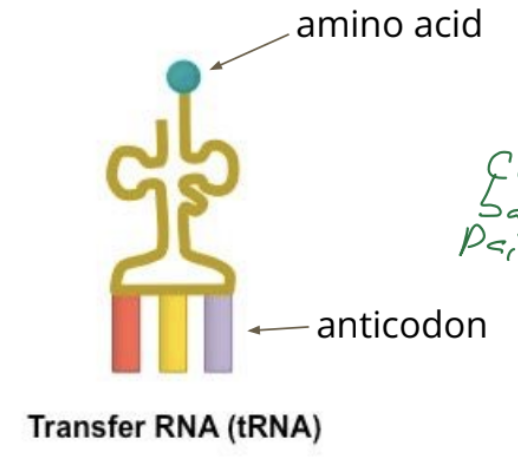
list and outline the 3 kinds of RNA
mRNA: messenger RNA that contains the code for building protein
tRNA: transfer RNA that brings amino acids to the ribosome
rRNA: ribosomal RNA is part of the ribosome itself
describe the function of mRNA
Messenger RNA (mRNA) carries the genetic code that determines the order of amino acids in a polypeptide sequence
describe the function of tRNA
Transfer RNA (tRNA) is responsible for transporting amino acids to the ribosome according to the mRNA sequence
describe the function and structure of a ribosome
composed of rRNA and protein with a complex, globular shape separated into a large and small subunit and are complex macromolecular machines, they consist of 2 distinct subunits
large subunit: 3 binding sites for tRNAs (E, P, A), but only 2 tRNA molecules can bind simultaneously at a time, creates peptide bonds between amino acids
small subunit: binds to the mRNA and decodes the genetic message
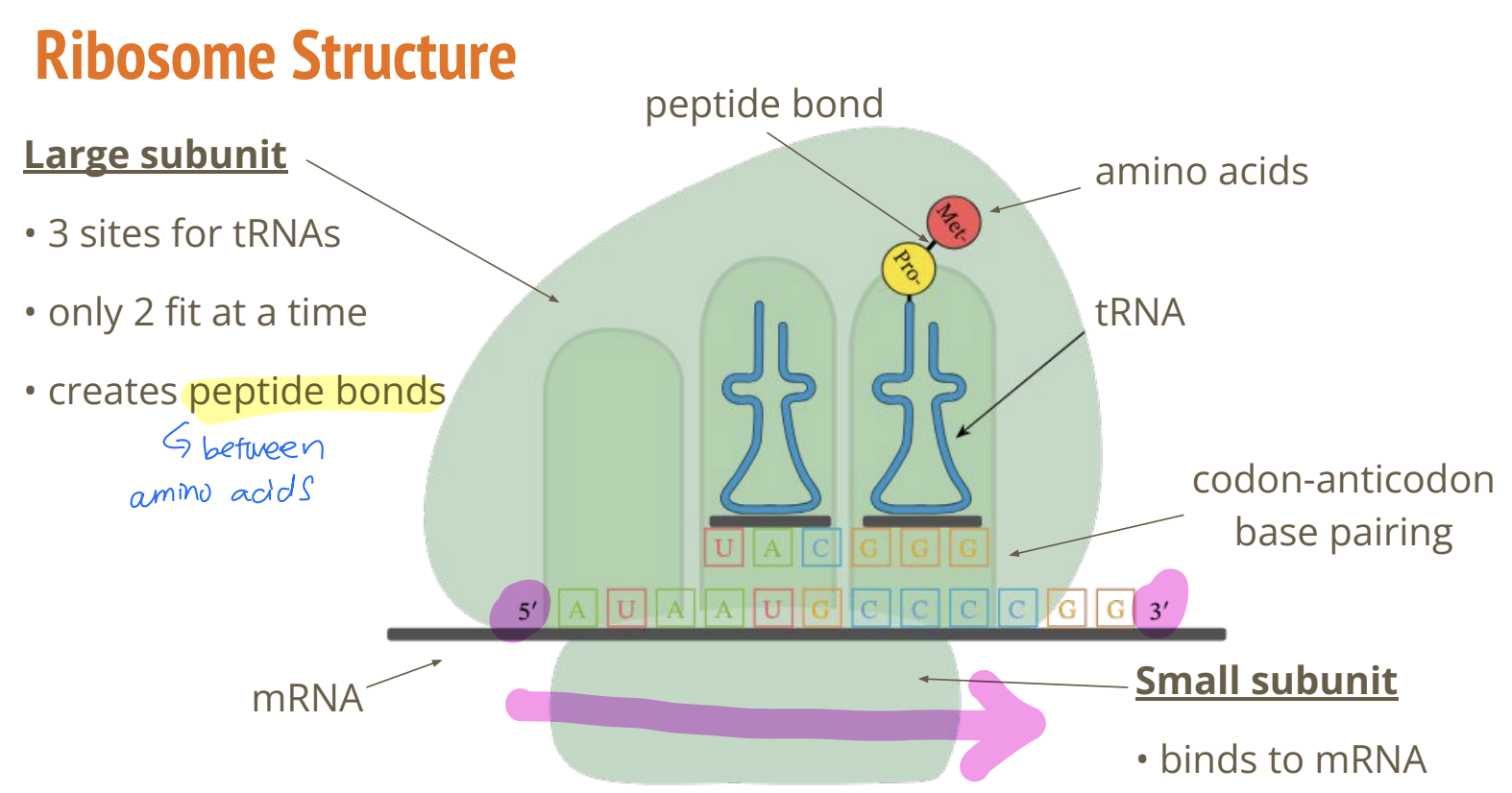
describe the function of rRNA
ribosomes are composed of both ribosomal RNA (which contributes to the catalytic activity) and protein (which provides structural stability)
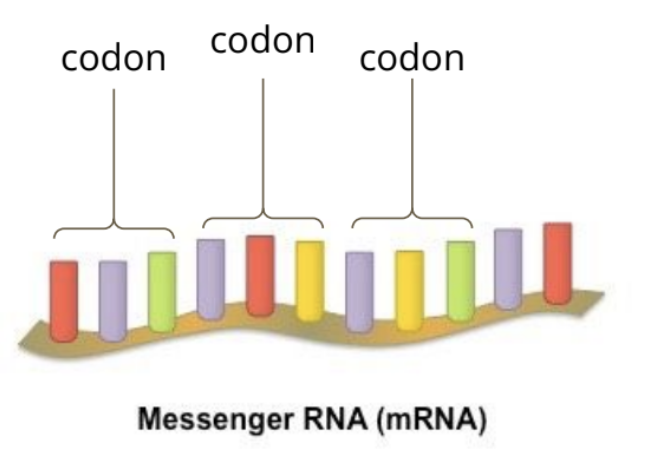
describe codon and anticodon
codon: a sequence of three mRNA bases that codes for a specific amino acid or a stop signal during protein synthesis
anticodon: a sequence of three nucleotides in tRNA that is complementary to a specific codon in mRNA
describe the function of codons and anticodons
codon function: codons which are read by the ribosome during the translation process which instructs the ribosome to start the creation of a polypeptide sequence, add a specific amino acid to the growing polypeptide chain, or to stop the creation of the polypeptide sequence
anticodon function: anticodons are a sequence of 3 tRNA bases that are complementary to a codon which carries the appropriate amino acid to be condensed onto the growing polypeptide chain during translation
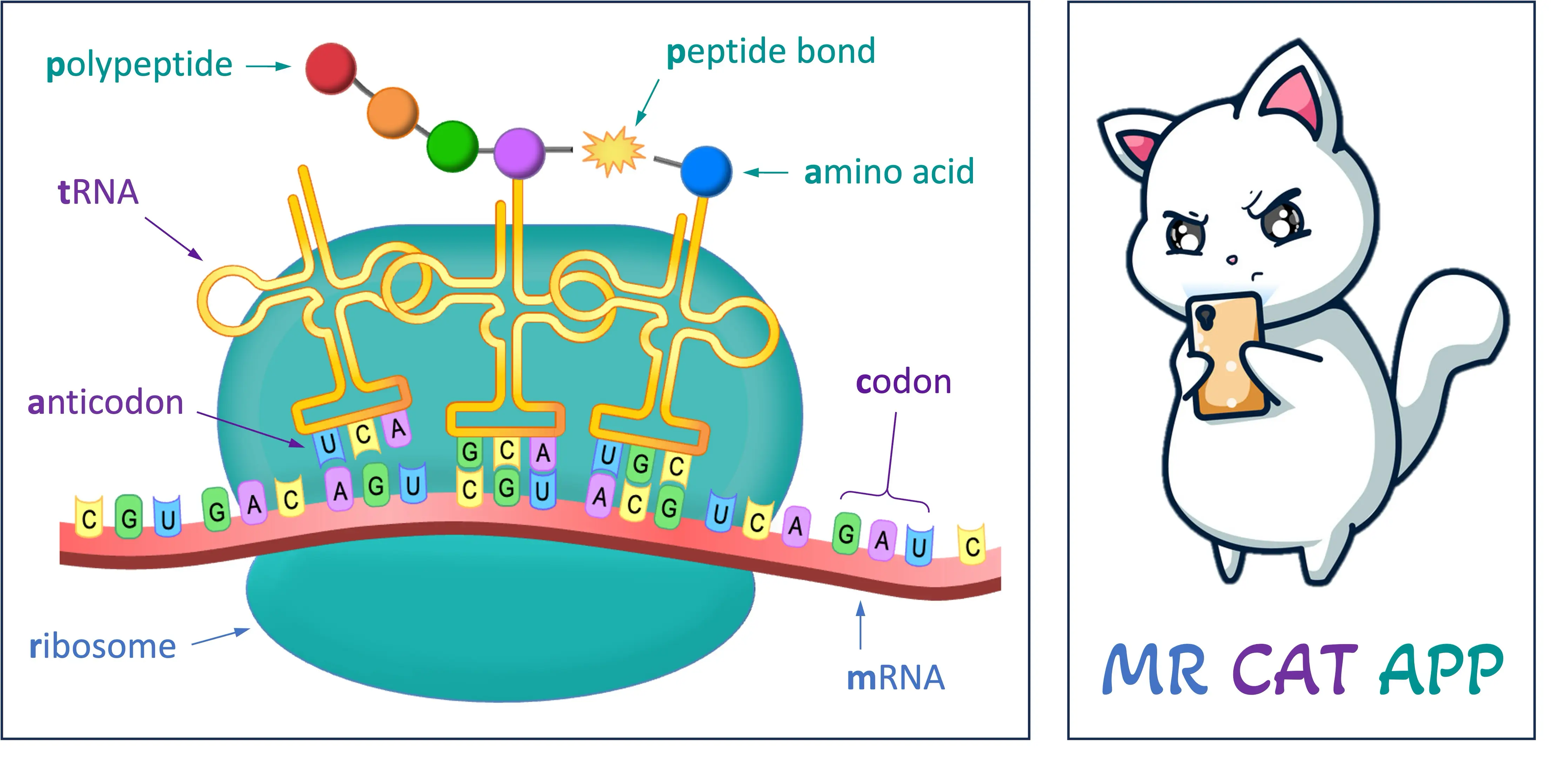
HOW TO REMEMBER TRANSLATION EASILY: Mr Cat App
MR: Messenger RNA binds to the Ribosome (via the small subunit)
CAT: Codons (on mRNA) are recognised by complementary Anticodons on Transfer RNA
APP: Amino acids are joined by the ribosome via Peptide bonds to form Polypeptides
what is a genetic code
the genetic code is a set of rules by which information encoded within mRNA sequences are converted into amino acid sequences (polypeptides) by living cells
represented by a table that identifies the corresponding amino acid for each codon combination (64 codon possibilities)
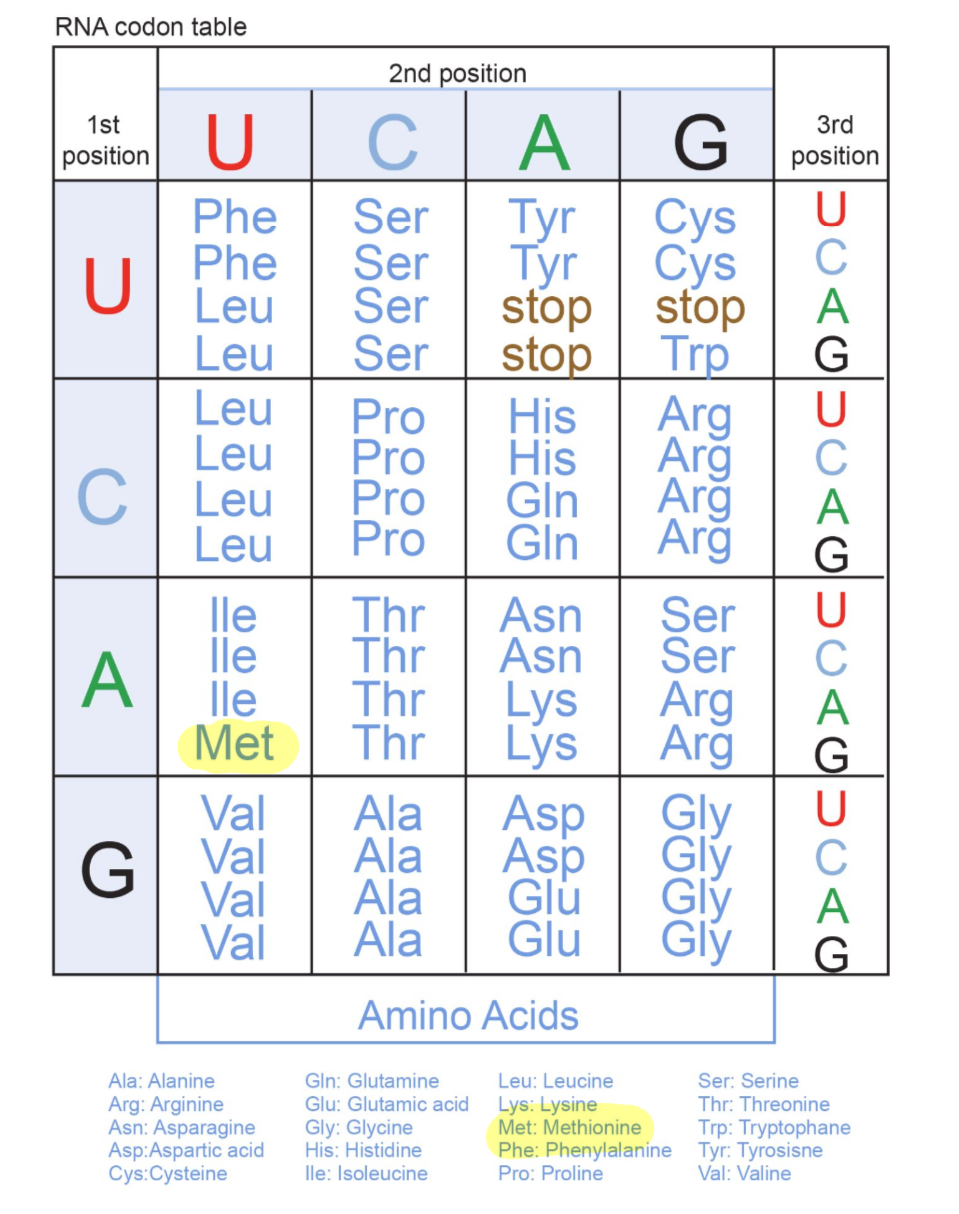
what are the two key features of genetic code
it is universal and it has degeneracy
universal: genetic code is universal as almost every living organism uses the same code (there are a few rare and minor exceptions)
degenerate: as the genetic code has around only 20 amino acids but 64 different codon combinations, more than one codon may code for a single amino acid
outline transcription and translation and describe the directionality
transcription is the process by which a gene sequence is copied into a complementary RNA sequence by RNA polymerase
translation is the process
explain the initiation process of transcription
promoter Binding and transcription factors: RNA polymerase binds to a specific region of the DNA known as the promoter with the help of transcription factors which are proteins as they guide the enzyme onto the promoter region which is located upstream of the gene to be transcribed
DNA strand separation: RNA polymerase unwinds and separates the DNA double helix, creating a transcription bubble which exposes the template strand of DNA that will be used for the synthesis of RNA
Free RNA nucleotides within the cell line up opposite their complementary base partner
RNA polymerase covalently binds the RNA nucleotides together via condensation reactions (forming phosphodiester bonds)
The 5’-phosphate is linked to the 3’-end of the growing mRNA strand, hence transcription occurs in a 5’ → 3’ direction
explain the elongation process of transcription
RNA synthesis: RNA polymerase synthesises a single strand of RNA by adding ribonucleotides that are complementary to the DNA template strand which occurs in the 5' to 3' direction.
Base Pairing:
adenine (A) - uracil (U)
thymine (T) - adenine (A)
guanine (G) - cytosine (C)
cytosine (C) - guanine (G)
explain the termination process of transcription
termination signals: RNA polymerase continues elongation until it reaches a specific sequence of nucleotides that signals the end of the gene (termination site)
release of RNA: once the RNA polymerase reaches a termination signal, the newly synthesised mRNA strand is released from the RNA polymerase and DNA template
explain what post transcriptional RNA modification is in eukaryotes and why they are essential
post transcriptional RNA modifications are crucial steps that occur after the initial synthesis of pre mRNA and before it is translated into protein
ensure the stability, transport, and proper function of the mRNA
explain post transcriptional RNA modifications in eukaryotes
capping: addition of methyl group at 5’-end
provides protection and allows transcript to be recognised by the ribosome
protecting mRNA from degradation
polyadenylation: addition of long chain of adenine at 3’-end (poly-A-tail)
improves stability of RNA and facilitates export
protecting mRNA from degradation
splicing: removal of non coding sequence (introns) and splicing together of coding sequences (exons) to form a continuous coding sequence
intron(thrown out/recycled): non-coding sequences of DNA within genes transcribed into RNA (doesn’t code for amino acids; have other functions)
exon(expressed, actually used to build protein): coding sequences of DNA or RNA within genes transcribed into RNA
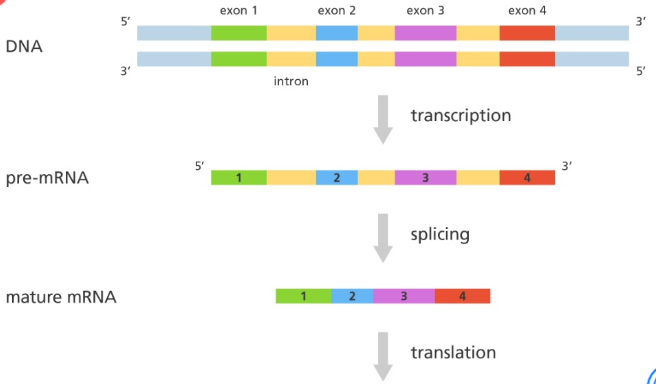
what are non-coding DNA
sequences that do not code for a protein which only occur in around 1.5% of a DNA sequence
regulatory regions: enhancer and silencer regions where activator and repressor proteins bind to help or hinder initiation
explain the the initiation process of translation
ribosomal assembly: the small subunit of the ribosome binds to the 5’ end to the 3’ end of the mRNA and slides across the molecule (in a 5’ to 3’ direction) until it reaches a start codon (AUG)
tRNA binding: the initiator tRNA carrying the amino acid methionine, recognises the start codon and binds to it by its anticodon (UAC)
large subunit joining: the large ribosomal subunit then attaches to the small ribosomal subunit to form a functional ribosome, creating the A (aminoacyl), P (peptidyl), and E (exit) sites
explain the the elongation process of translation
aminoacyl-tRNA binding: an aminoacyl-tRNA (charged tRNA carrying an amino acid) enters the ribosome and binds to the A site where its anticodon binds with corresponding codon on the mRNA
peptide bond formation: the amino acid in the a site is transferred into the growing polypeptide held in the P site where the ribosomal enzyme catalyses the formation of a peptide bond between the amino acid in the A site and the growing chain in the P site
translocation: as the ribosome slides along the mRNA by one codon, it shifts the tRNA holding the polypeptide chain that was in the A site into the P site and the tRNA with no amino acid into the E site where it exits the ribosome. after this happens, this opens the A site for a new tRNA with a new amino acid to bind to the A site and continue the elongation process
explain the the termination process of translation
stop codon recognition: when the ribosome reaches a stop codon (UAA, UAG, or UGA) on the mRNA, there is no corresponding tRNA
release factor binding: a release factor binds to the stop codon, prompting the ribosome to add a water molecule instead of an amino acid, which releases the polypeptide chain
disassembly: the ribosomal subunits, mRNA, and release factor disassemble, completing the translation process releasing all the enzymes involved
explain the modification of polypeptides into their functional state
proteins are modified after translation for functional state
formation of disulphite bridge between cysteine residues
conjugation with inorganic cofactors (haeme group)
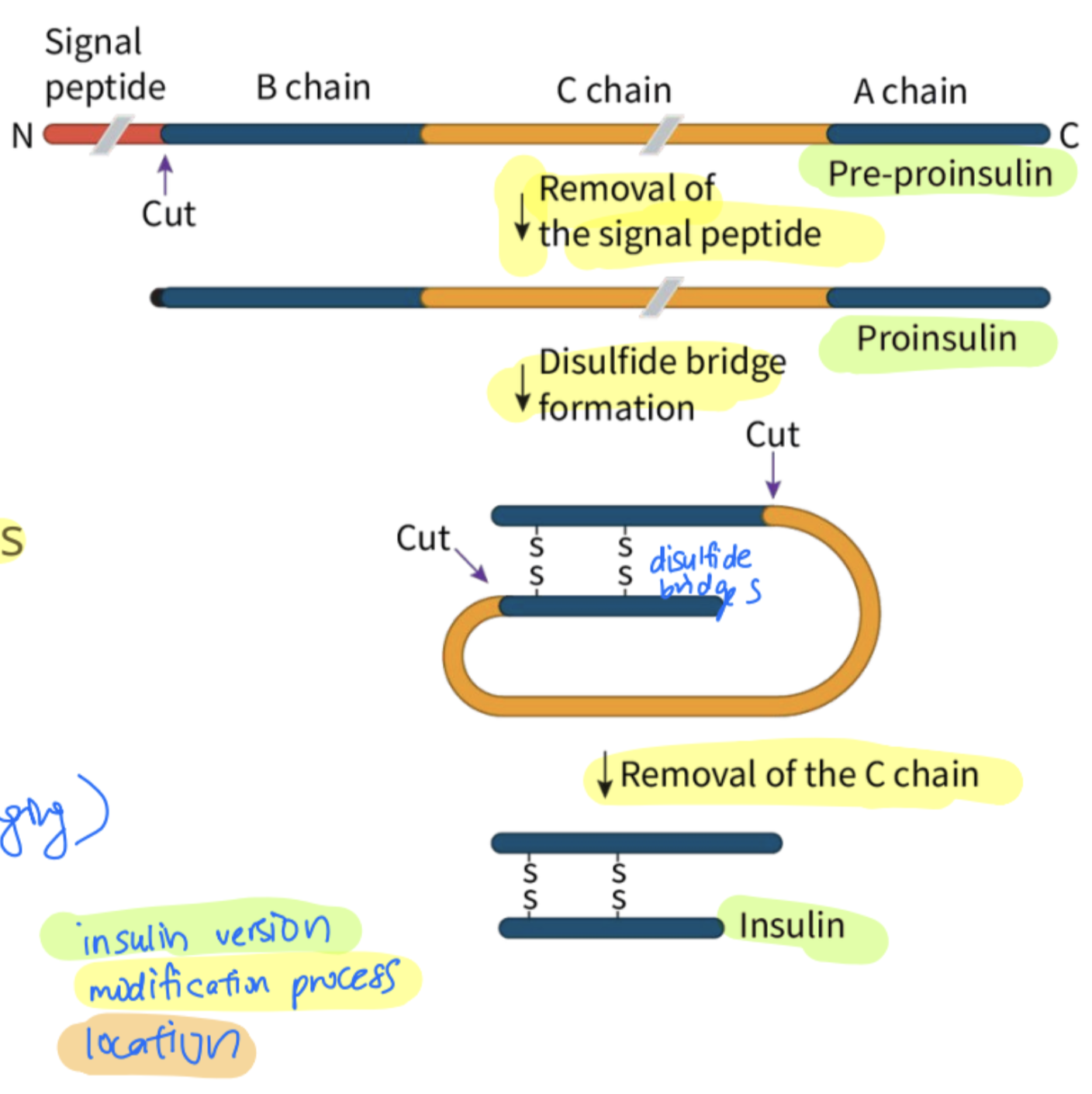
use insulin as an example and explain the modification of its polypeptides into its functional state
pre-insulin modified into insulin
pre-insulin consists of: signal sequence, A chain, B chain and C-peptide
signal sequence in preproinsulin is removed in the rER
proinsulin folds: the A chain and B chain of the preproinsulin are linked by disulphite bridges
converted into proinsulin
proinsulin is transported into the golgi complex where the c-peptide is removed
results in (mature) insulin which is then packaged into secretory vesicles
explain how proteasomes help recycle amino acids
proteasomes: they are complex structures within a cell that degrade uneeded proteins by breaking the peptide bonds within proteins
they help regulate protein expression levels and recycle amino acids
by controlling breakdown and synthesis of proteins, proteasome plays an important role in sustaining functional proteome
what is alternative splicing
regulatory mechanism in gene expression that allows a single gene to produce multiple mRNA variants
different arrangement of exons spliced together to produce a variety of proteins
explain how alternative splicing of exons are able to produce variants of a protein from a single gene
by selectively removing certain exons from polypeptides from a single gene sequence, cells can produce variants of proteins that may have different functional roles or regulatory mechanisms