BCM. 28 Oxidative phosphorylation
1/17
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
18 Terms
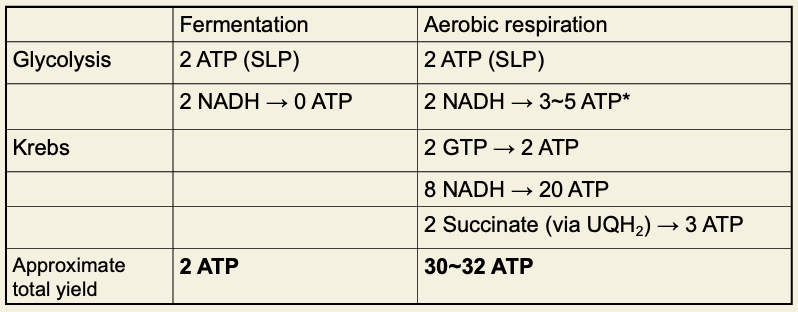
Structure of a mitochondrion
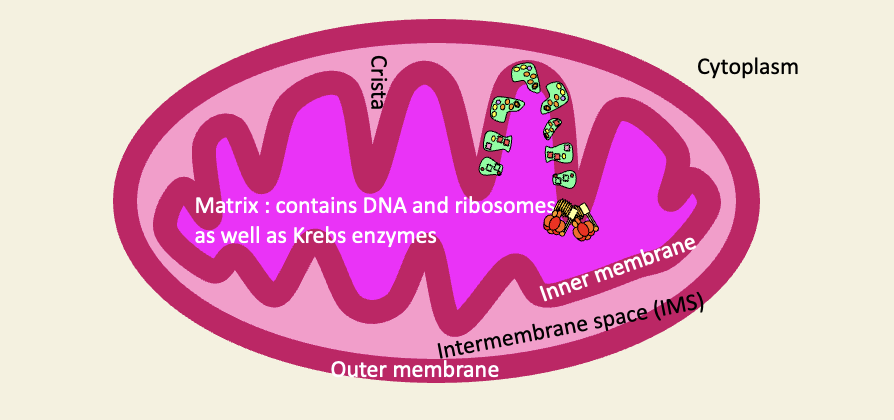
Structure of a proteobacterium
Topological equivalences
bacterial cytoplasm = mitochondrial matrix
inner cell membrane = inner mitochondrial membrane
Periplasmic space = IMS
Outer cell membrane = outer mitochondrial membrane
Extracellular space = cytoplasm of host cell
Structure of chloroplasts
chloroplasts are believed to derive from the cyanobacteria

Mitochondria have evolved by endosymbiosis - but margulis’s original theory has serious problems
ATP4- doesnt leak - they would need a specific transporter protein (if so desperate for ATP, how can it do phagocytosis)
Host: methanogen - made their living from the exergonic reduction of C02 to methane
Protomitochondrion - heterotroph (The waste-products of the proto-mitochondrion (hydrogen and CO2) were the food for the methanogenic proto-eukaryotic host

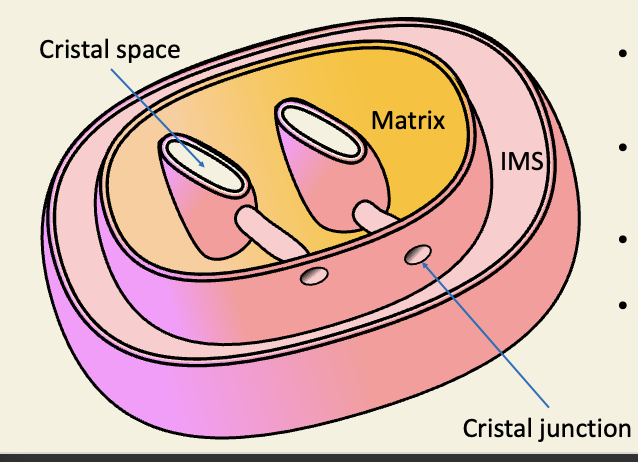
Real mitochondria vary
Mitochondria may be cigar-shaped, tubular, branched
cristae may be lamellar, discoidal, tubular
not everything is rate liver
even rat-liver cristae dont look like those in most text-books
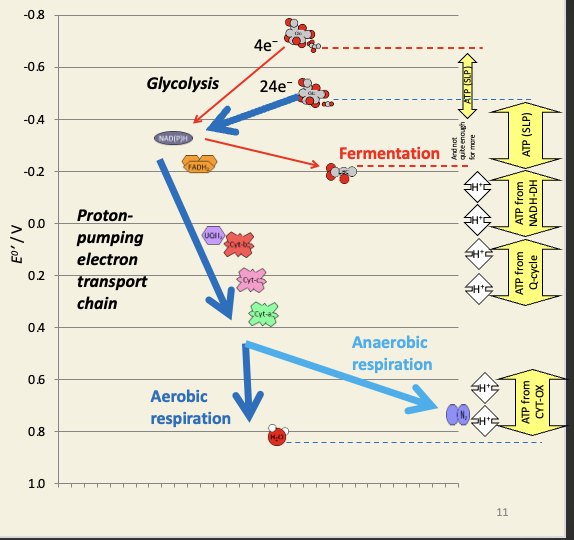
Oxidative phosphorylation is the process by which NADH is oxidised to generate ATP
redox-driven chemiosmosis
in mitochondria
and aerobic bacteria
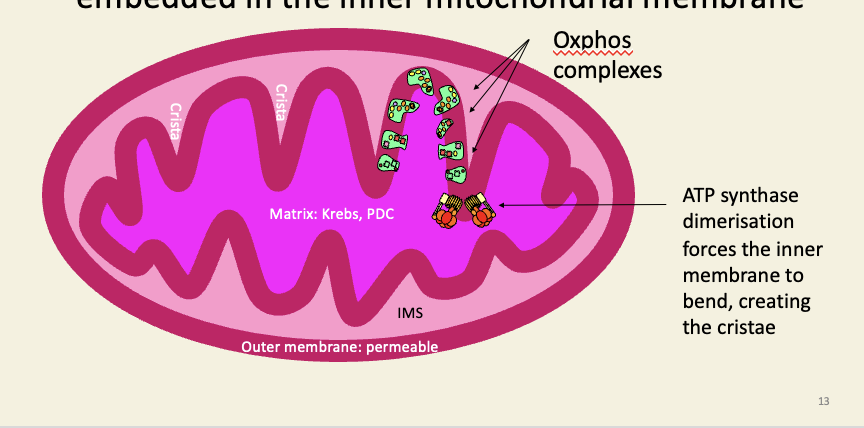
The complexes of oxidative phosphorylation are embedded in the inner mitochondrial membrane
Complex I is the flavoprotein NADH dehydrogenase
NADH from Krebs oxidation of a-ketoglutarate, malate, etc.
largest transmembrane protein → the structure only solved 6 years ago
When electrons pass into the hydrophilic head, they cause the bound quinol to shift upwards
One proton is pushed through the interface between the head and membrane-bound part at this point
The three subunits that compose the membrane-bound section are then warped by the movement of the headpiece: the one warped by the headpiece warps the middle subunit, and the middle subunit then warps the subunit at the end.
Each subunit pushes one more proton through
(the whole structure of the protein really does move like a piston in a steam engine)

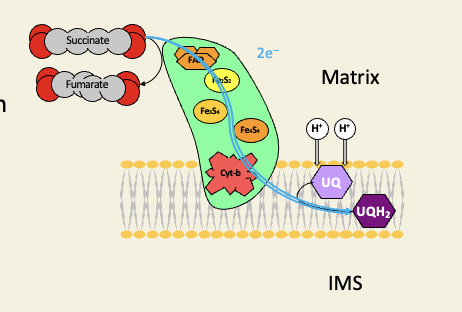
Complex II is the Krebs cycle enzyme succinate dehydrogenase
too small a change in free energy for any proton pumping
gets electrons from oxidation of succinate only
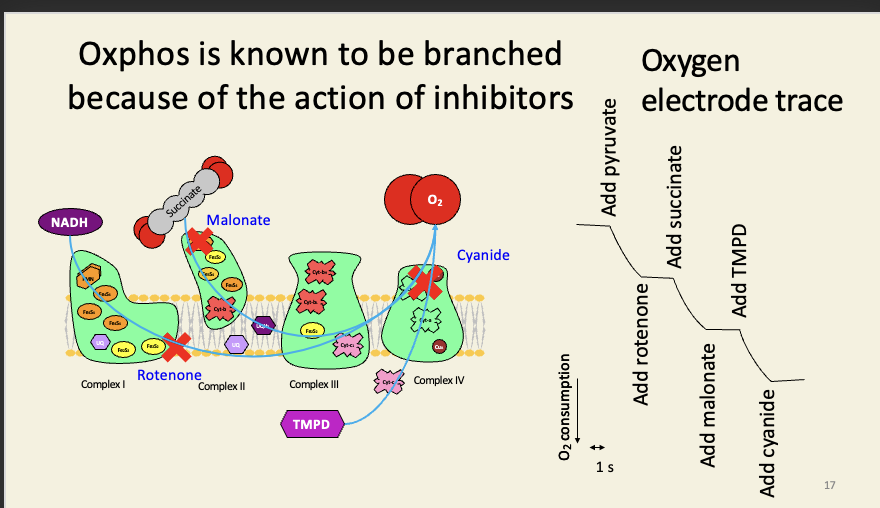
The oxygen electrode can be used to monitor oxygen consumption in cells and organelles
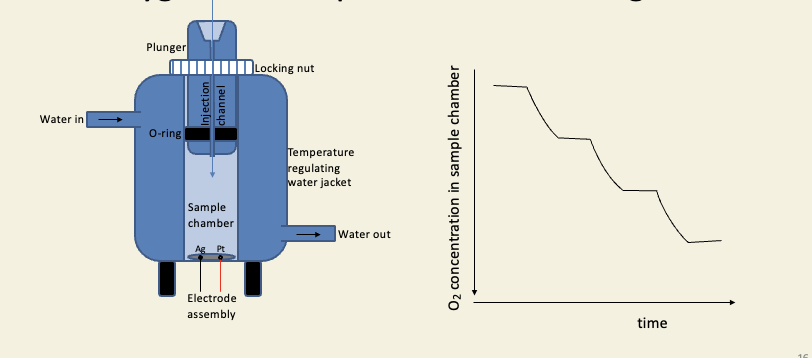
Oxphos is known to be branched because of the action of inhibitors
Tetramethyl-p-phenylenediamine is an artificial substrate that feeds electrons directly to cyt-c in the presence of ascorbic acid (vitamin C).
TMPD is also used as a test for a terminal cytochrome-c-oxidase: it turns blue as it is oxidised: this test (‘OXIDASE POSITIVE’) is part of the standard suite of tests used in identifying bacteria biochemically: e.g. Pseudomonas is oxidase positive; Escherichia is oxidase negative
Ubiquinol is a fat-soluble redox carrier dissolved in the inner membrane
Superoxide, hydroxyl radicals, peroxides, and other reactive oxygen species cause oxidative stress. One particularly lethal thing they can cause is lipid peroxidation, which is a free-radical chain-reaction that makes fats (e.g. membrane lipids) rancid. This would destroy the mitochondrion. Therefore mitochondria contain high concentrations of enzymes that scavenge and reduce these dangerous ROS molecules.

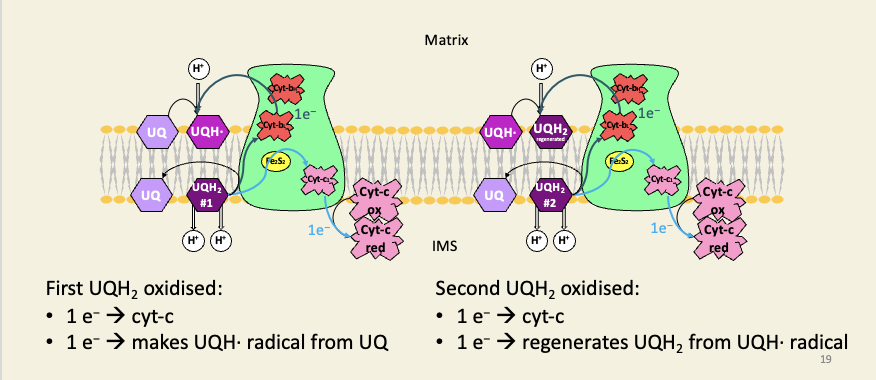
Complex III is a cyt-b/c complex running the Q-cycle, which doubles the efficiency of proton pumping
For each UQH2 the Q-cycle oxidises, one electron is passed onto the downstream carrier (cyt-c), which is always a 1-electron carrier.
The other electron is recycled. In the first-pass of the cycle, this recycled electron generates a UQH· free radical, which complex-III keeps a hold of; in the second pass, this radical is fully reduced to UQH2.
The two steps of the cycle therefore oxidise two molecules of UQH2, but they also regenerate a UQH2, so only one net UQH2 is oxidised.
Because of the positions of the Q-binding sites on complex III (oxidation on IMS-side, reduction on matrix-side), the protons are carried across the membrane from matrix to IMS, and for each UQH2 oxidised, the Q-cycle pumps four protons.
If the cycle didn’t exist, and both electrons were passed directly to cyt-c, the cycle would only pump two protons per UQH2 oxidised.
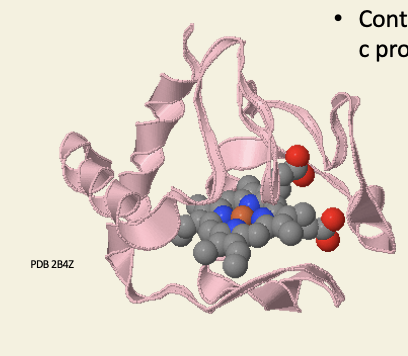
Cytochrome-c is a peripheral membrane haemoprotein
contains covalently bound haem-c prosthetic group
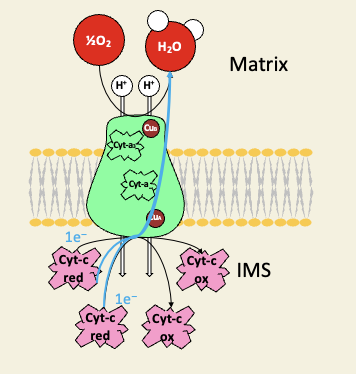
Complex IV is cytochrome-c oxidase
many famous inhibitors bind the cyt-a/a3 oxygen binding site
carbon monoxide
cyanide
azide
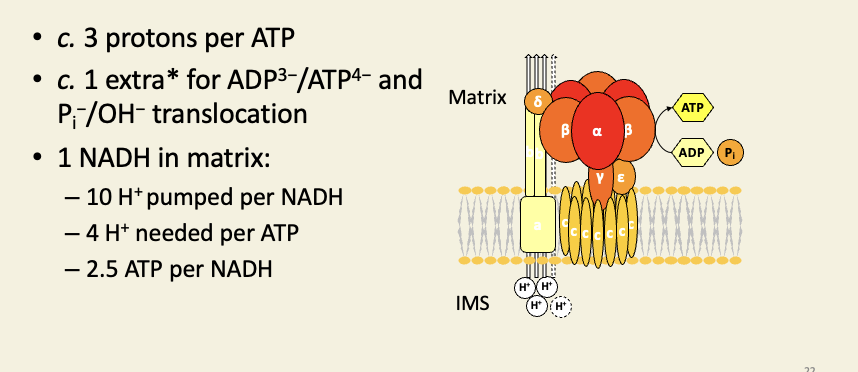
Complex V is the F-type/synthase
This extra proton is virtual: the ATP/ADP translocator does not translocate protons, but the movement of ATP into the IMS and ADP into the matrix results in a net charge of +1 moving into the matrix.
The movement of phosphate into the matrix and hydroxide into the IMS has no effect on ψ (it's an electroneutral exchange), but it does decrease the ∆pH component of the PMF by the equivalent of one proton (loss of one hydroxide = gain of one proton, as far as pH is concerned).
By the combination of both of these two processes, the PMF is decreased in both its components by the same amount as would be caused by one proton flowing into the matrix.
1 NADH oxidation (two electrons) results in 10 protons being moved to the IMS. At a ratio of 4 protons per ATP, we get 10/4 = 2.5 ATP per NADH.

Very approximately, aerobic respiration is 15 times as efficient as fermentation
The yield for cytosolic NADH depends on how it gets into the matrix: the malate/aspartate shuttle creates 1 matrix NADH for each cytosolic NADH, which has the same yield as matrix NADH (2.5 ATP per NADH), but the glycerol-3-phosphate shuttle creates 1 membrane UQH2 for each cytosolic NADH, which has the same yield as succinate (1.5 ATP per UQH2).
Different organs and organisms use different shuttles.