RRoy 7: Chromatin and Epigenetics - The Histone Code
1/24
Earn XP
Description and Tags
November 5th, 2025
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
25 Terms
How is DNA wound up in the nucleus and why?
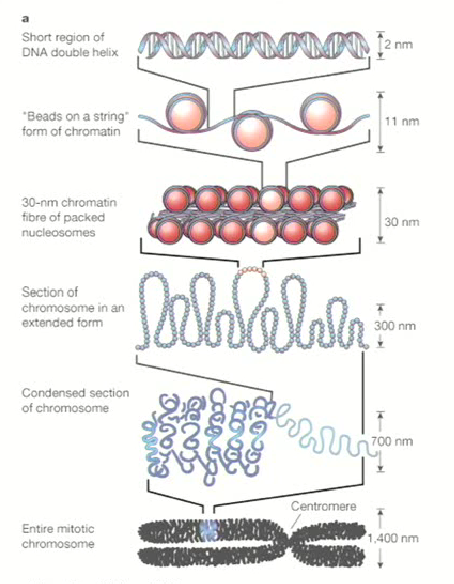
In the nucleus, DNA is wound up by histone proteins (H2A, H2B, H3 & H4) into nucleosomes that form tight coils to retain lots of information
Nucleosomes are necessary to package genomic DNA into the nucleus to form nucleosomes
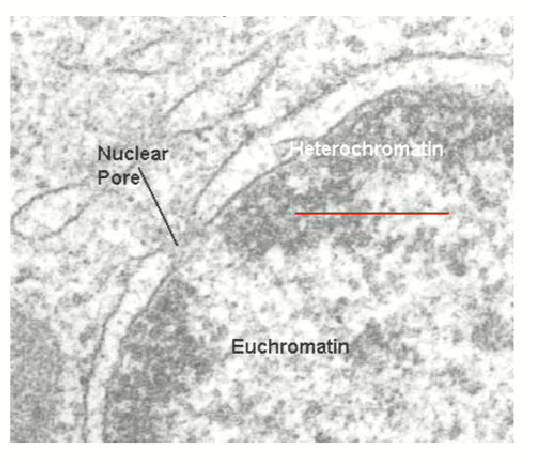
Two forms of chromatin and their differences
Heterochromatin
Made up of silenced DNA sequences (transcriptionally inactive) and very densely packed (dark on a micrograph)
Euchromatin
Made up of actively transcribing DNA sequences (transcriptionally active) and unwound to provide a transcriptional template
delicate
Higher order regulation of transcription (main purpose)
Find out why it is important to silence the genome
Mechanistically distinguish active and inactive regions - studied through yeast
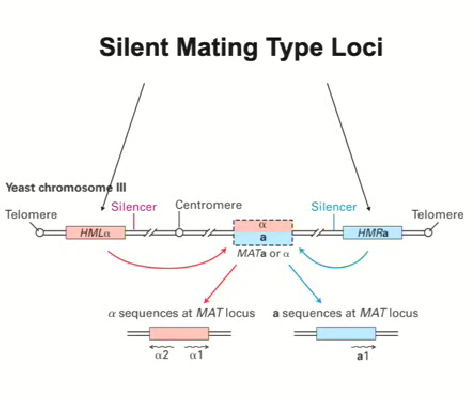
Yeast study explained (mating types)
Yeast can exist in both diploid and haploid form
Cells will bud off from one another (mother cell generates daughter cells)
When these daughter cells are formed, the mother will switch mating types (either a or alpha)
When conditions are not ideal for growth, the yeast need to mate and form a diploid organism
In order to do this, they must be the opposite mating type as the daughter cell, so they switch mating types
The region that controls the mating type is from the transcription on chromosome 3 (mating type loci)
When region alpha (left side of diagram) or region a (right side of diagram) is recombined into the mating locus at the center, it means that the gene will be expressed (2 possibilities)
These genes are only active when they get transcribed in the mating locus, otherwise they need to be silenced
Specific sequences in the HMLalpha and HMRa ensure that they are silenced which work outside of the context of mating so that they can block tRNA genes (RNA Pol III)
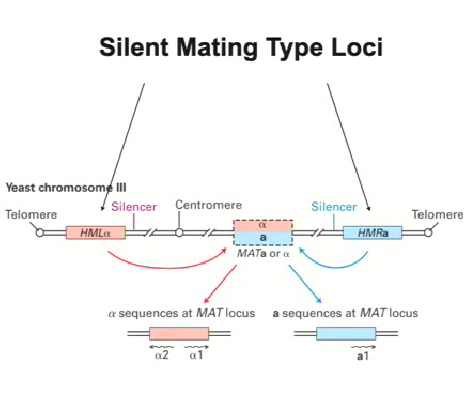
Silencing mechanism for HMLalpha and HMRa
There are specific sequences in the HMLalpha and HMRa to ensure that they are silenced which work in more than just mating (ex. to block tRNA genes (RNA Pol III)
Genetic experiments show that small changes in the histone tails (H2, H3) and telomeres are involved in the silencing process
If these are mutated, the genes are expressed again
Proteins Responsible for silencing process
RAP1
SIR1
SIR2, 3 and 4
RAP1
Protein involved in silencing process of HMLalpha and HMRa
binds to DNA in the region of the silencer
also binds to repetitive sequence in telomeres
SIR1
Protein involved in silencing process of HMLalpha and HMRa
Cooperates with RAP1 and is important for binding the silencer region in the silent mating type loci
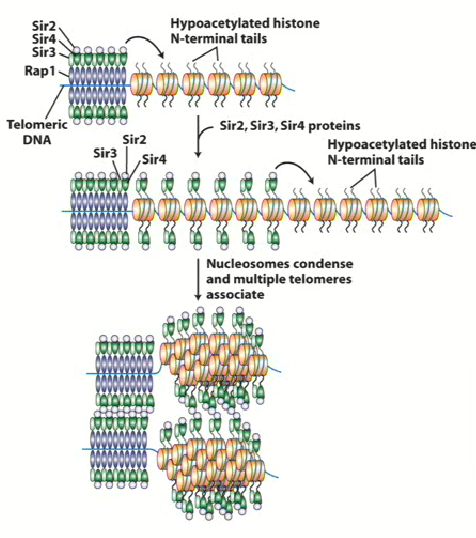
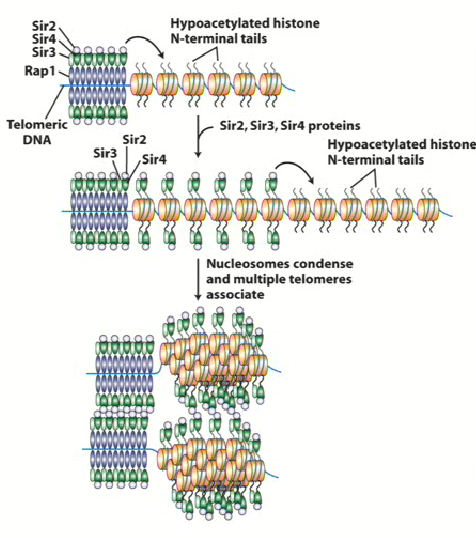
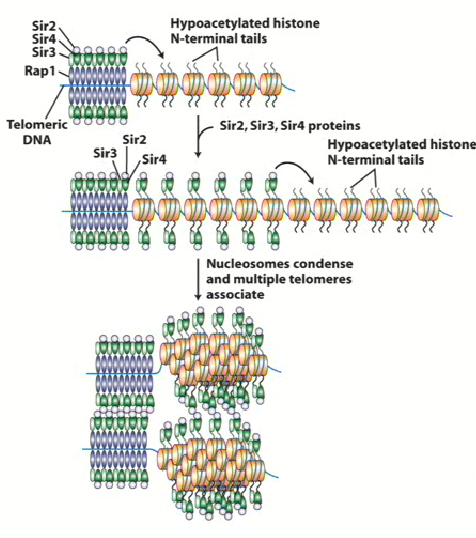
SIR2, 3 and 4
Stands for Silent Information Regulator
Proteins involved in silencing process of HMLalpha and HMRa
Binds to hypoacetylated histone tails of RAP1 (H3 and H4) and recruits SIR2 → changes to chromatin around histone 2
SIR
Form large complexes with telomeric DNA
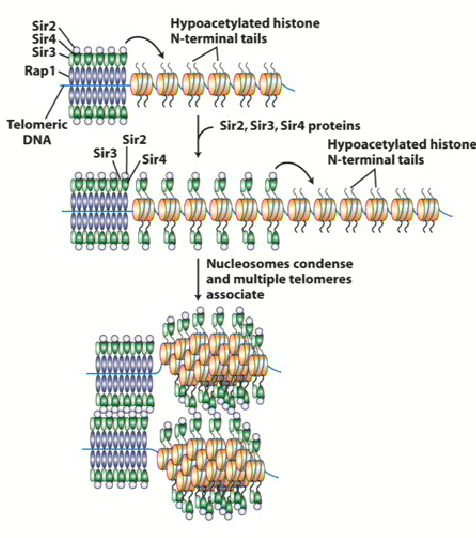
SIR2 & Histone Deacetylase
Histone tails are very positively charged due to increase in lysine residues = increase strongly with phosphate backbone in DNA
Removing acetyl groups from histone tails = neutralization = change in conformation in chromatin around DNA
This leads to spreading and the hypoacetylation of all histone tails around that region, blocking the ability for transcription factor or DNA methylase to interact with DNA
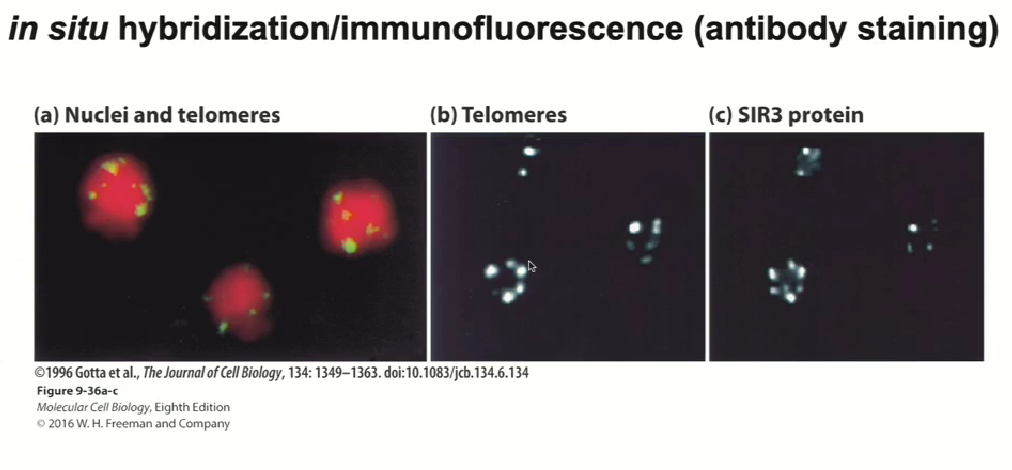
Immunofluorescence use to mark telomeres with nucleic acid probes
SIR3 is bound to telomeres, so silencing takes place at mating locus and telomeres
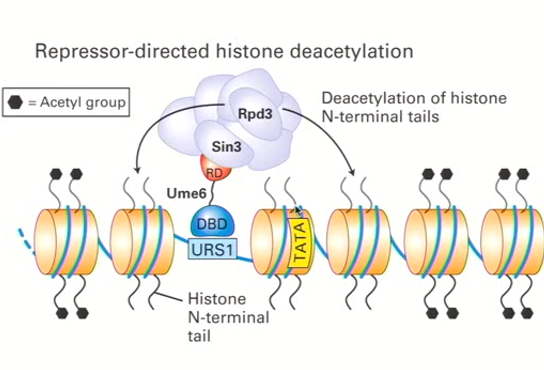
Histone Deacetylation Complexes (HDACs) function and how it works (Ume6 example)
Silencing specific sequences through transcriptional repressors
Positive charge of N-terminal histone tail interacts electrostatically with the DNA phosphate groups
Acetylation neutralizes the electrostatic interaction which makes the interactions less strong and permits complex formation (co-repressor complex)
For example, Ume6 (carries a repressor domain called RD) interacts with URS1 specifically and RD interacts with Sin3 and Rpd3
This ALTOGETHER forms a large protein complex which is important for the repression of a particular sequence by changing the conformation of the chromatin since it is now hypoacetylated (all acetyl groups are removed)
Chromatin therefore becomes more tightly compacted and it is no longer accessible to transcription factors
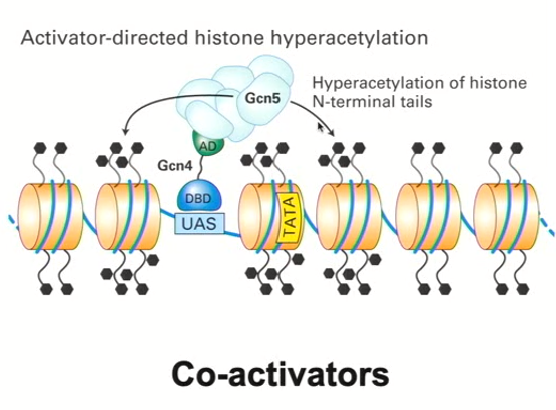
Similarity of DNA binding transcriptional activator similarities with histone deacetylation complexes (recruiting Histone Acetyl Transferases)
Gcn4 interacts with its upstream activation sequence
The activation domain mediates the interaction with another protein complex (coactivator) called Gcn5 which adds acetyl groups instead of removing them (histone acetyl transferases)
This changes the conformation of the histones and chromatin so that it becomes more open and has access to DNA transcription factors
What do activation domains do? What experiment was done?
Can trigger the decondensation of chromatin in order to activate genes
An experiment was done to show this used heterologous factors, which affect the configuration of the chromatin and examining how these heterologous factors can actually affect configuration of the chromatin in the cell
Researchers want to put in specific sequences that are not normally present in the organism
They introduced the lac operon DNA binding sites (where lac repressor will recognize)
They can now express the lac repressor which can be seen with an antibody tag
in diagram a, the chromatin is very compressed
They take the lac repressor and add on a transcriptional activation domain called VP16
in diagram b, the chromatin is decondensed and all over the cell → this proves the idea that just by putting a transcriptional activation domain on a DNA binding factor can change the configuration of chromatin
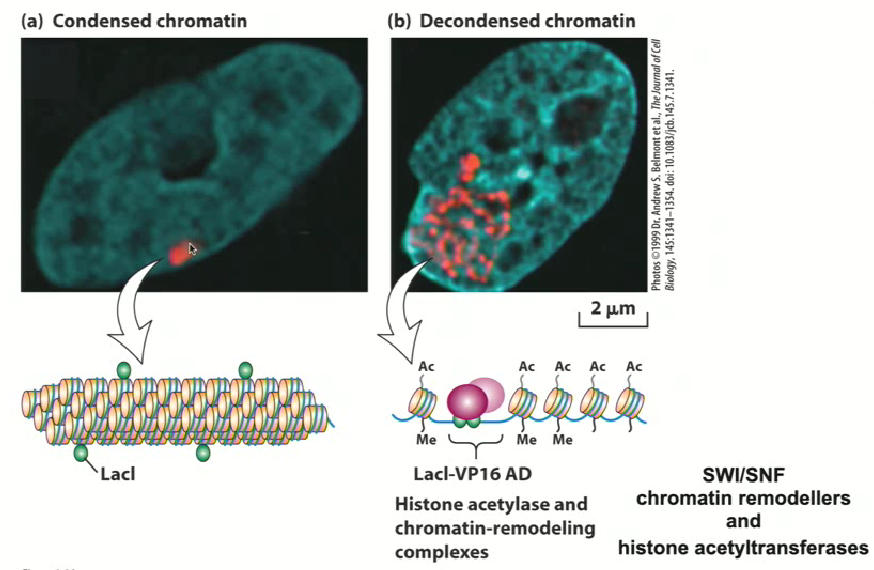
Histone acetyl transferase
it is recruited by activation domains
it adds on acetyl groups so that chromatin conformation is changed and becomes open
This allows for strong gene expression since chromatin remodelers (SWI and SNF, which are ATP dependent enzymes that push nucleosomes from one side to another) can change chromatin
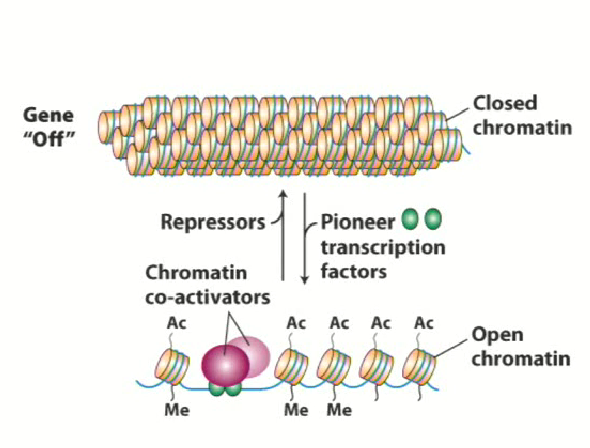
What are pioneer transcription factors?
Transcription factors that bind to DNA in the bound up form of chromatin in order to open it up
“Pave the way” to allow other transcription factors to work
Can interact with the outer portions of the nucleosome and bind to DNA to interact with histones to change the conformation of chromatin
Why are histone tails important?
Provide information on what type of gene expression has to take place on the chromatin
Histone tails of H2, H3 and H4 are important as various types of gene expression (code) can be assigned to either heterochromatin or euchromatin
This happens through post translational modifications
4 types of post-translational modifications
Methylation
Phosphorylation
Acetylation
Ubiquitination
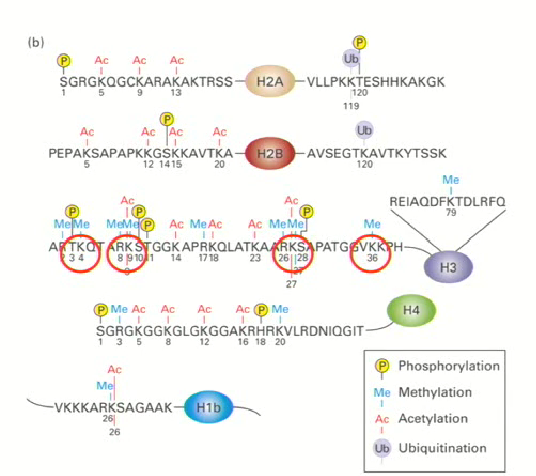
2 main methylations
Methylation of H3 on K4 → active (uncompacted)
Methylation of H3 on K9 → inactive (compact)
For this reason, you cannot generalize any mutations aside from acetylation. They are highly specific
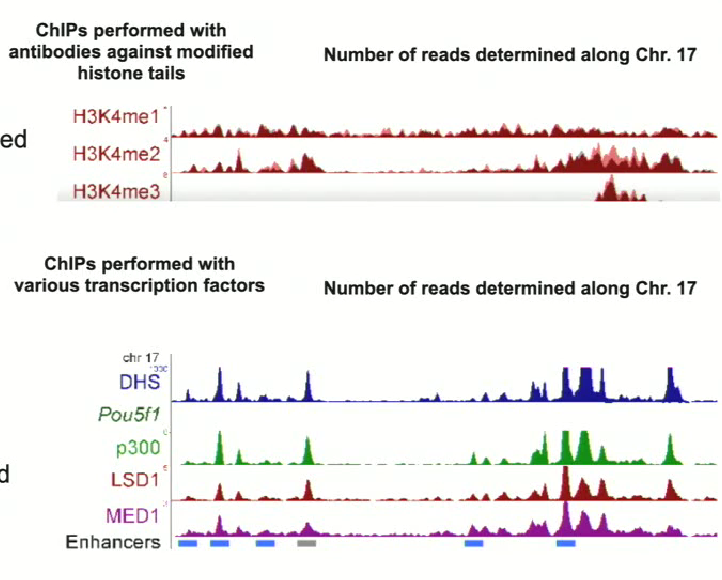
What technique can we use to figure out what regions of the genome have the H3 K4 methylation mark? How do we do it?
ChIP (Chromatin Immunoprecipitation)
We can get a genome wide representation of where these marks are occuring. By using reversible crosslinking agents, proteins bound to chromatin can be isolated using antibodies and the sequence of the bound DNA can be determined
1. Using known primers if you want to know whether a specific gene is affectedBy using NGS (next generation sequencing) the entire genome can be analyzed to determine what regions of the genome are being affected
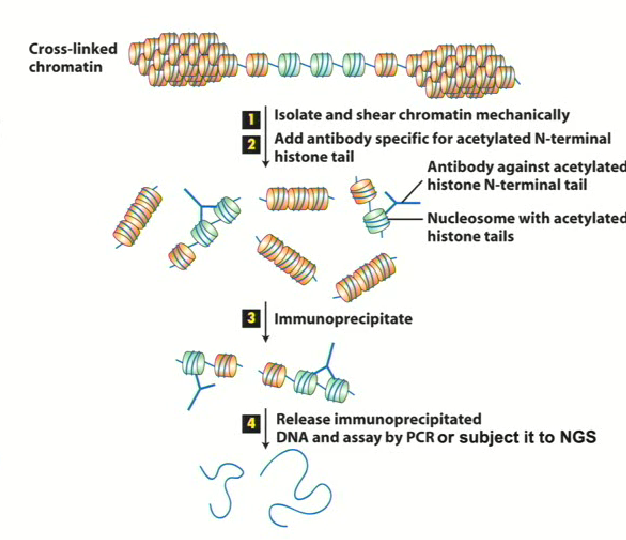
Example of using ChIP to detect H3K4 methylation marks
On chromosome 17
Using NGS, the entire genome can be surveyed to provide info on the genes (loci) affected by the marks
For the three cases pictured (mono, di and trimethylation), the pattern isn’t the same → suggested that the signal that they confer are different
ChIPs performed with antibodies against transcription factors help to determine enhancer positions and other key regulatory elements
This data for different antibodies can be combined with the data from the ChIP with histone tails
IN CONCLUSION: dimethylation and trimethylation have specific patterns that are associated with enhancers. Trimethylation is always associated with the transcriptional start site, where major changes in the promoter configuration occur (since RNA Pol II starts transcription here)
Imprints (DNA methylation)
example of epigenetic traits
DNA marks (with methylation) are read by specific proteins then used to modify histones through mSIn3 recruitment
What are epigenetic traits?
genes that are transmitted independently of the DNA sequence itself, but rather silenced or activated due to effects that happen on chromatin proteins
ex. all biological women have one x chromosome be inactive since they dont need two. This is done by histone methylation and heterochromatin spreading.
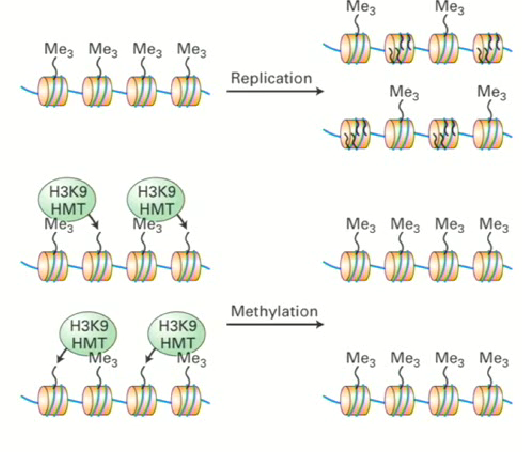
Propagation of epigenetic marks (readers and writers)
Cell has specific proteins that recognize histone tail modifications associated with activation and repression (epigenetic readers)
following cell division, they will recruit other proteins that will write the same mark/modification on the histones in the daughter cell (epigenetic writers)
How can epigenetic readers also be epigenetic writers?
A specific histone methyltransferase (HMT) recruits H3K9me3 to methylate neighbourhouring histones that have not been marked yet, and this cell information is conserved through different generations.