RNA translation into proteins and protein targeting
1/11
Earn XP
Description and Tags
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
12 Terms
Outline the basic process of translation
RNA acts as a template for the production of proteins in the cytoplasm
Ribosomes synthesise proteins from the RNA
Proteins are synthesised by the linking of amino acids to form peptides
What is the genetic code?
The genetic code is a set of rules that specifies the correspondence between the nucleotide sequence in DNA or RNA and the amino acids in proteins.
It represents the fundamental basis for the transmission of genetic information from DNA to RNA and, ultimately, to the synthesis of proteins during translation.
the genetic code is a triplet code: a three-nucleotide codon codes for one amino acid
codons are successive and non-overlapping
the genetic code is almost universal (between all the species, the codon bias may be different- as to which codon is used for the same amino acid)
there are 20 different amino acids and 64 different codons
the genetic code is degenerate- as some amino acids are represented by more than one codon
Methionine- AUG is the start codon (in RNA)
UAA, UAG, and UGA are the stop codons (in RNA)
What is the reading frame?
The reading frame is defined by the start codon.
There are three possible reading frames in a given RNA sequence, as codons are read in groups of three nucleotides.
The reading frame is determined by the starting point of the codon sequence.
Shifting the reading frame by one or two nucleotides alters the grouping of codons, potentially leading to a completely different amino acid sequence.
This concept is particularly important for understanding how the correct amino acid sequence is maintained during translation.
The reading frames are always read from the 5’ to the 3’ end
What are the 3 main types of RNA? What are their functions?
Messenger RNA (mRNA):
Function: Carries the genetic information from DNA to the ribosome during protein synthesis (translation). It serves as a template for the synthesis of a specific protein.
Characteristics: Typically single-stranded and carries the genetic code in the form of codons.
Transfer RNA (tRNA):
Function: Transfers amino acids to the ribosome during protein synthesis. Each tRNA molecule has an anticodon region that pairs with a specific mRNA codon, and it carries the corresponding amino acid (the attachment of the amino acid is catalysed by aminoacyl tRNA synthase)
Characteristics: Folded into a cloverleaf structure with three loops, one of which contains the anticodon.
Ribosomal RNA (rRNA):
Function: A structural and catalytic component of ribosomes, the cellular machinery responsible for protein synthesis. It helps in the alignment and bonding of tRNA and mRNA during translation.
Characteristics: Forms the core structure of ribosomes and is composed of both protein and RNA.
What is aminoacyl RNA transfer synthase?
Aminoacyl-tRNA synthases are enzymes responsible for attaching the correct amino acid to their corresponding tRNA molecules.
The accurate pairing of amino acids with tRNAs is essential for the faithful translation of the genetic code.
Error checking:
Specificity of Aminoacyl-tRNA Synthases:
Each aminoacyl-tRNA synthase is highly specific for its cognate amino acid and tRNA.
These enzymes recognise specific structural features of both the amino acid and the tRNA to ensure accurate pairing.
Editing Domains:
Some aminoacyl-tRNA synthases have editing domains that function to proofread and correct errors in amino acid attachment. T
he editing domain recognises mis-charged tRNAs and removes the incorrect amino acid, preventing it from being incorporated into the growing polypeptide chain.
Second Binding Site:
Aminoacyl-tRNA synthases often have a second, separate binding site called the editing site.
This site provides an additional opportunity for the enzyme to detect errors.
If a mis-charged tRNA binds to the editing site, the incorrect amino acid can be hydrolysed and released.
Discrimination Against Similar Amino Acids:
Aminoacyl-tRNA synthases are designed to discriminate between similar amino acids.
Even amino acids with similar chemical structures are often recognised differently by these enzymes, ensuring the correct amino acid is attached to the appropriate tRNA.
Proofreading by tRNA:
tRNA molecules themselves contribute to error checking.
The structure of tRNA includes regions that can form specific base pairs with the correct amino acid and reject incorrect amino acids.
This "identity element" within the tRNA helps ensure the accuracy of amino acid selection.
Cellular Quality Control Mechanisms:
Cellular quality control mechanisms monitor and regulate the accuracy of protein synthesis.
For example, the ribosome has a peptidyl transferase centre that can detect mis-paired amino acids and trigger the release of the incorrect amino acid.
Can a single tRNA recognise more than one codon?
Yes, a single tRNA can recognise more than one codon.
Once the first two bases are paired, there is wobble at the 3rd position, allowing for that base to pair with a purine or a pyrimidine as appropriate.
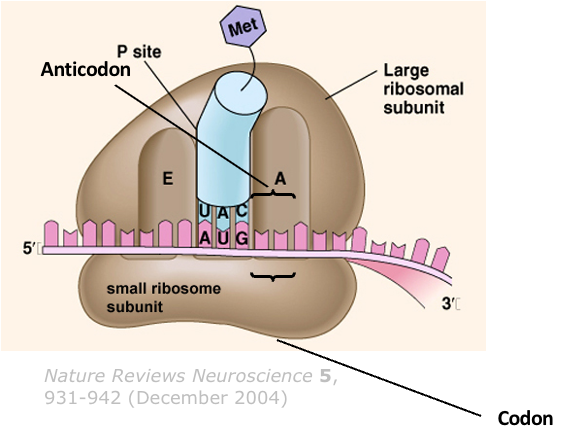
Give details about the initiation stage of translation
The small subunit of a ribosome binds to the AUG (start codon)
A specific initiator transfer RNA (tRNAi Met) carrying a methionine binds to the AUG start codon
The large 60S subunit of a ribosome binds to this complex such that the tRNA occupies the P site
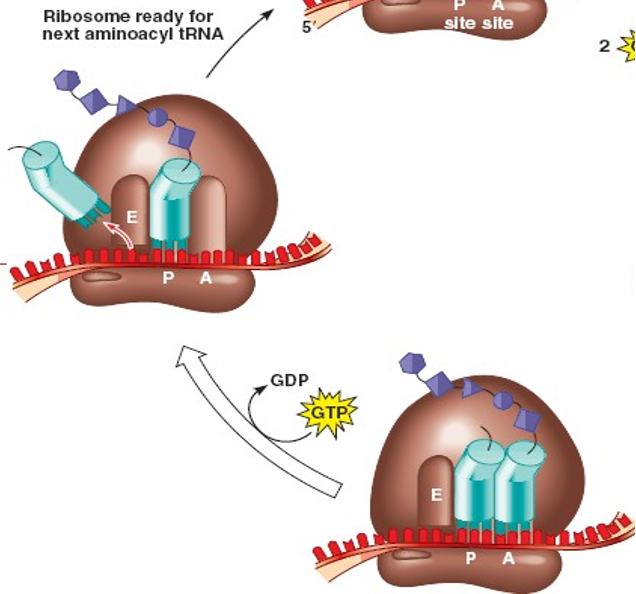
Give details about the elongation stage of translation
A tRNA with the complementary anti-codon binds to the mRNA, occupying the A site in the ribosome
A peptide bond forms between the two amino acids via a condensation reaction
The bond between the amino acid and the first tRNA is cleaved
The ribosome moves one codon further along the mRNA (translocation) in 5’ to 3’ direction
The empty tRNA moves from the P site to the E site
The A site is empty and ready for another tRNA
The first, now uncharged tRNA is released from the E site
This process requires energy which we get from the breakdown of GTP into GDP (a phosphate is released during the exothermic reaction)
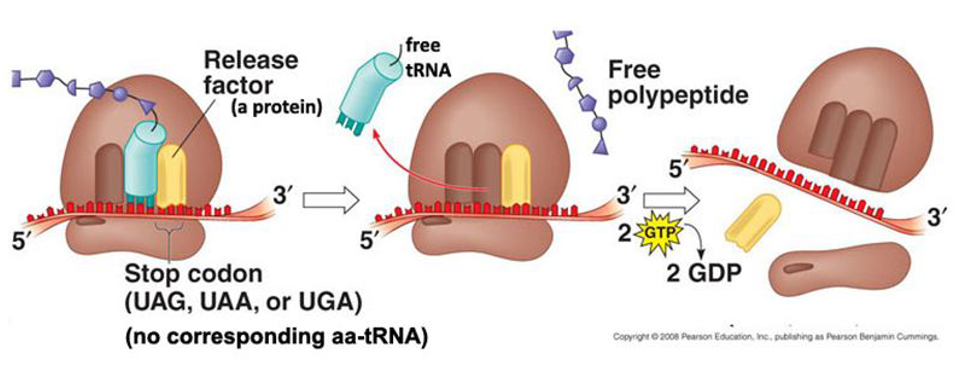
Give details about the termination stage of translation
The ribosome reaches a stop codon (UAG, UAA or UGA)
The polypeptide is released from the ribosome
The tRNA is released from the ribosome
The ribosomal subunits dissociate from the mRNA
What is protein targeting?
Protein targeting, also known as protein targeting or protein localisation, refers to the process by which proteins are directed to specific cellular compartments, organelles, or extracellular spaces within a cell.
Proper protein targeting is crucial for the functioning and regulation of various cellular processes.
Cells have intricate mechanisms to ensure that proteins are delivered to their designated locations, where they can perform their intended functions.
Eukaryotic cell is made multi compartments
Proteins can be targeted using a ‘unique address’
Mitochondrial and nuclear proteins targeted to correct compartment
Membrane and secreted proteins inserted into the ER:
the signal sequence (only 4 amino acids long) on the protein will bind to the signal recognition particle (SRP)
the SRP will bind to a transmembrane protein in the membrane of the ER
this will direct the protein to a channel protein on the membrane
the newly processed protein can then be translocated into the lumen of the ER
the signal is then cleaved off, and the protein can then be correctly folded within the ER (the SRP is recycled)
Give details on bacterial and drug interference
Transcription and translation are different in prokaryotes
Different RNA Pol
Different ribosome structure
Therefore these are used for targets for antibiotics- the protein synthesis in bacteria can be inhibited
Conversely it is why some drugs work on eukaryotes and not bacteria
Give details on how anti-virals can be developed
Viruses are unable to replicated without taking over the host machinery– so they have developed mechanisms to allow this to occur.
Aim of antivirals is to stop host gene expression and to hijack the host translation initiation
This is complex and will depend on whether the viruses are DNA or RNA