Bio 230
0.0(0)
Card Sorting
1/109
Last updated 9:51 PM on 5/28/23
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
110 Terms
1
New cards
Prokaryotic organism
* Eubacteria and archaea (which live in more extreme environments)
* single-celled (but not only this group are single-celled the other is too)
* lack nucleus and organelles
* single-celled (but not only this group are single-celled the other is too)
* lack nucleus and organelles
2
New cards
Eukaryotic organisms
* plants, fungi, animal, humans
* single-celled or multicellular
* have nuclei and organelles
* much larger
* typically much large genomes
* single-celled or multicellular
* have nuclei and organelles
* much larger
* typically much large genomes
3
New cards
Cell wall
Tough protective outercoat, optional not in all prokaryotes
4
New cards
Microbiota (singular microbiome)
* the assembly of living microorganisms present in a defined environment (for example our body)
Example:
* residue on skin, lungs, mouth, GI tract
* microbial cell: Human cell ratio is at least 1:1
* up to 200 times more microbial genes than humans in the body
* an essential role in human health (digestion, immune system, etc.)
Example:
* residue on skin, lungs, mouth, GI tract
* microbial cell: Human cell ratio is at least 1:1
* up to 200 times more microbial genes than humans in the body
* an essential role in human health (digestion, immune system, etc.)
5
New cards
Genomes
* all known life forms possess one
* encodes the info to construct and maintain an organism
* most are made of DNA
* except some viruses have ones made of RNA (viruses are not living things (not made up of cells) but they have genetic material either RNA or DNA genomes
* encodes the info to construct and maintain an organism
* most are made of DNA
* except some viruses have ones made of RNA (viruses are not living things (not made up of cells) but they have genetic material either RNA or DNA genomes
6
New cards
Transcriptome
The repertoire of RNA molecules present in a cell at a particular time
7
New cards
DNA microarray
* rows are the genes and the columns are the samples
* red - a lot of RNA present for this gene
* green - very little
* black - a moderate amount
* red - a lot of RNA present for this gene
* green - very little
* black - a moderate amount
8
New cards
Proteome
* collection of all the proteins in a cell
* defines the biochemical functions of the cell
* defines the biochemical functions of the cell
9
New cards
2D gel electrophoresis
* y axis is the molecular weight
* and the x axis is the isoelectric point (acidic or basic) in the same spot = shared protein
* and the x axis is the isoelectric point (acidic or basic) in the same spot = shared protein
10
New cards
Central dogma
Genome (DNA) → transcriptome → proteome
11
New cards
RNA polymerase
* transcribes DNA
* adds on to the 3’ end
* there’s a DNA RNA hybrid (helix)
* goes 5’ → 3’
* there’s an active site Mg2+
* takes in a ribonucleoside triphosphate uptake channel
* adds on to the 3’ end
* there’s a DNA RNA hybrid (helix)
* goes 5’ → 3’
* there’s an active site Mg2+
* takes in a ribonucleoside triphosphate uptake channel

12
New cards
Transcription (prokaryotes)
* the promoter positions the RNA polymerase and indicates the transcription start site
* 1 part of the sigma factor binds to the early sequence while the other binds to the later and stabilizes the transcription bubble
* holoenzyme = RNA + pol and sigma factor
* RNA associates with the active site
* RNA pol unwinds DNA
* transcription begins at the active site after the promoter
* sigma factor is released once \~10 nucleotides synthesized
* transcription elongation (transcription is now going faster)
* enzyme reaches a terminator sequence where it halts and releases both the RNA and the template
* 1 part of the sigma factor binds to the early sequence while the other binds to the later and stabilizes the transcription bubble
* holoenzyme = RNA + pol and sigma factor
* RNA associates with the active site
* RNA pol unwinds DNA
* transcription begins at the active site after the promoter
* sigma factor is released once \~10 nucleotides synthesized
* transcription elongation (transcription is now going faster)
* enzyme reaches a terminator sequence where it halts and releases both the RNA and the template
13
New cards
E. coli
* unicellular prok
* one chromosome of circular DNA
* encodes about 4300 proteins
* many genes are transcriptionally regulated by food availability
Prokaryotic feature:
* multiple genes can be transcribed into a single RNA molecule
* this system is called an operon
* one chromosome of circular DNA
* encodes about 4300 proteins
* many genes are transcriptionally regulated by food availability
Prokaryotic feature:
* multiple genes can be transcribed into a single RNA molecule
* this system is called an operon
14
New cards
operon
multiple genes can be transcribed into a single RNA molecule
15
New cards
Tryptophan (Trp) operon and how it operates
* contains 5 genes encoding enzymes for trp biosynthesis (forms one long mRNA molecule)
* transcription regulated by a single promoter
* when bound by RNA pol → trp gene expression ON
* bound by a tryptophan repressor protein → Trp gene expression OFF
* the tryptophan repressor binds a specific DNA sequence of the promoter called an operator (aka cis-regulatory sequence)
* the repressor blocks promoter access so RNA pol can’t bind (negatively regulates Trp expression) BUT the repressor must bind 2 molecules of trp to bind to DNA)
* therefore the repressor and operator provide a simple switch to control trp synthesis based on trp availability
* transcription regulated by a single promoter
* when bound by RNA pol → trp gene expression ON
* bound by a tryptophan repressor protein → Trp gene expression OFF
* the tryptophan repressor binds a specific DNA sequence of the promoter called an operator (aka cis-regulatory sequence)
* the repressor blocks promoter access so RNA pol can’t bind (negatively regulates Trp expression) BUT the repressor must bind 2 molecules of trp to bind to DNA)
* therefore the repressor and operator provide a simple switch to control trp synthesis based on trp availability
16
New cards
The structure of the Tryptophan Repressor
* Tryptophan repressor contains a helix-turn helix DNA binding motif
* Binds in the major groove of the DNA double helix
* Trp binding induces a conformational change and the protein fits into the major groove
* Binds in the major groove of the DNA double helix
* Trp binding induces a conformational change and the protein fits into the major groove
17
New cards
The Lac Operon function
* 3 genes are required for the transport of lactose into the catabolism
* enables the use of lactose in the absence of glucose
Dual regulation: both positive and negative control
* 1) Activator (is needed because pol binding is inefficient to the operon): catabolite activator protein (CAP) (which helps pol bind as it has a helix-turn-helix DNA binding domain) promotes Lac expression: low glucose/ high lactose
* decreasing glucose levels increases the levels of the signal molecule cyclic AMP (cAMP)
* cAMP binds CAP protein:
* conformation change and increases binding activity and it can bind to CAP site and it can recruit the RNA pol
* 2) Repressor: Lac repressor protein inhibits Lac expression: low lactose and lac operon gene expression is off and increased lactose removes the repressor as it increases allolactose; requires b-galactosidase
* the allolactose binds to the Lac repressor (causes a change in shape and decreases binding activity and releases from the operator)
* Note: 1st choice is to use glucose, when there’s low glucose and high lactose it will use lactose (must be both conditions) and turn on the Lac operon.
* enables the use of lactose in the absence of glucose
Dual regulation: both positive and negative control
* 1) Activator (is needed because pol binding is inefficient to the operon): catabolite activator protein (CAP) (which helps pol bind as it has a helix-turn-helix DNA binding domain) promotes Lac expression: low glucose/ high lactose
* decreasing glucose levels increases the levels of the signal molecule cyclic AMP (cAMP)
* cAMP binds CAP protein:
* conformation change and increases binding activity and it can bind to CAP site and it can recruit the RNA pol
* 2) Repressor: Lac repressor protein inhibits Lac expression: low lactose and lac operon gene expression is off and increased lactose removes the repressor as it increases allolactose; requires b-galactosidase
* the allolactose binds to the Lac repressor (causes a change in shape and decreases binding activity and releases from the operator)
* Note: 1st choice is to use glucose, when there’s low glucose and high lactose it will use lactose (must be both conditions) and turn on the Lac operon.
18
New cards
The lac operon schematic
there’s a sequence for CAP binding and one where the lac repressor binds
* the 1st gene of Lac operon encodes beta-galactosidase; breaks down lactose to glucose and galactose
* the 1st gene of Lac operon encodes beta-galactosidase; breaks down lactose to glucose and galactose
19
New cards
NtrC protein (prok)
* transcriptional activator
* DNA looping allows it directly interact with RNA pol to activate transcription from a distance
* DNA looping allows it directly interact with RNA pol to activate transcription from a distance
20
New cards
Bacteriophage Lambda
* A virus that infects bacterial cells and has a - and + regulatory mechanism that works together to regulate lifestyle
* there’s the prophage pathway where the RNA is incorporated into the hosts DNA and there’s rapid cloning. here the lambda repressor occupies the operator and blocks the synthesis of Cro and it activates its own synthesis and most of the viruses’ DNA is not transcribed (the viral proteins) (positive feedback)
* there’s a lytic pathway where there’s damage (inactivates repressor) to the cell and the RNA induces out of the DNA and there are viral proteins made to make new viruses and find new hosts.
* The Cro occupies the operator
* blocks the synthesis of the lambda repressor and allows its own synthesis
* viral genes are transcribed
* DNA is replicated, and packaged, and new bacteriophage released by
* there’s the prophage pathway where the RNA is incorporated into the hosts DNA and there’s rapid cloning. here the lambda repressor occupies the operator and blocks the synthesis of Cro and it activates its own synthesis and most of the viruses’ DNA is not transcribed (the viral proteins) (positive feedback)
* there’s a lytic pathway where there’s damage (inactivates repressor) to the cell and the RNA induces out of the DNA and there are viral proteins made to make new viruses and find new hosts.
* The Cro occupies the operator
* blocks the synthesis of the lambda repressor and allows its own synthesis
* viral genes are transcribed
* DNA is replicated, and packaged, and new bacteriophage released by

21
New cards
Feed forward loops
* both a and b are required for the transcription of z
* Brief input b does not accumulate → z not transcribed
* prolonged input B accumulates→ z is transcribed
* Brief input b does not accumulate → z not transcribed
* prolonged input B accumulates→ z is transcribed
22
New cards
Synthetic biology
Scientists can construct artificial circuits and examine their behaviour in cells.
23
New cards
The repressilator
a created gene oscillator using a delayed negative feedback circuit, over time it is amplified

24
New cards
Circadian gene regulation
* the TIM gene is degraded during the day (like a light sensor) and is then synthesized during the day) and form a heterodimer with the per gene and is then phosphorylated to repress per
* the heterodimer degrades and per is active again and wakes up
* the heterodimer degrades and per is active again and wakes up
25
New cards
Transcriptional attenuation
* in both proks and euks there can be a premature termination of transcription
* RNA adapts a structure that interferes with RNA pol
* regulatory proteins can bind to RNA and interfere
* prok, plants and some fungi also use riboswitches to regulate gene expression (not always the same as this)
* RNA adapts a structure that interferes with RNA pol
* regulatory proteins can bind to RNA and interfere
* prok, plants and some fungi also use riboswitches to regulate gene expression (not always the same as this)
26
New cards
Riboswitches
* Short RNA sequences that change conformation when bound by a small molecule
* example: purine biosynthesis
* low guanine levels → transcription of purine biosynthetic gene is on
* high guanine levels → guanine binds, conformation change, RNA pol terminates transcription gene is off
* example: purine biosynthesis
* low guanine levels → transcription of purine biosynthetic gene is on
* high guanine levels → guanine binds, conformation change, RNA pol terminates transcription gene is off
27
New cards
Eukaryotic Transcription overview
* RNA pol II transcribes protein-coding genes
* requires 5 TF’s: TFIID, TFIIB, TFIIF, TFIIE, and TFIIH and transcriptional activation required many gene regulatory proteins
* euk genomes lack operons and their DNA is packed into chromatin which allows for more regulation
* the mediator acts as an intermediate btw regulatory proteins and RNA pol
* thus expression can be controlled by many different activators and repressors and some repressors can act over very large distances (ie. DNA looping)
* there are also corepressor and coactivators (don’t bind directly to DNA) but on regulatory proteins
* requires 5 TF’s: TFIID, TFIIB, TFIIF, TFIIE, and TFIIH and transcriptional activation required many gene regulatory proteins
* euk genomes lack operons and their DNA is packed into chromatin which allows for more regulation
* the mediator acts as an intermediate btw regulatory proteins and RNA pol
* thus expression can be controlled by many different activators and repressors and some repressors can act over very large distances (ie. DNA looping)
* there are also corepressor and coactivators (don’t bind directly to DNA) but on regulatory proteins
28
New cards
EUK transcription steps
1. general transcription factor = TBP (tata binding protein) and the TFIID (transcription factor for DNA polymerase which binds to the TATA box that is recognized by TBP causing a distortion of DNA @ TATA
2. TFIIF binds to the RNA pol and with a mediator the TFIIH and TFIIE binds to RNA pol
3. TFIIH hydrolyzes ATP pulling apart the DNA strands at the start site, exposes the template strand
29
New cards
Activator proteins
* recognizes specific DNA seq → DNA binding domain (DBD)
* Activation domain → accelerates freq/rate of transcription
* can mix and match DBDs and ADS
* Attract, position and modify
* general TFs
* mediator
* RNA pol2
* can do this directly (by acting on these components) or indirectly (modifying chromatin structure)
1. Activator proteins can bind directly to transcriptional machinery or other mediator and attract them to proteins
2. Activator proteins can alter chromatin structure via nucleosomes to increase promoter accessibility
* Activation domain → accelerates freq/rate of transcription
* can mix and match DBDs and ADS
* Attract, position and modify
* general TFs
* mediator
* RNA pol2
* can do this directly (by acting on these components) or indirectly (modifying chromatin structure)
1. Activator proteins can bind directly to transcriptional machinery or other mediator and attract them to proteins
2. Activator proteins can alter chromatin structure via nucleosomes to increase promoter accessibility
30
New cards
Nucleosomes
* basic structure of euk chromatin
* DNA winds around a histone octamer (DNA is about 147 bps)
* (H2A, H2B, H3, and H4) X 2 and they’re connect by linker DNA (10 - 80 bps)
* DNA winds around a histone octamer (DNA is about 147 bps)
* (H2A, H2B, H3, and H4) X 2 and they’re connect by linker DNA (10 - 80 bps)

31
New cards
Activator proteins and Nucleosome Sliding
* ATP dependent chromatin remodeling complex moves DNA and winds it tighter around the octamer
32
New cards
Activators modifying chromatin with Histone chaperones
* a histone chaperone can replace the H2A-H2B dimer and modify it accordingly
* the histone chaperone helps and replace the whole octamer
* the histone chaperone helps and replace the whole octamer

33
New cards
histone modifications
* enzymes produce specific patterns of histone modifications. These modifications are a “histone code”
* addition of phosphate group: phosphorylation (with kinase)
* addition of acetyl group: acetylation (acetyltransferase)
* addition of methyl group: methylation (methyltransferase)
* These modifications occur on histone tails
\
* addition of phosphate group: phosphorylation (with kinase)
* addition of acetyl group: acetylation (acetyltransferase)
* addition of methyl group: methylation (methyltransferase)
* These modifications occur on histone tails
\
34
New cards
Histone Code
* specific modifications to histone tails by histone modifying enzymes called writers
* reader proteins can recognize specific modifications and provide meaning
* reader proteins can recognize specific modifications and provide meaning
35
New cards
Human interferon gene regulation as a histone code
1. activator protein binds to chromatin and attracts a histone acetyltransferase (HAT)
2. HA acetylates lysine 9 of histone H3 and lysine 8 of histone H4
3. activator proteins attracts a histone kinase (HK)
4. HK phosphorylates serine 10 of histone H3. can only occur after acetylation of lysine 9.
5. serine modification signals HAT to acetylate lys 14 of H3
6. TFIID and a chromatin remodeling complex binds to modified histone tails and initiate transcription
36
New cards
Repressor proteins ( euks)
* rarely compete with RNA pol for DNA access
1. competitive DNA binding (it covers part of the site for the activator)
2. masking the activation surface (stops the activation domain)
3. direct interaction with the general transcription factors (loops and “kisses” the general transcription factor and inhibits it)
4. the recruitment of chromatin remodeling complexes (makes histones more compact)
5. recruitment of HAT and methyl transferase to write a code to stop transcription)
1. competitive DNA binding (it covers part of the site for the activator)
2. masking the activation surface (stops the activation domain)
3. direct interaction with the general transcription factors (loops and “kisses” the general transcription factor and inhibits it)
4. the recruitment of chromatin remodeling complexes (makes histones more compact)
5. recruitment of HAT and methyl transferase to write a code to stop transcription)
37
New cards
Stopping transcription using both writers and readers
* the writer writes a code and attracts a reader which attracts another writer and this continues and makes a complex that makes the chromatin more compact
* spreads the histone code along chromatin carried out by reader-writer complexes
* DNA methylase enzyme is attracted by the reader and methylates nearby cytosines DNA
* DNA methyl-binding proteins bind methyl groups and stabilize the structure
* methylation and therefore gene expression patterns can be inherited
* a process called epigenetic inheritance
* spreads the histone code along chromatin carried out by reader-writer complexes
* DNA methylase enzyme is attracted by the reader and methylates nearby cytosines DNA
* DNA methyl-binding proteins bind methyl groups and stabilize the structure
* methylation and therefore gene expression patterns can be inherited
* a process called epigenetic inheritance

38
New cards
RNA capping
addition of modified guanine nucleotide to the 5’ end of pre-mRNA (3 enzymes involved). The cap is bound by cap-binding (CBC)
functions:
1. helps in RNA processing and export from the nucleus
2. important role in translation of mRNAs in the cytosol
3. protects mRNA from degradation
functions:
1. helps in RNA processing and export from the nucleus
2. important role in translation of mRNAs in the cytosol
3. protects mRNA from degradation
39
New cards
RNA splicing
* the removal of introns from the RNA
* carried out by an enzyme complex of RNA and proteins termed the splicesome
* sites of proper splicing are the then bound exon junction complexes (EJCs) (serve as a marker)
* carried out by an enzyme complex of RNA and proteins termed the splicesome
* sites of proper splicing are the then bound exon junction complexes (EJCs) (serve as a marker)
40
New cards
Alternative splicing regulation

41
New cards
Drosophila sex determination
* ratio of X chromosomes: autosomal chromosomes
* X:A 0.5 Male (default)
* X:A 1.0 Female
* 3 genes involved: sex lethal: splicing repressor, Transformer: splicing activator, doublesex: regulates sex gene expression all 3 have regulated splice sites
* Male:
* the sex lethal site is not occurring and the splice product is nonfunctional
* then the transformer without the sex lethal is able to be regulated so the product is also nonfunctional
* finally the splice site is regulated for the Doublesex gene and it produces the Dsx protein which represses femal gene expression
* Female:
* a special Sxl is produced that is functional when spliced out (transient) blocks the splice site in the Sxl gene
* this produces a functional Sxl protein which again represses the site of the sxl and the Tra
* a function Tra protein is formed and activates the splicing of the DsX and a functional Dsx is fored and represses male gene expression
* X:A 0.5 Male (default)
* X:A 1.0 Female
* 3 genes involved: sex lethal: splicing repressor, Transformer: splicing activator, doublesex: regulates sex gene expression all 3 have regulated splice sites
* Male:
* the sex lethal site is not occurring and the splice product is nonfunctional
* then the transformer without the sex lethal is able to be regulated so the product is also nonfunctional
* finally the splice site is regulated for the Doublesex gene and it produces the Dsx protein which represses femal gene expression
* Female:
* a special Sxl is produced that is functional when spliced out (transient) blocks the splice site in the Sxl gene
* this produces a functional Sxl protein which again represses the site of the sxl and the Tra
* a function Tra protein is formed and activates the splicing of the DsX and a functional Dsx is fored and represses male gene expression

42
New cards
3’ polyadenylation
* more complex than transcription termination in prok
* signals encoded in genome
* RNA pol transfers protein complexes to RNA
* CstF (cleavages stimulating factor)
* CPSF (cleavage and polyadenylation specificity factor)
* RNA is cleaved
* transcription terminates
* Poly-A polymerase (PAP) adds \~200 A nucleotides to the 3’ end of RNA from ATP
* not genome encoded
* poly- A tail is bound by poly-A binding proteins
* aid in…..
* RNA export
* translation
* mRNA stability
* signals encoded in genome
* RNA pol transfers protein complexes to RNA
* CstF (cleavages stimulating factor)
* CPSF (cleavage and polyadenylation specificity factor)
* RNA is cleaved
* transcription terminates
* Poly-A polymerase (PAP) adds \~200 A nucleotides to the 3’ end of RNA from ATP
* not genome encoded
* poly- A tail is bound by poly-A binding proteins
* aid in…..
* RNA export
* translation
* mRNA stability
43
New cards
Coupling transcription and RNA processing
* during transcription elongation, the C-terminal domain of RNA pol binds RNA processing proteins and transfers them to RNA at the appropiate time
* the binding of RNA processing proteins is regulated by phosphorylation of RNA polymerase
* the binding of RNA processing proteins is regulated by phosphorylation of RNA polymerase
44
New cards
RNA transport out of the nucleus
The cell selectively transports mature mRNA from the nucleus
\
markers of mature mRNA must be acquired for export:
* cap binding complex (CBC)
* exon junction complexes (EJC)
* poly-A-binding proteins
These proteins travel with the mRNA to the cytosol
\
markers of immature mRNA must be lost for export
* proteins involved in RNA splicing (snRNPs)
improperly processed mRNAs will eventually by degraded in the nucleus by the exosome
\
markers of mature mRNA must be acquired for export:
* cap binding complex (CBC)
* exon junction complexes (EJC)
* poly-A-binding proteins
These proteins travel with the mRNA to the cytosol
\
markers of immature mRNA must be lost for export
* proteins involved in RNA splicing (snRNPs)
improperly processed mRNAs will eventually by degraded in the nucleus by the exosome
45
New cards
translation
* mRNA message is decoded in ribosomes made up of >50 different proteins and several RNA molecules
* AA’s are added to the c-terminal end of the growing polypeptide chain → therefore, synthesized from N- to C- terminus
* AA’s are added to the c-terminal end of the growing polypeptide chain → therefore, synthesized from N- to C- terminus
46
New cards
Translation initiation
* translation initiation machinery recognizes the 5’-cap and poly-A tail:
* euk initiation factors (eIFs)
* 5’ cap bound by eIF4E
* poly-A binding protein bound by eIF4G
* Recruit small ribosomal complex which will initiate translation at first AUG downstream of 5’ cap
* ensures that both ends of mRNA are intact
* EJC’s also stimulate translation which ensures proper slicing
* euk initiation factors (eIFs)
* 5’ cap bound by eIF4E
* poly-A binding protein bound by eIF4G
* Recruit small ribosomal complex which will initiate translation at first AUG downstream of 5’ cap
* ensures that both ends of mRNA are intact
* EJC’s also stimulate translation which ensures proper slicing
47
New cards
Nonsense-mediated mRNA decay
* prominent mRNA surveillance system
* surveys for nonsense (STOP) codons in the wrong place
* indicator of improper splicing
* Normal splicing:
* the ribosome binds mRNA as it emerges from the nuclear pore
* EJCs are displaced by the moving ribosome
* the stop codon is in the last exon
* no EJCs remain bound when the ribosome reaches the stop codon
* mRNA is released in the cytosol
* Abnormal splicing:
* there’s a premature stop codon so EJCs remain on the mRNA when the ribosomes reach the stop codon
* mRNA is degraded (mediated by Upf (binds to the EJC))
* surveys for nonsense (STOP) codons in the wrong place
* indicator of improper splicing
* Normal splicing:
* the ribosome binds mRNA as it emerges from the nuclear pore
* EJCs are displaced by the moving ribosome
* the stop codon is in the last exon
* no EJCs remain bound when the ribosome reaches the stop codon
* mRNA is released in the cytosol
* Abnormal splicing:
* there’s a premature stop codon so EJCs remain on the mRNA when the ribosomes reach the stop codon
* mRNA is degraded (mediated by Upf (binds to the EJC))
48
New cards
mRNA quality control in prok
* ribosomes stall on broken or incomplete mRNAs and do not release
* a special RNA tmRNA is recruited to the A site
* carries an alanine amino acid
* acts as both tRNA and mRNA
* broken mRNA is released
* alanine is added onto the polypeptide from the tmRNA, which acts like a tRNA but with no anticodon-codon binding
* the ribosome translates 10 codons from the tmRNA, which now acts as an mRNA
* the 11 AA tag is recognized by proteases that degrade the entire protein
* a special RNA tmRNA is recruited to the A site
* carries an alanine amino acid
* acts as both tRNA and mRNA
* broken mRNA is released
* alanine is added onto the polypeptide from the tmRNA, which acts like a tRNA but with no anticodon-codon binding
* the ribosome translates 10 codons from the tmRNA, which now acts as an mRNA
* the 11 AA tag is recognized by proteases that degrade the entire protein
49
New cards
mRNA stability with poly-A-tail
* in proks, exonucleases rapidly degrade most mRNAs in euks, mRNAs are more stable and degradation is regulated
* there are two main mechanisms: involve poly-a tail shortening
* processes carried out by an exonuclease (deadenylase) when mRNA reaches the cytoplasm → acts as a timer of mRNA lifetime
* once the tail reaches a critical length
* there’s a decapping followed by degradation
* no poly-A tail results in degradation
* both mechanisms can occur on the same mRNA
* cytoplasmic poly-A elongation can also occur to stabilize mRNA
* proteins can also interfere poly-A shortening
* there are two main mechanisms: involve poly-a tail shortening
* processes carried out by an exonuclease (deadenylase) when mRNA reaches the cytoplasm → acts as a timer of mRNA lifetime
* once the tail reaches a critical length
* there’s a decapping followed by degradation
* no poly-A tail results in degradation
* both mechanisms can occur on the same mRNA
* cytoplasmic poly-A elongation can also occur to stabilize mRNA
* proteins can also interfere poly-A shortening
50
New cards
Transferrin
another example of protein-regulated mRNA stability
* imports iron into the cell
* needed when cellular iron is low
* mRNA stabilized by cytosolic aconitase
* binds 3’ UTR
* aconitase binds iron and undergoes a conformational change
* mRNA released
* exposes 3’UTR endonucleolytic cleavage site (polyA removed; mRMA is degradedd
* imports iron into the cell
* needed when cellular iron is low
* mRNA stabilized by cytosolic aconitase
* binds 3’ UTR
* aconitase binds iron and undergoes a conformational change
* mRNA released
* exposes 3’UTR endonucleolytic cleavage site (polyA removed; mRMA is degradedd

51
New cards
Deadenylase
* shortens the poly-A tail and binds to the 5’ cap like eIF’s

52
New cards
miRNAs
* non-coding RNAs
* base-pair with specific mRNAs
* synthesized by RNA pol II and get a 5’ cap and poly A tail
* after special processing (it folds and forms a ladder association and is then cleaved of the cap and poly-A tail and the dicers (while In cytosol) further cleave to form two strands (one of which is degraded) it associates with a protein complex called an RNA-induced silencing complex (RISC)
* RISC seeks mRNa with complementary nucleotide seq
* a protein of RISC called argonaute plays a critical role in base-pairing with it
* two possibilities:
* with an extensive match the RISC hydrolyzes ATP and cleaves the mRNA which causes degradation
* less extensive match: blocks the ribosome which results in rapid translational repression, deadenylation and in most cases eventual degradation of mRNA
* base-pair with specific mRNAs
* synthesized by RNA pol II and get a 5’ cap and poly A tail
* after special processing (it folds and forms a ladder association and is then cleaved of the cap and poly-A tail and the dicers (while In cytosol) further cleave to form two strands (one of which is degraded) it associates with a protein complex called an RNA-induced silencing complex (RISC)
* RISC seeks mRNa with complementary nucleotide seq
* a protein of RISC called argonaute plays a critical role in base-pairing with it
* two possibilities:
* with an extensive match the RISC hydrolyzes ATP and cleaves the mRNA which causes degradation
* less extensive match: blocks the ribosome which results in rapid translational repression, deadenylation and in most cases eventual degradation of mRNA

53
New cards
RNA interference
* double-stranded RNAs that suppress the gene expression of other RNAs in a sequence-specific manner
* example miRNA and these proteins can fight foreign RNA molecules
* found in euks, like fung, plants, and worms
* example miRNA and these proteins can fight foreign RNA molecules
* found in euks, like fung, plants, and worms
54
New cards
RNAi (6) with siRNAs (5)
* many viruses (and transposable elements) produce double-stranded RNA as part of their life cycles
* (6) destroys the double-stranded RNA
* initiated by a dicer protein complex forming small interfering RNAs (5)
* (5) can interact with argonaute and RISC proteins and follow the miRNA route to destroy double-stranded RNA or…
* (5) can also regulate transcription
* (5) interact with argonaute and the RNA-induced transcriptional silencing complex (RITS) → which interacts with newly transcribed RNA (in the nucleus) and recruiter chromatin-modifying enzymes (makes it very condense)
* (6) destroys the double-stranded RNA
* initiated by a dicer protein complex forming small interfering RNAs (5)
* (5) can interact with argonaute and RISC proteins and follow the miRNA route to destroy double-stranded RNA or…
* (5) can also regulate transcription
* (5) interact with argonaute and the RNA-induced transcriptional silencing complex (RITS) → which interacts with newly transcribed RNA (in the nucleus) and recruiter chromatin-modifying enzymes (makes it very condense)

55
New cards
CRISPR-Cas immunity
* short fragments of viral DNA integrate in the CRISPR region of the genome and become templates to produce crRNAs (CRISPR RNAs)
* viral DNAs complementary to CRISPR regions are directed for degradation by Cas (Crispr associated) proteins
* similarly to argonaute: use of small sing-stranded RNA
* viral DNAs complementary to CRISPR regions are directed for degradation by Cas (Crispr associated) proteins
* similarly to argonaute: use of small sing-stranded RNA
56
New cards
Shine-Dalgarno sequence (only in proks)
* a six nucleotide sequence upstream of the AUG start codon on mRNA
* correctly positions AUG in the ribosome and provides translation control mechanisms
* correctly positions AUG in the ribosome and provides translation control mechanisms
57
New cards
Translation regulation mechanism in prokaryotes
1. a specific RNA binding protein blocks access the shine Dalgarno (SD sequence)
2. temperature regulated RNA structure (change in shape blocks SD
3. Riboswitch→ small molecule causes structural rearrangement of RNA blocking SD
4. Antisense RNA→ produced elsewhere in the genome base-pairs with mRNA and blocks SD
58
New cards
Ferritin
* binds iron and releases it in a controlled manner
* not needed when iron is low
* aconitase binds to the _____ RNA near the start site and block translation
* Translated when iron is in excess
* aconitase binds iron
* conformational change
* ferritin RNA released
* not needed when iron is low
* aconitase binds to the _____ RNA near the start site and block translation
* Translated when iron is in excess
* aconitase binds iron
* conformational change
* ferritin RNA released
59
New cards
Regulation of euk initiation (eIFs)
* eIF2 plays a crucial role in translation initiation
* eIF2 forms a complex with GTP and recruits the initiator tRNA (methionyl) to the small ribosomal subunit
* the small ribosomal subunit binds the 5’ end of mRNA and scans for the 1st AUG
* when aug is recognized it hydrolyzes GTP to GDP and causes a change in eIF2
* eIF2 bound to GDP is released and is inactive
* reactivation of eIF2 requires eIF2B which is a guanine nucleotide exchange factor (GEF)
* BUT phosphorylated eIF2 sequesters eIF2B as an inactive complex
* since there’s more eIF2 then eIF2B is sequestered a translation is dramatically reduced
* not all mRNAs are equally affected by eIF2 phosphorylation
* eIF2 forms a complex with GTP and recruits the initiator tRNA (methionyl) to the small ribosomal subunit
* the small ribosomal subunit binds the 5’ end of mRNA and scans for the 1st AUG
* when aug is recognized it hydrolyzes GTP to GDP and causes a change in eIF2
* eIF2 bound to GDP is released and is inactive
* reactivation of eIF2 requires eIF2B which is a guanine nucleotide exchange factor (GEF)
* BUT phosphorylated eIF2 sequesters eIF2B as an inactive complex
* since there’s more eIF2 then eIF2B is sequestered a translation is dramatically reduced
* not all mRNAs are equally affected by eIF2 phosphorylation
60
New cards
steps for a protein to be functional
1. proteins must fold properly to adopt their 3d structure
2. proteins are covalently modified with chemical groups
3. proteins interact with other proteins and small molecules (cofactors)
61
New cards
Protein folding
* hydrophobic amino acids are buried in the interior core
* for some proteins, folding begins as they emerge from ribosomes; some are completely folded after synthesis
* for some proteins, folding begins as they emerge from ribosomes; some are completely folded after synthesis
62
New cards
Heat shock proteins (Hsp)
* synthesized in high amounts by cells at elevated temperatures
* ___70 and ()60 assist protein folding:
* both interact ith exposed hydrophobic regions of misfolded proteins
* both use NRG from ATP hydrolysis to promote proper folding
* ___70 and ()60 assist protein folding:
* both interact ith exposed hydrophobic regions of misfolded proteins
* both use NRG from ATP hydrolysis to promote proper folding
63
New cards
misfolded proteins
* can aggregate and become toxic
* cause of many inherited diseases
* process is closly regulated by a protein degrading apparatus called the proteasome
* exposed hydrophobic resides mark protein for degradation by the proteasome (competes with chaperones)
* longer time to fold more chance for degradation
* cause of many inherited diseases
* process is closly regulated by a protein degrading apparatus called the proteasome
* exposed hydrophobic resides mark protein for degradation by the proteasome (competes with chaperones)
* longer time to fold more chance for degradation
64
New cards
Proteasome
* abundant protein complex found in the cytosol and the nucleus
* hollow cylinder with a cap at each end and an active site in the core
* caps protect cellular proteins from degradation
* acts on proteins that have been marked for destruction by the addition of a small protein tag named ubiquitin
* hollow cylinder with a cap at each end and an active site in the core
* caps protect cellular proteins from degradation
* acts on proteins that have been marked for destruction by the addition of a small protein tag named ubiquitin
65
New cards

Ubiquitin
* added to proteins by a ___-conjugating system made up of 3 enzymes
* E1 (a couple of different ones in humans): an ATP-dependent ____-activating enzyme creates an activated E1-bound ubiquitin (bound to SH)
* E2: ___-conjugating enzyme accepts *from E1 and exists as a complex with E3 a* __ligase that selects substrates
* E3 binds to specific degradation sequences in substrates. ___ is added to a lysine residue of the target protein and the process is repeated to form a poly_ chain
* the chain is recognized by a specific receptor in the proteasome
* depending on number of ubiquitin molecules and type of linkage
* E1 (a couple of different ones in humans): an ATP-dependent ____-activating enzyme creates an activated E1-bound ubiquitin (bound to SH)
* E2: ___-conjugating enzyme accepts *from E1 and exists as a complex with E3 a* __ligase that selects substrates
* E3 binds to specific degradation sequences in substrates. ___ is added to a lysine residue of the target protein and the process is repeated to form a poly_ chain
* the chain is recognized by a specific receptor in the proteasome
* depending on number of ubiquitin molecules and type of linkage
66
New cards

Regulation of protein destruction
* activating:
* E3 phosphorylated by protein kinase
* unmasking protein by dissociation
* creation of destabilizing N-terminus
* E3 phosphorylated by protein kinase
* unmasking protein by dissociation
* creation of destabilizing N-terminus
67
New cards
examples of gene regulatory proteins

68
New cards
PKA (protein kinase A)
* activated by numerous extracellular stimuli result in increased levels of the small molecule cyclic AMP( cAMP)
* has two regulatory subunits and two catalytic subunits
* binding of cAMP to the regulator subunits causes a conformational change and release of the active catalytic subunits
* its substrates include enzymes involved in glycogen metabolism in skeletal muscle and liver
* ligand = adrenaline
* response = to promote glucose release
* when activated it has two effects
* promote the breakdown of glycogen
* inhibits glycogen synthesis
* glycogen is broken down into glucose-1-phosphate (which is converted to glucose-6-phosphate → glycolytic pathway)
* inactive PKA is located in the cytosol
* activated PKA catalytic subunits translocate to the nucleus
* PKA catalytic subunits phosphorylate specific substrate proteins
* activation of target genes with cAMP-responsive elements (CRE)
* has two regulatory subunits and two catalytic subunits
* binding of cAMP to the regulator subunits causes a conformational change and release of the active catalytic subunits
* its substrates include enzymes involved in glycogen metabolism in skeletal muscle and liver
* ligand = adrenaline
* response = to promote glucose release
* when activated it has two effects
* promote the breakdown of glycogen
* inhibits glycogen synthesis
* glycogen is broken down into glucose-1-phosphate (which is converted to glucose-6-phosphate → glycolytic pathway)
* inactive PKA is located in the cytosol
* activated PKA catalytic subunits translocate to the nucleus
* PKA catalytic subunits phosphorylate specific substrate proteins
* activation of target genes with cAMP-responsive elements (CRE)

69
New cards
Activation of target genes with cAMP responsive elements (CRE)
* Activated PKA phosphorylates CREB (CRE binding protein)
* CREB recruits CBP coactivator ( CREB binding protein)
* target genes are transcribed
* CREB recruits CBP coactivator ( CREB binding protein)
* target genes are transcribed

70
New cards
Post-translational process
* proteins fully synthesized in the cytosol before sorting
* unfolded: mitochondria, plastids (remain unfolded with hsp70)
* folded: nucleus, peroxisomes
* unfolded: mitochondria, plastids (remain unfolded with hsp70)
* folded: nucleus, peroxisomes
71
New cards
Co-translational process
* in ER
* proteins with ER signal seq
* associated with ER during protein synthesis
* proteins with ER signal seq
* associated with ER during protein synthesis
72
New cards
Gated transport
* proteins moving between cytosol and nucleus
* nuclear pore complex (made up of nucleoporins)
* selective transport of macromolecules
* free diffusion of small molecules (
* nuclear pore complex (made up of nucleoporins)
* selective transport of macromolecules
* free diffusion of small molecules (
73
New cards
Nuclear import receptors
* binds to specific NLS (nuclear localization signal) (rich in Lys and Arg)
* binds to nucleoporins in NPC
* transport into nucleus
Cargo proteins have a NLS
* binds to nucleoporins in NPC
* transport into nucleus
Cargo proteins have a NLS

74
New cards
Nuclear export receptors
* structurally related to the nuclear import receptor
* binds to NES
* binds to nucleoporins in NPC
* cargo proteins have a nuclear export signal (NES)
* newly assembled ribosomal subunits, RNA, proteins with regulated nuclear import and export
* binds to NES
* binds to nucleoporins in NPC
* cargo proteins have a nuclear export signal (NES)
* newly assembled ribosomal subunits, RNA, proteins with regulated nuclear import and export

75
New cards
regulation of Ran() GTPase in nucleus and cytosol
* cycles between:
* GDP-bound
* GTP-bound
* regulated by:
* ()-GAP (GTP-ase-Activating protein) (in the cytosol)
* stimulates GTP hydrolysis by ()
* ()-GEF (guanine nucleotide exchange factor)
* promotes the exchange of GDP for GTP by Ran
1. ()-GTP: to cytosol
* with nuclear import/ export receptors
2. ()-GDP: to the nucleus, transported by NTF2 (nuclear transport factor 2)
* GDP-bound
* GTP-bound
* regulated by:
* ()-GAP (GTP-ase-Activating protein) (in the cytosol)
* stimulates GTP hydrolysis by ()
* ()-GEF (guanine nucleotide exchange factor)
* promotes the exchange of GDP for GTP by Ran
1. ()-GTP: to cytosol
* with nuclear import/ export receptors
2. ()-GDP: to the nucleus, transported by NTF2 (nuclear transport factor 2)

76
New cards
Nuclear import of cargo proteins
1. nuclear import receptor (binds cargo in cytosol)
2. receptor + cargo move to nucleus
3. RAN-GTP binding (causes cargo release)
4. Empty import receptor + RAN-GTP move to cytosol
5. RAN binding protein and RAN-GAP promote: GTP hydrolysis and release of import receptor

77
New cards
Nuclear export of cargo proteins
1. nuclear export receptor (binds RAN-GTP + cargo in nucleus)
2. Receptor + cargo + Ran-GTP move to cytosol
3. Ran Binding Protein and Ran-GAP promote: GTP hydrolysis, release of cargo, release of export receptor
4. empty export receptor returns to nucleus

78
New cards
NFAT (nuclear factor of activated T cells)
1. in high calcium 2+ in activated T cell the calcineurin removes the phosphate from the () and blocks the export signal and causes a change to reveal the import signal resulting in the activation of gene transcription
2. in low \[ Ca2+\] in resting T cell the cap comes off and ATP + active protein kinase phosphorylates the NFAT which exposes the export signal and it returns to the cytosol
![1. in high calcium 2+ in activated T cell the calcineurin removes the phosphate from the () and blocks the export signal and causes a change to reveal the import signal resulting in the activation of gene transcription
2. in low \[ Ca2+\] in resting T cell the cap comes off and ATP + active protein kinase phosphorylates the NFAT which exposes the export signal and it returns to the cytosol](https://knowt-user-attachments.s3.amazonaws.com/3a2ef02f476d4c1cad89e62ea30e7f8e.jpeg)
79
New cards
Transmembrane Transport
* unidirectional
* ER, mitochondria, plastids, peroxisomes
* protein translocators: transport of protein across membrane, protein usually unfolded
* ER, mitochondria, plastids, peroxisomes
* protein translocators: transport of protein across membrane, protein usually unfolded
80
New cards
Importing proteins to the mitochondrial matrix
* protein translocators:
* TOM: Translocator of the outer membrane
* TIM23: Translocator of the inner membrane
* precursor protein has a mitochondrial signal sequence (peptide)
* N-terminal amphipathic helix which binds to import receptors that help feed it through the TOM then through the TIM23
* in the matrix space the signal sequence is cleaved
* proteins are further sorted
* TOM: Translocator of the outer membrane
* TIM23: Translocator of the inner membrane
* precursor protein has a mitochondrial signal sequence (peptide)
* N-terminal amphipathic helix which binds to import receptors that help feed it through the TOM then through the TIM23
* in the matrix space the signal sequence is cleaved
* proteins are further sorted
81
New cards
Importing proteins to the chloroplast
* there’s a TIC and TOC translocator
* precursor protein has a chloroplast signal sequence
* N-terminal amphipathic alpha helix
* signal sequence is cleaved inside
* different from mitochondrial signal sequence for correct targeting in plants
* if targeting to thylakoid
* hydrophobic thylakoid signal sequence
* unmasked when chloroplast signal seq. cleaved
* precursor protein has a chloroplast signal sequence
* N-terminal amphipathic alpha helix
* signal sequence is cleaved inside
* different from mitochondrial signal sequence for correct targeting in plants
* if targeting to thylakoid
* hydrophobic thylakoid signal sequence
* unmasked when chloroplast signal seq. cleaved
82
New cards
Sorting proteins to the peroxisome
* precursor protein
* peroxisomal targeting signal
* 3 AA’s at C-terminus (SKL)
* protein folded
* transported across membrane by large translocator complex (SKL binds to soluble receptor and the binds to a docking protein and its fed through a translocator)
* peroxisomal targeting signal
* 3 AA’s at C-terminus (SKL)
* protein folded
* transported across membrane by large translocator complex (SKL binds to soluble receptor and the binds to a docking protein and its fed through a translocator)

83
New cards
SRP (signal recognition particle)
* () and () receptor have GTPase domains that bind GTP
* SRP + ribosome → low affinity
* SRP + ribosome + ER signal sequence
* high affinity binds SRP receptor
* ribosome forms a tight seal with the translocator → prevents diffusion of ions, small molecules
* SRP+ SRP receptor and GTP is hydrolyzed and the complex dissociates
* SRP released
* SRP + ribosome → low affinity
* SRP + ribosome + ER signal sequence
* high affinity binds SRP receptor
* ribosome forms a tight seal with the translocator → prevents diffusion of ions, small molecules
* SRP+ SRP receptor and GTP is hydrolyzed and the complex dissociates
* SRP released

84
New cards
Protein sorting to the ER: soluble proteins
* the ER signal seq is an N-terminal start-transfer seq → bound to the translocator
* a signal peptidase cleaves the ER signal seq
* the ER signal sequence laterally diffuses in to the lipid bilayer → translocator is gated in a 2nd direction
* Translocated proteins is released into the ER
* a signal peptidase cleaves the ER signal seq
* the ER signal sequence laterally diffuses in to the lipid bilayer → translocator is gated in a 2nd direction
* Translocated proteins is released into the ER

85
New cards
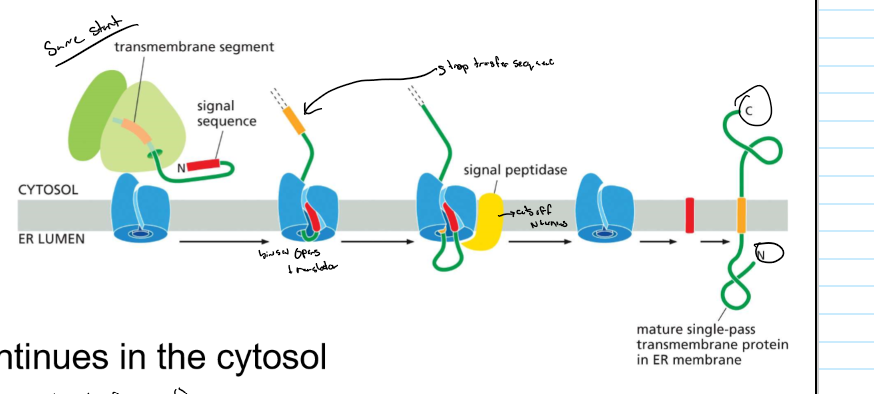
Protein sorting into the ER: transmembrane proteins (example 1)
* ER signal sequence: N-terminal start transfer
* TM domain is a stop transfer sequence → laterally diffuses into lipid bilayer
* protein synthesis continues in the cytosol → COOH in cytosol
* TM domain is a stop transfer sequence → laterally diffuses into lipid bilayer
* protein synthesis continues in the cytosol → COOH in cytosol
86
New cards
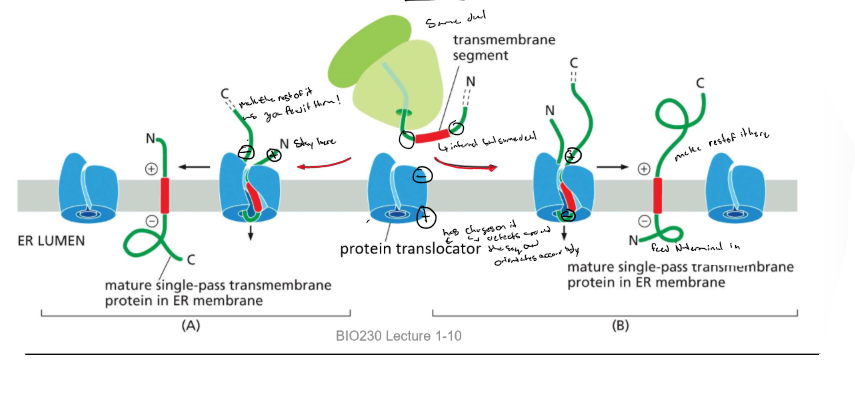
Protein sorting into the ER: transmembrane proteins (example 2 + 3)
* TM domain is an internal start transfer sequence and is not cleaved → laterally diffuses into the lipid bilayer
* orientation is determined by AA’s flanking the internal start transfer sequence → more positive = cytosolic side
* orientation is determined by AA’s flanking the internal start transfer sequence → more positive = cytosolic side
87
New cards
Examples of protein sorting to the ER: Multipass TM proteins
1. 1st TM domain: internal start transfer seq and the 2nd TM domain: stop transfer sequence
2. rhodopsin: 1st TM→ start-transfer; (+) AA’s, cytosolic, 2nd TM→ start transfer, 3rd TM→ stop transfer, 4th TM → start- transfer
* note: these sequences are specific hydrophobic sequences

88
New cards
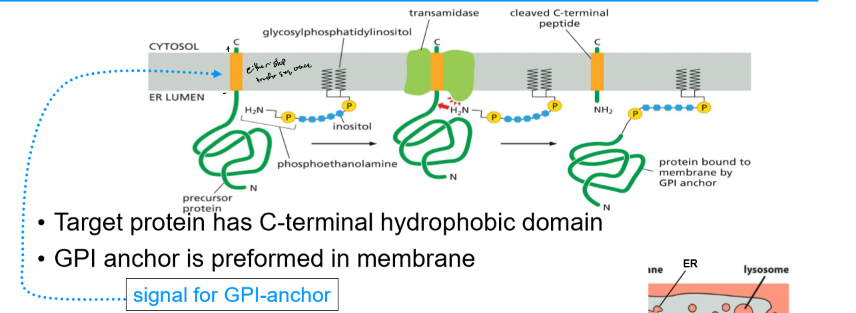
Formation of Glycosylphosphatidylinositol (GPI)-anchored proteins
* target protein has C-terminal hydrophobic domain (signal for GPI anchor)
* GPI anchor is preformed in membrane
* ER enzyme transfers protein to GPI anchor
* GPI-anchored protein ends up on ER luminal side and can go to cell exterior surface
* GPI anchor is preformed in membrane
* ER enzyme transfers protein to GPI anchor
* GPI-anchored protein ends up on ER luminal side and can go to cell exterior surface
89
New cards
Glycosylation
* most soluble and transmembrane proteins in the ER are glycosylated on the ER side
* two types:
* O-linked glycosylation → added on to the oxygen of a side group (\~10%)
* N-linked glycosylation → added to the nitrogen of an asparagine side chain (\~90%)
* N-linked oligosaccharide precursor is preformed in the ER → linked to the target proteins in the ER
* two types:
* O-linked glycosylation → added on to the oxygen of a side group (\~10%)
* N-linked glycosylation → added to the nitrogen of an asparagine side chain (\~90%)
* N-linked oligosaccharide precursor is preformed in the ER → linked to the target proteins in the ER
90
New cards
N-linked glycosylation in the ER
* In the ER lumen, an N-linked oligosaccharide precursor is transferred by an oligosaccharide transferase to an Asn on a protein being synthesized
* Asn-X-Ser or Asn-X-Thr where X is any amino acid except proline
* proteins are only glycosylated on the ER lumen side
* Asn-X-Ser or Asn-X-Thr where X is any amino acid except proline
* proteins are only glycosylated on the ER lumen side
91
New cards
Processing of N-linked Oligosaccharides in the ER
After transfer of the N-linked oligosaccharide to the protein:
1. 3 glucoses removed
* linked to proper folding of the protein
2. 1 mannose removed by ER mannosidase
3. Glycosylated protein is transported via vesicles to the Golgi
\
1. 3 glucoses removed
* linked to proper folding of the protein
2. 1 mannose removed by ER mannosidase
3. Glycosylated protein is transported via vesicles to the Golgi
\
92
New cards
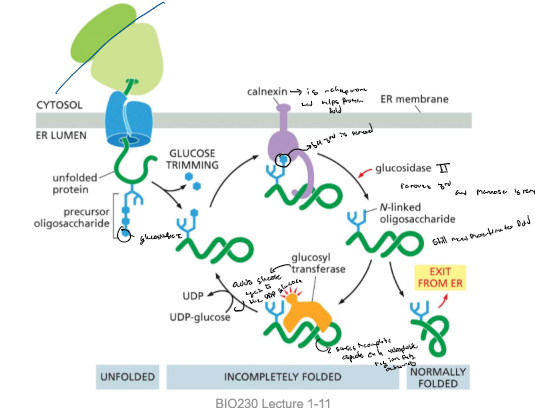
Glycosylation as a mark for the state of protein folding
* (on N-linked) one glucose is removed by glucosidase I and another is removed by glucosidase II and the last glucose binds to the calnexin (chaperone)
* last glucosidase II removes the last and mannosidase removes the mannose glucose if it has no revealing hydrophobic regions it exits from the ER or else glycosyl transferase binds to the area and the enzyme adds glucose again using a UDP-glucose
* and the cycle repeats until it is folded properly
* last glucosidase II removes the last and mannosidase removes the mannose glucose if it has no revealing hydrophobic regions it exits from the ER or else glycosyl transferase binds to the area and the enzyme adds glucose again using a UDP-glucose
* and the cycle repeats until it is folded properly
93
New cards
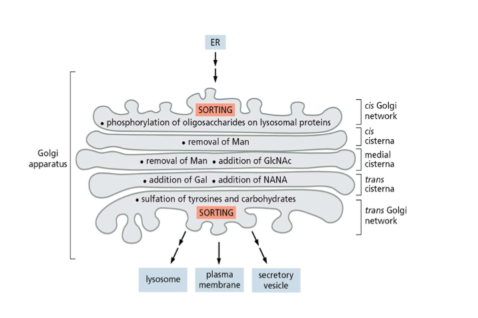
Golgi apparatus structure
* cis, medial, and trans cisternae in Golgi each with different enzymes
* remove or add sugars and result in diff modifications to diff proteins
* vesicles kept close to golgi by tethering proteins
* remove or add sugars and result in diff modifications to diff proteins
* vesicles kept close to golgi by tethering proteins
94
New cards
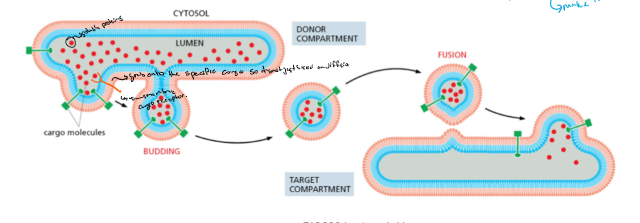
Vesicular transport
* vesicles move cargo between compartments
* budding with cargo
* fusion to target
* release cargo
* cargo is delivered by transport vesicles
* cargo proteins:
* transmembrane proteins
* soluble proteins
* some are bound by transmembrane cargo receptors
* budding with cargo
* fusion to target
* release cargo
* cargo is delivered by transport vesicles
* cargo proteins:
* transmembrane proteins
* soluble proteins
* some are bound by transmembrane cargo receptors
95
New cards
protein coats in vesicle budding
* nascent (new) transport vesicles have protein coats
* purpose:
* select cargo for vesicles
* give curvature
* promote vesicle budding
* COPI-Coated vesicles: from golgi to ER, btw different golgi cisternae
* COPII-Coated vesicles: from ER to Golgi
* Clathrin-coated vesicles: from GA and plasma membrane to endosome
* purpose:
* select cargo for vesicles
* give curvature
* promote vesicle budding
* COPI-Coated vesicles: from golgi to ER, btw different golgi cisternae
* COPII-Coated vesicles: from ER to Golgi
* Clathrin-coated vesicles: from GA and plasma membrane to endosome
96
New cards
Monomeric GTPases
* cycles between GDP-bound (OFF) and GTP-bound (ON)
* regulated by:
* GEF (mediates exchange and activates it)
* GAP (activates the ___ to cut the TP but that essentially turns the protein off
* regulated by:
* GEF (mediates exchange and activates it)
* GAP (activates the ___ to cut the TP but that essentially turns the protein off
97
New cards
General steps in coat assembly and vesicle formation
1. GEF at site of membrane budding → recruits GTPase → GTP bound
2. GTP-GTPase recruits coat proteins
3. vesicle bud formation, cargo selected
4. vesicles buds off
5. vesicle uncoating:
* COPI and COPII- coated vesicles involved GAPs (cuts the GTP)
* different mechanism for clathrin-coated vesicles
finally the vesicle is ready for transport the target compartment
98
New cards
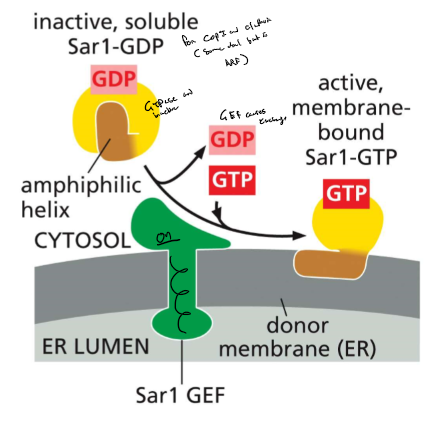
Monomeric GTPase recruit coat proteins
* COPI, clathrin-coated vesicles: ARF GTPase
* COPII-coated vesicle : SAR1 GTPase
* formation of COPII coated:
* SAR1-GEF in ER membrane: recruits Sar1
* SAR-GTP
* amphipathic a helix exposed
* interacts with membrane
* recruits coat protein subunits
* COPII-coated vesicle : SAR1 GTPase
* formation of COPII coated:
* SAR1-GEF in ER membrane: recruits Sar1
* SAR-GTP
* amphipathic a helix exposed
* interacts with membrane
* recruits coat protein subunits
99
New cards
Protein coats and cargo selection
coat:
1. inner layer
* binds to membrane and selects cargo
2. outer layer
* associates with the inner layer to promote polymerization of the coat (sometimes selects cargo as well)
3. coat proteins need to select:
* cargo (TM proteins)
* TM cargo receptors (bind soluble cargo proteins)
* SNARES
1. inner layer
* binds to membrane and selects cargo
2. outer layer
* associates with the inner layer to promote polymerization of the coat (sometimes selects cargo as well)
3. coat proteins need to select:
* cargo (TM proteins)
* TM cargo receptors (bind soluble cargo proteins)
* SNARES
100
New cards
COPI coated vesicles
* inner 4 subunits (β, γ, δ, ζ )
* outer: 3 subunits (a, β’, ε)
* select specific cargo
Uncoating:
* y-COP binds to Arf GAP
* GTP hydrolysis (Arf-GTP → Arf-GDP)
* Arf-GDP detaches from the membrane and the coat is released
* outer: 3 subunits (a, β’, ε)
* select specific cargo
Uncoating:
* y-COP binds to Arf GAP
* GTP hydrolysis (Arf-GTP → Arf-GDP)
* Arf-GDP detaches from the membrane and the coat is released