BIOL 200 (DNA)
1/50
Earn XP
Description and Tags
RL7-RL9
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
51 Terms
What is the purpose of DNA replication?
to duplicate DNA before cell division so each daughter cell inherits the full genome
Parental strand serves as?
a template for the formation of a new daughter strand (complementary to the daughter strand)
In which direction does the DNA replication does it follow?
from 5’ to 3’
DNA replication proceeds through which type of mechanism?
semi-conservative mechanism (each DNA molecule has one old strand and new strand)
Explain to me the DNA polymerization (process itself), what is required?
catalyzed by DNA polymerase, requires deoxynucloside 5’ - triphosphates, requires primer, proceeds from 5’ to 3’
What are the different steps of DNA synthesis?
DNA unwiding
Primer formation
Elongation
Primer removal
Ligation
Key enzymes in DNA unwiding?
enzymes: DNA Helicase (to open and seperate the two strands), topoisomerase (relieve tension, supercoils)
Key enzymes in Primer formation?
enzymes: primase
What is a primer?
a primer is a short RNA molecule = complementary to a single-stranded region of unwounded DNA
Key enzymes in Elongation?
DNA polymerase
What will the DNA polymerase do?
extend the primer by adding complementary nucleotides
What are the two strands present in DNA synthesis?
leading DNA synthesis
lagging strand DNA synthesis
What is the leading strand? What is the lagging strand?
follows movement of the replication fork (5’ to 3’)
must be formed in the opposite direction of the movement of replication fork
How many primers are necessary for each strand?
leading strand (1 primer = synthesis will continue)
lagging strand (primase going to produce primer every few bases = synthesis discontinued) = done in fragments
What do we call those fragments in the lagging strand?
Okazaki fragments
These Okazaki fragments are made of?
RNA and DNA (RNA needs to be removed and replaced by DNA) = primer removal step
What enzymes are necessary for ligation?
DNA ligase = joining the DNA fragments together
What is the replisome?
complex molecular machinery that carries out DNA replication
Enzyme that start the process of replication?
CMG helicase = binds and surrounds leading strands = move on to the DNA = separates the 2 strands
contains accessory subunits Cdc45 and GINS complex
What is a problem that lagging strands have that leading strands do not have?
the DNA in lagging can be single-stranded which base-pairing could occur, because of that it would be difficult for the polymerase to carry on it’s acitivity
What protein would be useful that resembles topoisomerase in a way
RPA (replication protein A) = binds single-stranded DNA, keeps single-stranded DNA template in optimal conformation for DNA polymerase
What synthesizes the leading strand in DNA?
DNA polymerase epsilon
3 types of polymerases in eukaryotic replisome
PCNA
Primase/polymerase alpha
Polymerase beta/PCNA complex
What is PCNA?
homotrimetric protein that encircles DNA and prevents polymerase epsilon or beta complex from disassociating from the template = keep synthesizing for longer
What about Primase/polymerase alpha
has primase activity = primase forms the RNA component of the primer
DNA polymerase activity = use of actual polymerase alpha (extends the primer with short DNA segments
What about polymerase beta/PCNA complex
it replace the polymerase alpha/Primase complex and completes the synthesis of an Okazaki fragments = synthesis of lagging strand
What is RFC/PCNA complex?
RFC is the PCNA loader, it opens the PCNA ring and loads it on a primer = polymerase binds to PCNA to start synthesis
Additional activities?
ribonuclease H and FEN-1 = removes RNA primers
polymerase beta = replaces the RNA with DNA
DNA ligase = ligates the DNA fragments together
So in order what are the steps?
Helicase (unwind the two strands)
RPA (stabilize separated strands)
primase + poly alpha (synthesize RNA-DNA primers)
pol epsilon (synthesize DNA continuously)
pol. beta (synthesize Okazaki fragments)
PCNA + RFC (clamp and loader to keep polymerases attached)
DNA ligase (join fragments)
What are origins of replication?
specific DNA sequences where replication begins
What is ORC and 2 functions?
ORC = origin recognition complex = six subunit protein that
mark replication origins
load helicase aided by other initiation factors
MCM helicase is loaded in? and activated in?
loaded in G1 and activated in the S phase = ensuring replication occurs one per cell cycle
This is the overall steps all together, to review
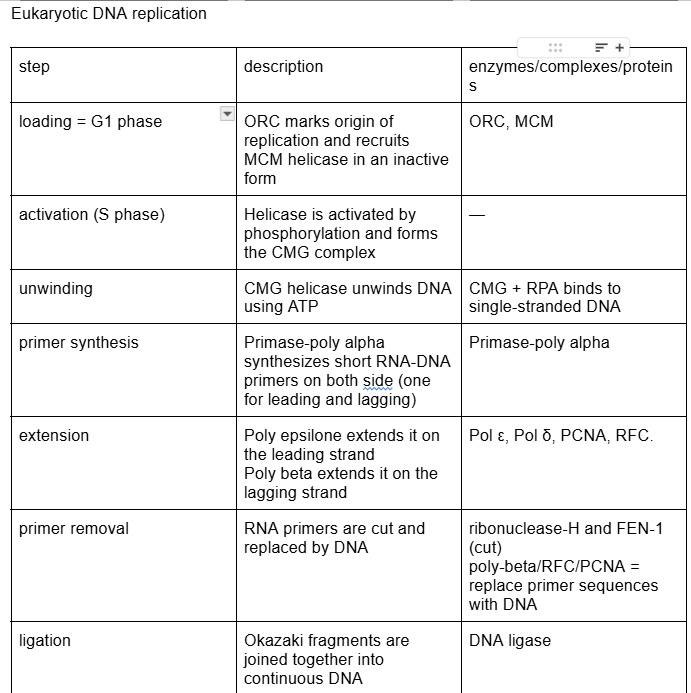
//RL8
DNA is subject to constant change = damage, examples?
hydrolytic depurination (removal of the purine), cytosine deamination, guanidine oxidation, methylation of adenine
These modifications produce many different modified bases, what could that lead to?
blocking of the activity of DNA polymerases during DNA replication
Define for me mutations?
permanent, transmissible changes (to another generation) to the genetic material of a cell
can occur spontaneously by transposable elements, by errors during replication
What are mutagens?
chemical compounds, ultraviolet (UV) radiation or ionizing radiation that increase the frequency of mutations
Example of mutagen?
carcinogen = agent that causes cancer
What are the implications of mutation rates in health
fathers who had accumulation of mistakes in DNA (add up) = higher probability having children with schizophrenia, autism
rate of mutations in most viruses is much higher than in cells = higher probability of selection of more successful variants
Name me the different mechanisms to repair DNA
repair of mismatches
repair of small modifications (base excision repair)
repair of bulky lesions (nucleotide excision repairs)
Describe DNA polymerase in terms of making mistakes?
DNA polymerases have high fidelity = meaning they don’t a lot of mistake (1 error every 10,000), still pretty very frequent
What activity can undergo to fix those errors?
proofreading exonuclease activity and mismatch repair
Which DNA polymerase have this exonuclease or proofreading activity
DNA polymerase epsolin and beta
Explain to me the mismatch repair mechanism?
MSH2 + MSH6 = binds to daughter strand
triggers the activity of protein MLH1 endonuclease + PMS2 = makes cuts
DNA helicase = peels off the segment generated by MHL1 + PMS2
DNA exonuclease = digest the fragments
gap repair by polymerase beta and dna ligase
Explain to me the process of Base excision repair
DNA glycosylase hydrolyzes the bond between the mispaired base and the sugar phosphate backbone (ribose) = detects the wrong base and remove it
provides specificity in repair
APE1 cuts inside the DNA backbone where there is no base
Ap lyase (pol.beta) = removes the deoxyribose phosphate
DNA polymerase beta fills the gap and DNA ligase seals the nick in the sugar-phosphate backbone
Explain to me the process of bulky lesions (nucleotide excision repair) = NER
complex formed by 23B and XP-C = initial recognition of DNA lesion
recruit of transcript factor (TFIIH) = unwinding DNA, which then recruits RPA and XP-G (also unwinding activity)
XP-F is recruited and XP-F and XP-G both have endonuclease activities = generates cuts = fragments removed and degraded
gap is filled by DNA polymerase and continuously joined by DNA ligase
What is one common thing in the 3 mechanism DNA repair?
required that DNA is in a double-stranded form
When DNA lesions are not repaired, what will the DNA polymerase do?
it will stop it’s activity after encoutering the lesion = preventing completion of DNA replication
What mechanism do we use in that case? what does it involve?
translesion synthesis (TLS) uses translesion polymerases, by pass the lesion using the damaged DNA as a template
Benefit and consequences?
benefit = the DNA replication can continue
consequences = translesions polymerases will often insert incorrect base at the position opposite to the lesion, don’t have proofreading exonuclease activity, error prone