lec 18 - nutrient scavenging autophagy (zong)
1/31
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
32 Terms
lysosomes
characteristics
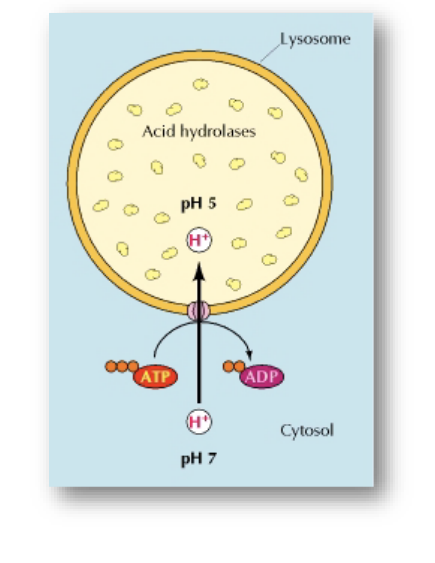
acidic interior → pH = 5
contains hydrolytic enzymes (catalyze the breakdown of molecules using water)
surrounded by membrane
lysosomes are analogous to human stomach
pH is very acidic
enzymes within work effectively in this environment
pump H+ into lysosome expends energy
ATP → ADP
lysosome functions
intracellular digestion
recycling cellular organelles
breakdown of viruses and other cellular invaders
single celled organisms such as amoebas use lysosomes to digest their food since they have no process for extracellular digestion
lysosomes and cell death
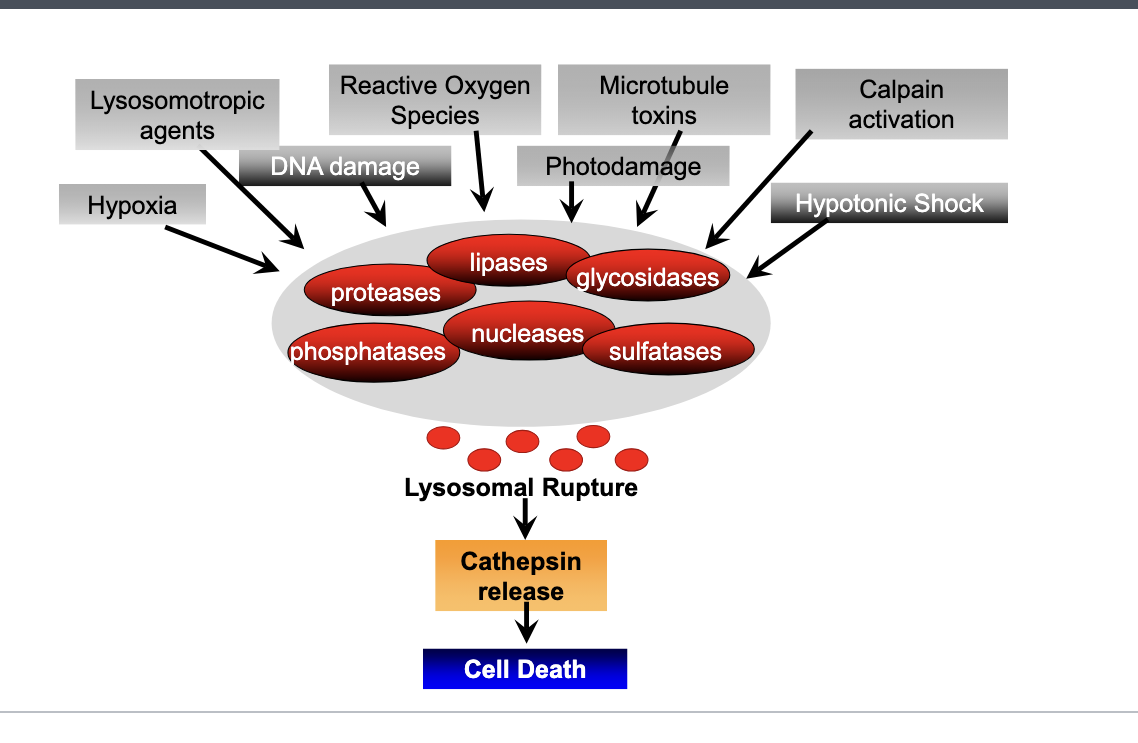
lysosomes are full of hydrolytic enzymes such as
proteases
phosphatases
lipases
nucleases
glycosidases
sulfatases
triggers of lysosomal rupture
hypoxia
lysosomotropic agents
DNA damage
reactive oxygen species
microtubule toxcins
photodamage
calpain activation
hypotonic shock → causes swelling on lysosomal mebranes
trigger of lysosomal rupture → lysosomal rupture → enzymes released → cathepsin release (proteolytic enzyme that degrades proteins once released in cytosol) → cell death
lysosomal storage diseases
undegraded material accumulates within the lysosomes of affected individuals
most of these diseases result from deficiencies in single lysosomal enzymes
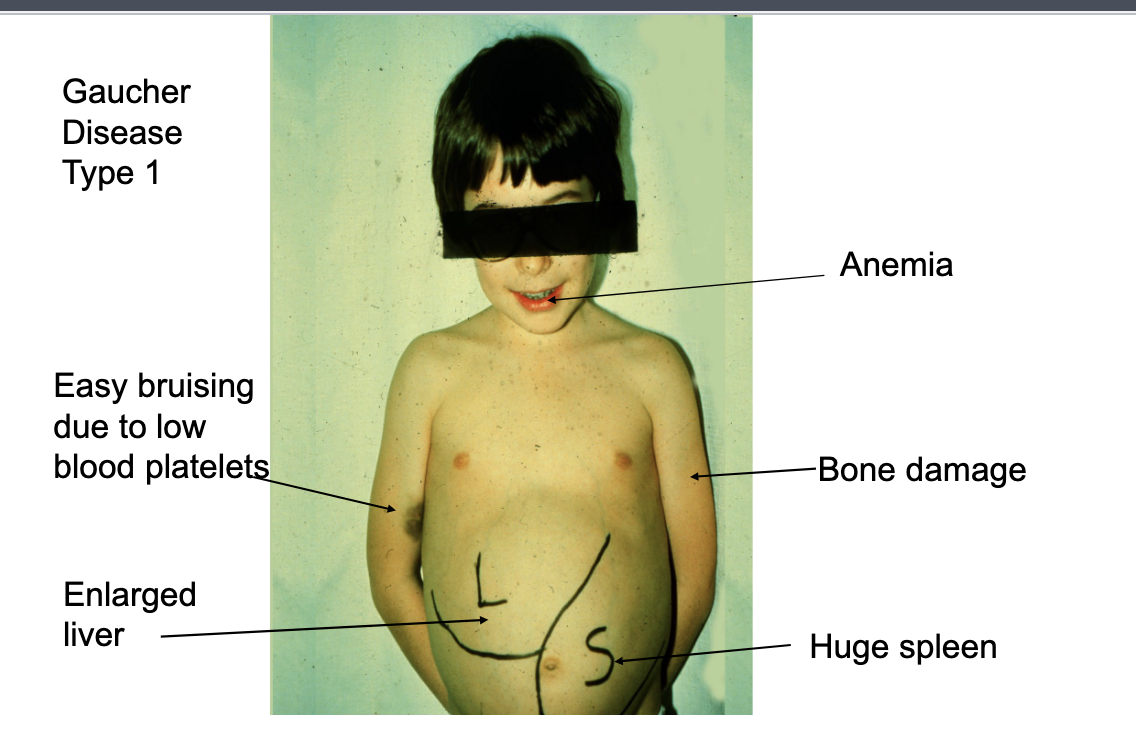
example = gaucher’s disease
gauchers disease
most common of lysosomal storage diseases
1 in 50,000 to 100,000 people
genetic disorder where theres accumulation of lipids in cells and organs
results from a mutation in the gene that encodes a lysosomal enzyme required for the breakdown of glycolipids → glucocerebrosidase: Gcase
gaucher disease type 1
anemia
easy bruising due to low blood platelets
bone damage
enlarged liver
huge spleen
autophagy
greek
auto = self
phagein = to eat
autophagy = the process by which cells recycle cytoplasm and dispose of excess or defective organelles
autophagy can be induced by cellular stress such as nutrient deprivation
autophagy promotes cell survival
excessive autophagy promotes cell death
micro-autophagy
by invagination of the lysosome membrane, cytosolic components are directly taken up by the lysosome itself
it may be selective or nonselective
engulfs material like:
proteins
lipids
organelles
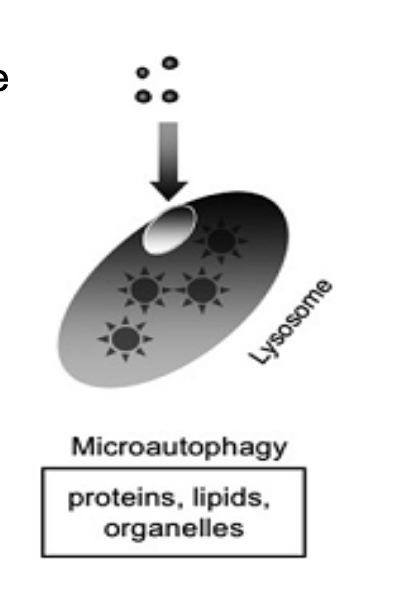
chaperon-mediated autophagy (CMA)
targeted proteins are translocated across the lysosomal membrane in a complex with chaperone proteins (such as Hsc-70) that are recognize by the lysosomal membrane receptor lysosomal-associated membrane protein 2A (LAMP-2A) → results in their unfolding and degradation
KFERQ binds to chaperone protein hsc70 → complexbinds to LAMP2A receptor → protein transported into lysosome
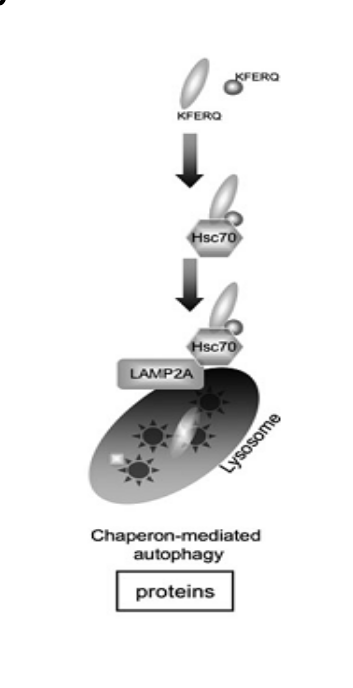
macro-autophagy
delivers cytoplasmic cargo to the lysosome through autophagosome (double membrane bound vesicle)
autophagosome fuses with the lysosome to form an autolysosome
could be selective or non selective
most important type is macroautophagy → referred to as autophagy
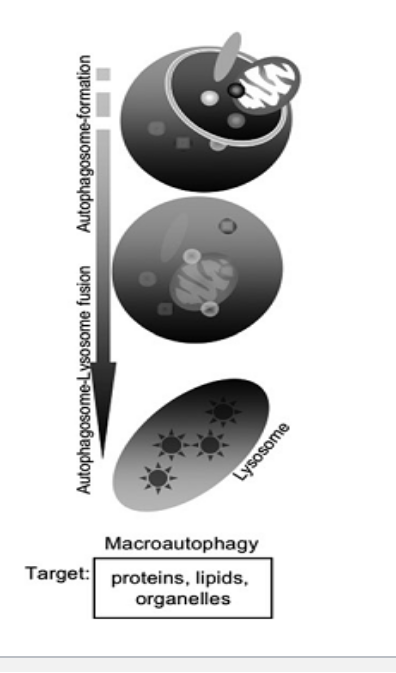
types of autophagy
microautophagy
constitutive → happens regularly under normal conditions
vesicle-mediated
proteins/organelles
selective/nonselective
macroautophagy
inducible
vesicle mediated
proteins/organelles
selective/nonselective
chaperon-mediated
inducible
direct transport
proteins NO organelles
selective
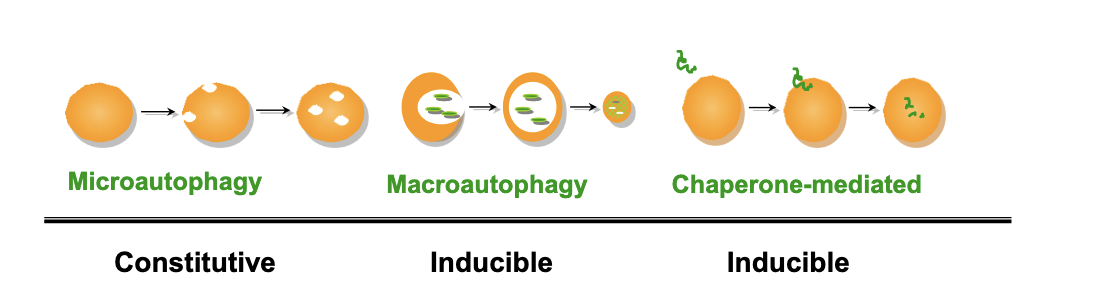
autophagy is commonly used to refer to
macroautophagy
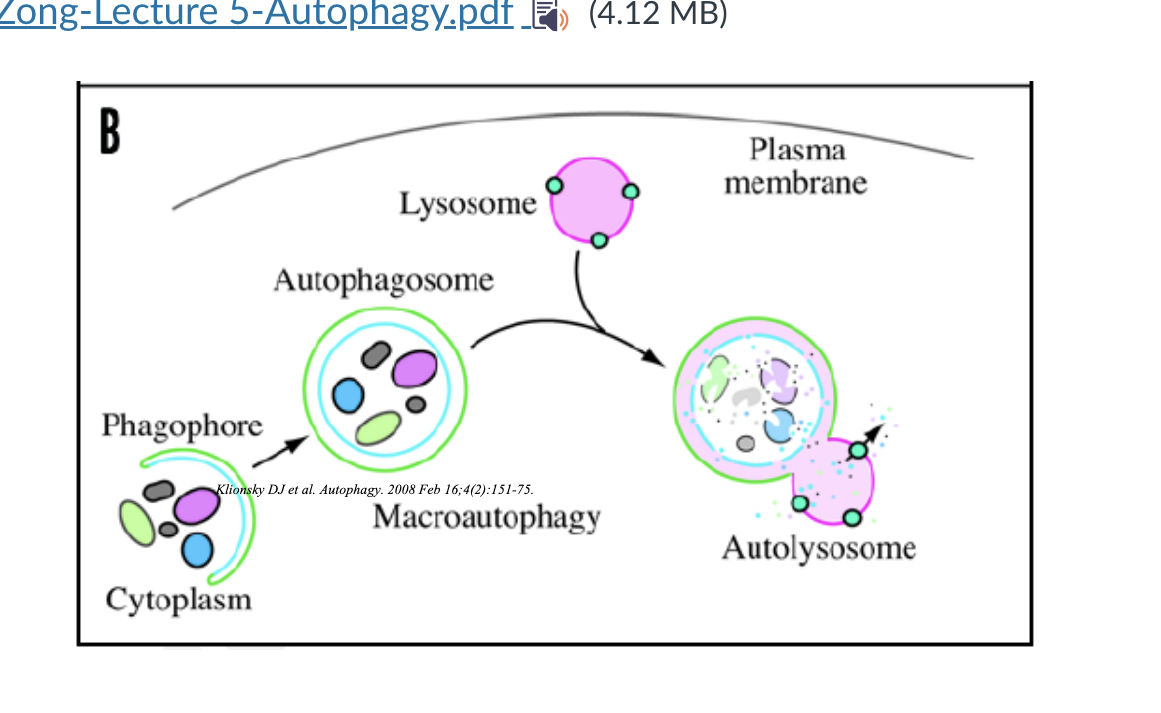
macroautophagy diagram
phagospore formation → starts engulfing cytoplasmic material → phagospore closes into a double membraned vesicle called autophagosome → lysosome fuses with autophagosome and forms an autolysosome → material degraded
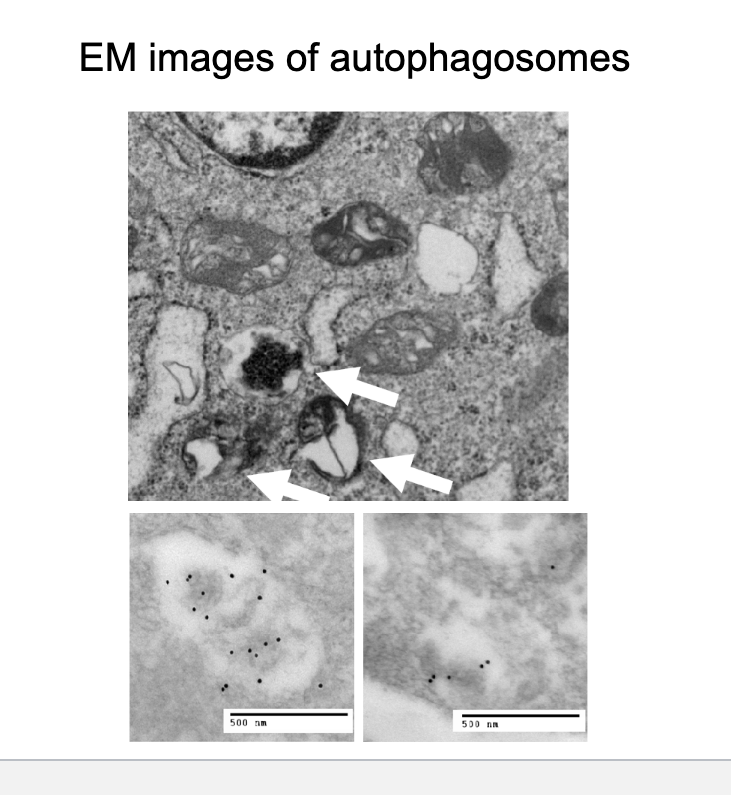
EM images of autophagosomes
electron microscopy images
the arrows are pointing to autophagosomes → double membrane
autophagy timeline
1955 → christian de duve discovered the lysosome
1963 → christian de duve coined the term autophagy
1992 → yoshinori ohsumi studied autophagy in yeast and later identified ATG1
1999 → beth levine cloned beclin 1 in mammalian cells
2002 → protection role of autophagy in huntington disease
2003 → tumor suppression role of autophagy in cancer
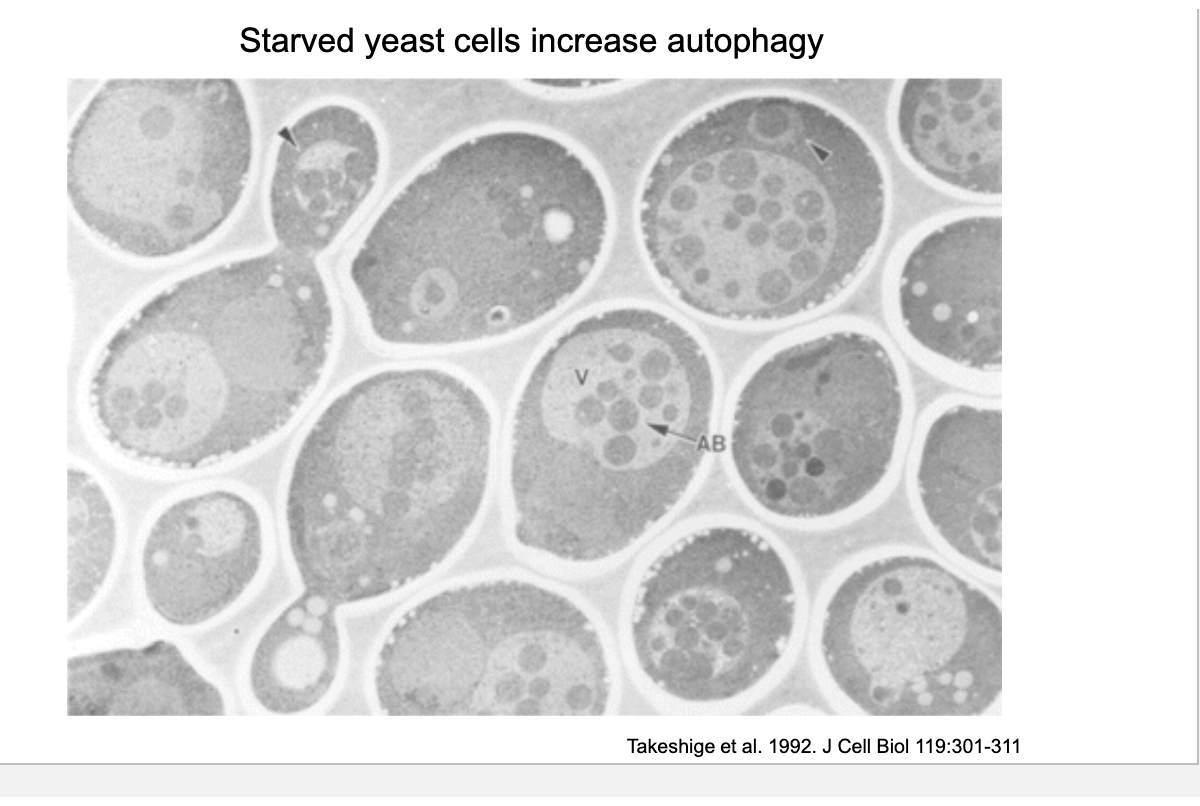
starved yeast cells increased autophagy
nutrient deprivation induces autophagy → break down own proteins, lipids and organelles for energy
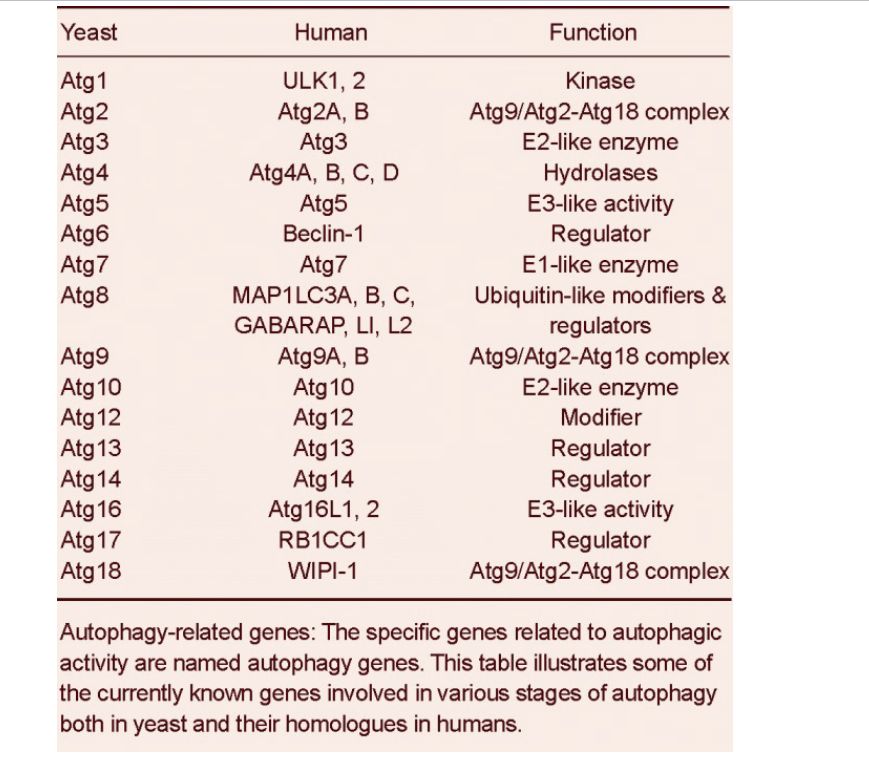
autophagy related genes
specific gene related to autophagic activity are named autophagy genes
table shows some currently known genes involved in various stages of autophagy both in yeast and homologues in humans
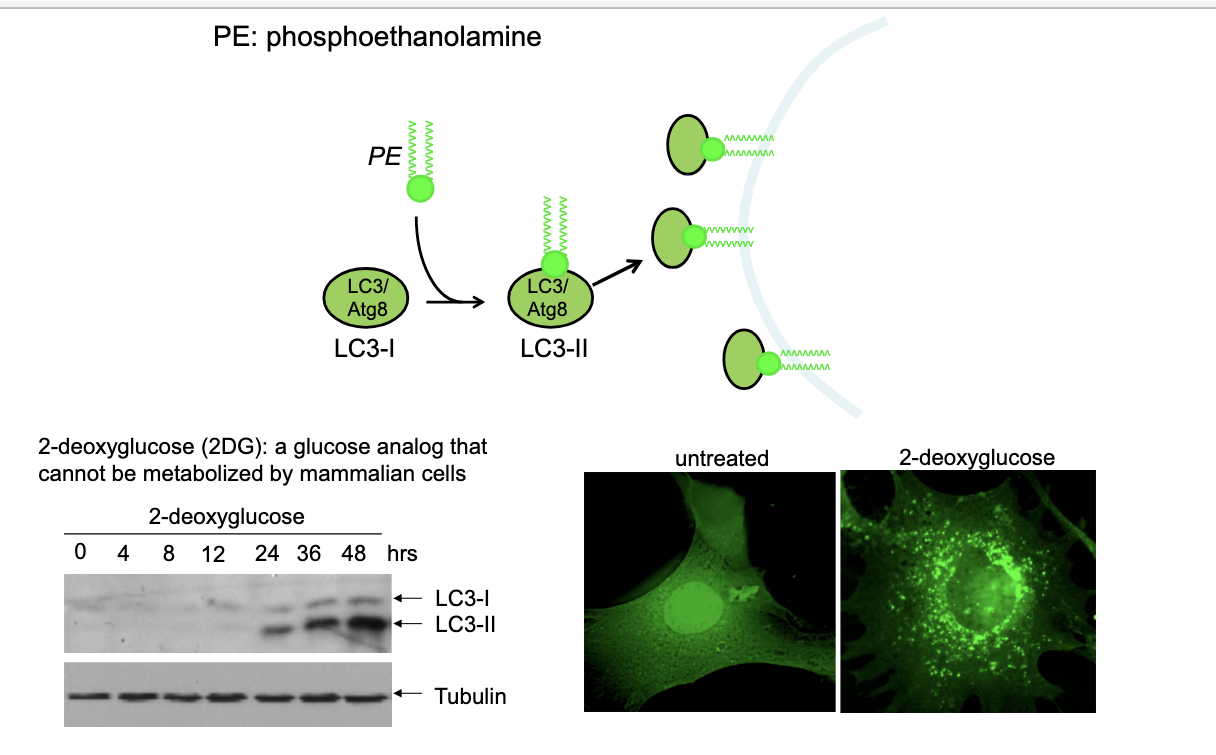
LC3 as marker for autophagy
LC3 → microtubule-associated protein (MAP)-1 light chain 3
first identified protein localized in the autophagosome
28% identified as yeast Apg8 which is essential for formation of autophagosome
LC3-I = cytosolic form
LC3-II = membrane bound form (phosphatidylethanoamine conjugation of LC1)
LC3 diagram
LC3-1 + PE → LC3-II → localizes to autophagosomal membrane; serving as marker to visualize autophagy
western blot
shows the effect of treating cells with 2DG (glucose analog that CANNOT be metabolized by mammalian cells) → causes energy stress and activates autophagy
increasing LC3-I levels over time indicate autophagic activity
tubulin is shown as loading control to confirm equal protein loading
untreated vs 2DG
untreated → minimal LC3 dots
treated with 2DG → more LC3 staining → accumulating of autophagosomes due to activated autophagy
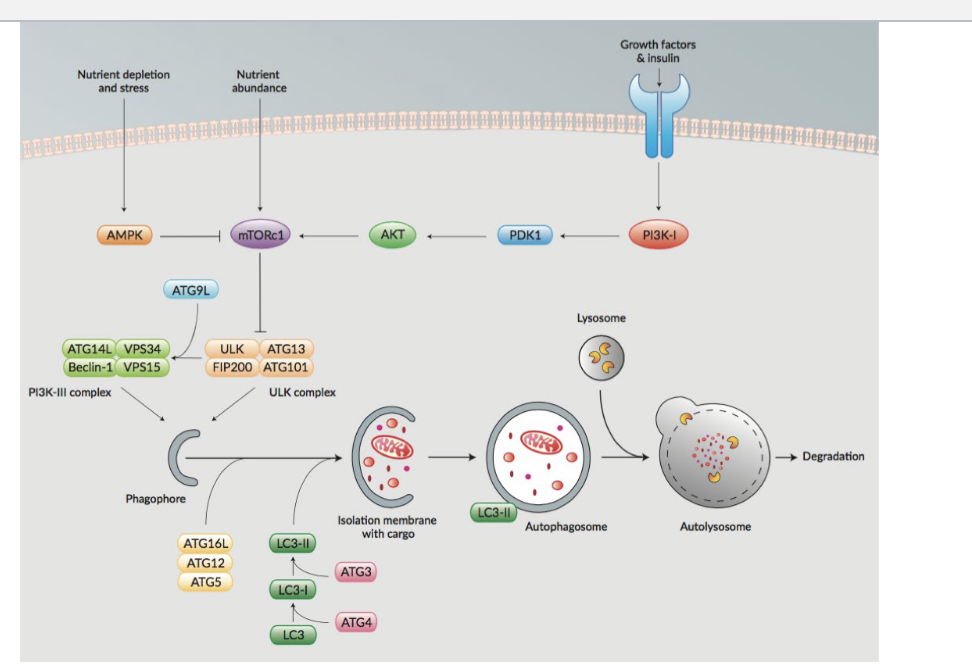
signaling pathway and autophagy
nutrient abundance
activates mTORc1 → inhibits ULK complex which suppresses autophagy
nutrient depletion and stress
activates AMPK → inhibits mTORc1 → since mTORc1 is NO LONGER SUPRESSES ULK complex, it activates autophagy directly or activates PI3K-III complex which also activates autophagy
growth factors & insulin
activates PI3K-1 → activates PDK1 → activates AKT → activates mTORc1 which suppresses autophagy
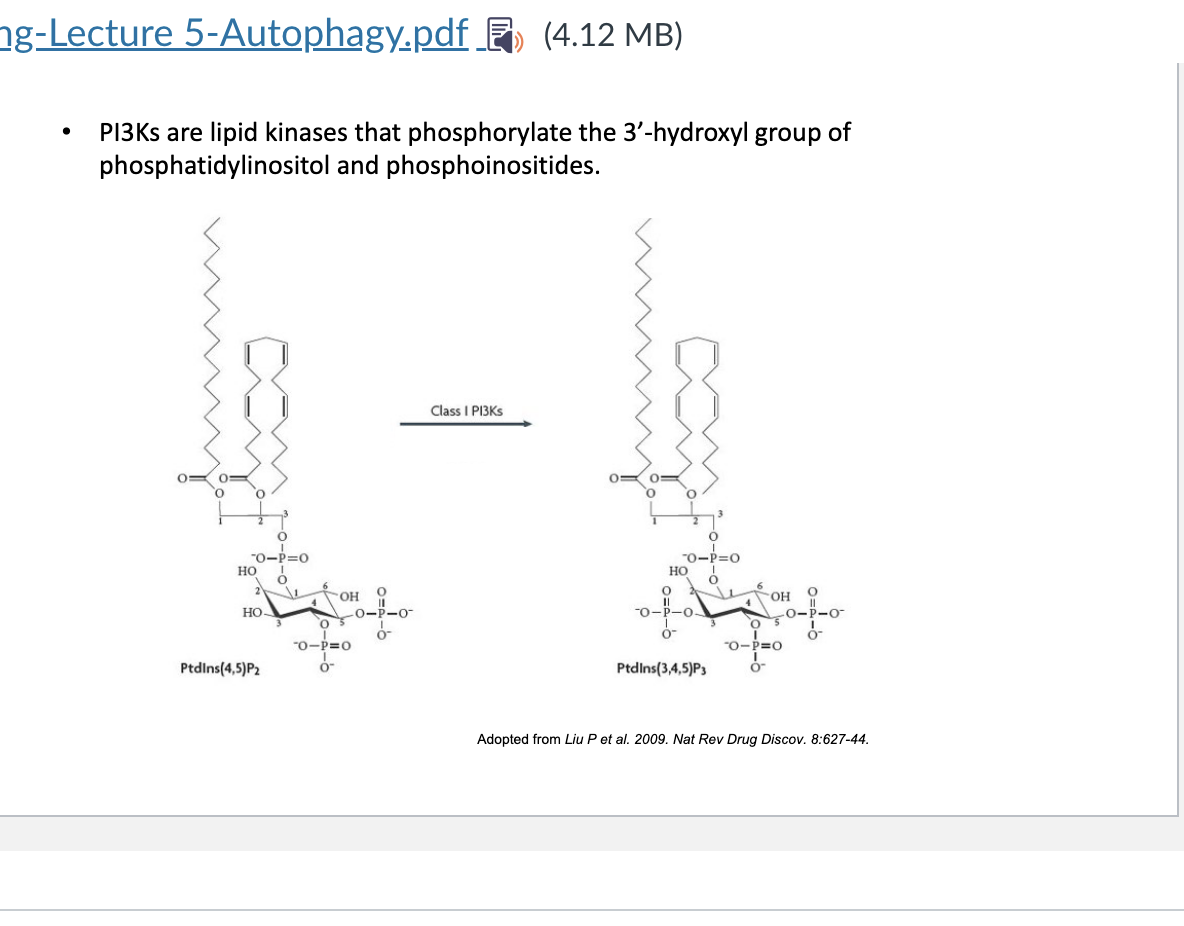
PI3K
PI3K = lipid kinases that phosphoryate the 3-’OH group of phosphatidylinositol and phosphoinositides → they are critical signaling lipid
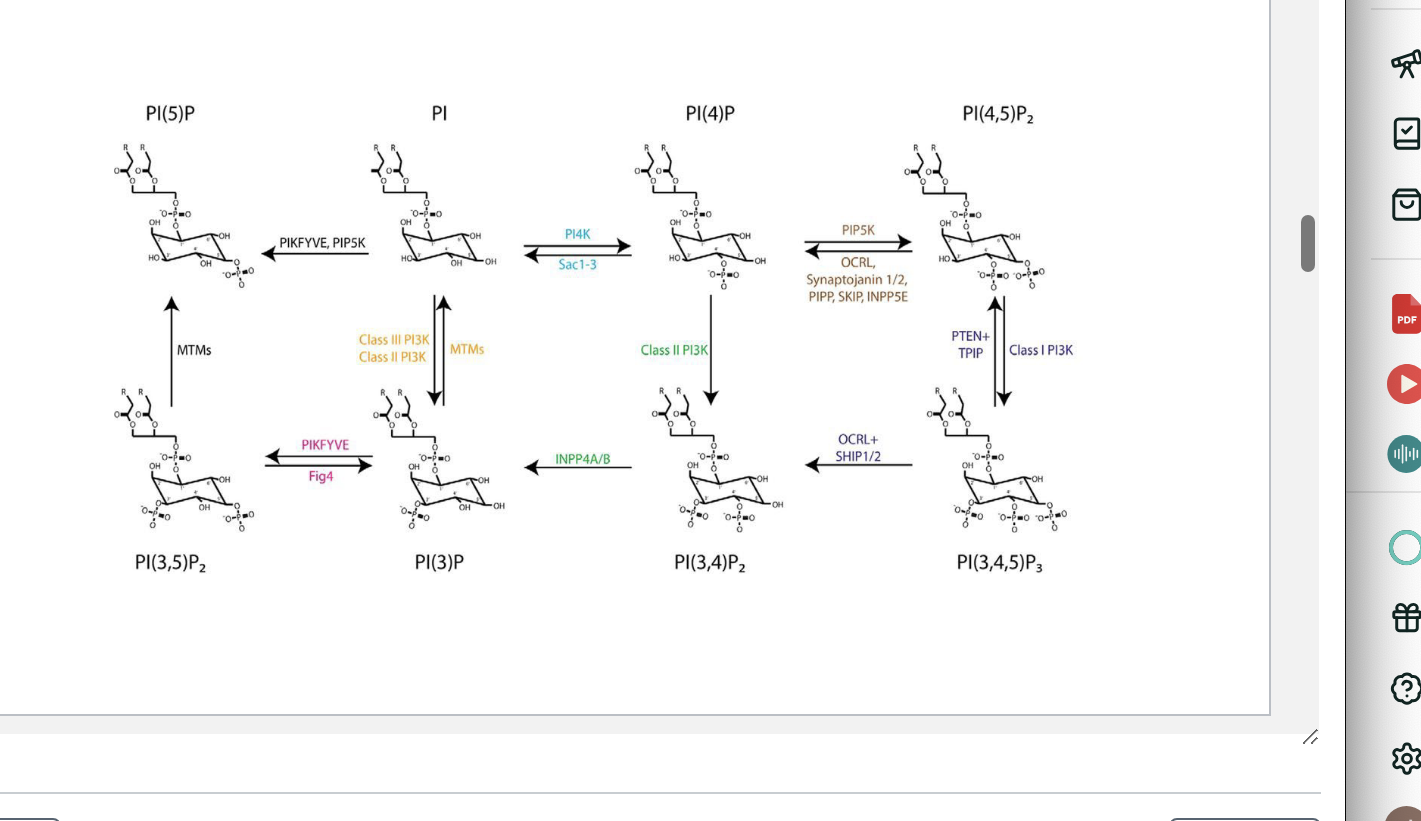
PIs
kinases add phosphate groups to specific positions
phosphatases remove phosphate groups
class III PI3K converts PI → PI(3)P → essential step in forming autophagosomal membranes → recruits autophagic machinery and contributes to me,brane trafficking
PI3K and autophagy
growth factors cross into cell → activates class IA PI3Ks → converts PI(4,5)P2 to PI(3,4,5)P3 → activates AKT → activates mTOR → promotes translation, proliferation, survival and inhibits autophagy
class III PI3Ks convert PI → PI(3)P → autophagy
so class IA = inhibits autophagy
class III = induces autophagy
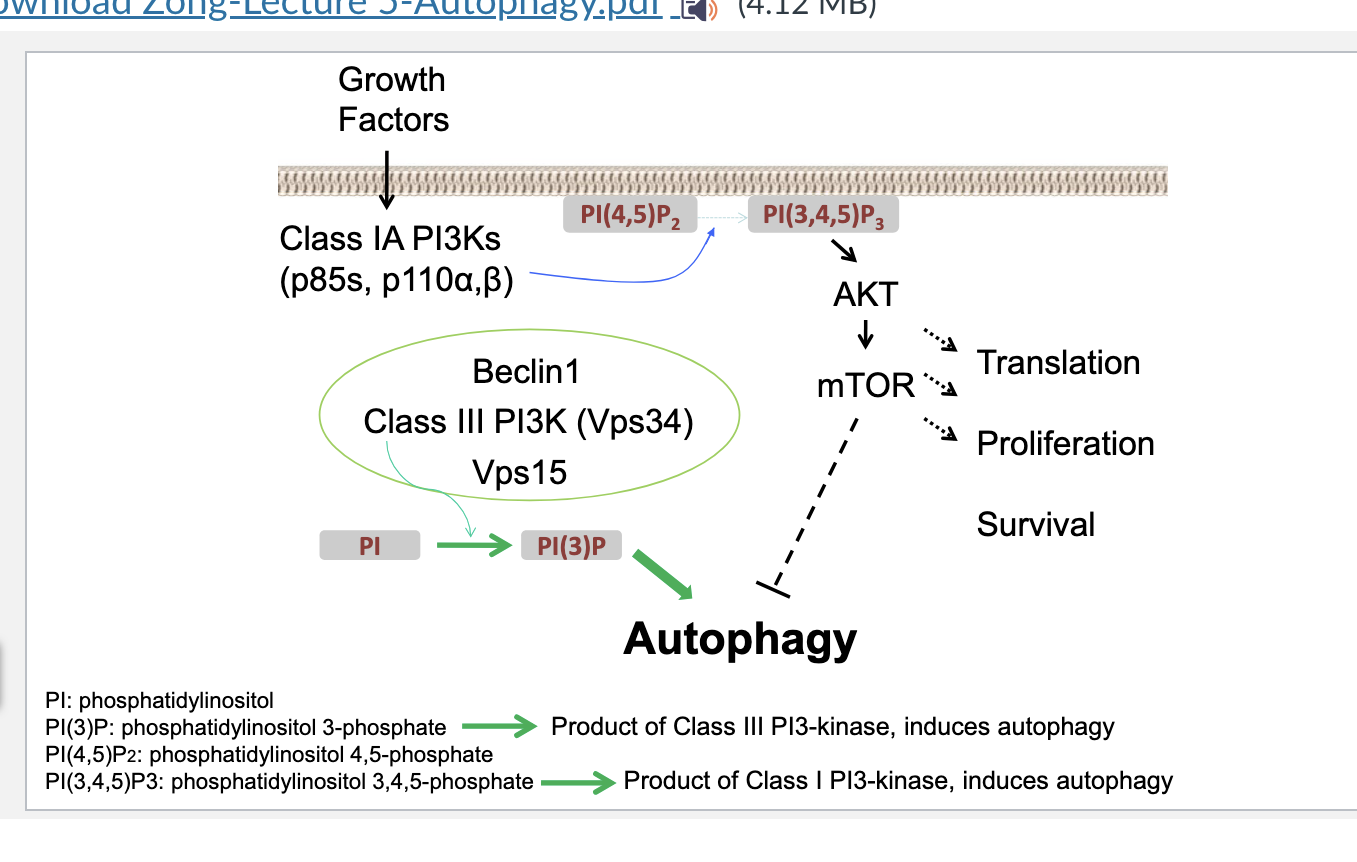
vps34 deficiency
vps34 deficiency leads to enlarged fatty liver
deficient H&E staining → clear structural disruption
oil red O staining → significant increase in red lipid drops → confirming fatty liver
western blot → VPS34 deficient show increased p62 and ubiquinated proteins → they are markers that are degraded via autophagy but since VPS34 is not really there they arent getting degraded
liver to body mass comparison in fed vs fasted
normally, fasting decreases liver mass slightly due to increased autophagy mediated breakdown
VPS34-deficient animals fail to signifiantly reduce liver mass → impaired autophagy
VPS34 = class III PI3K critical for starting autophagy → deficiency = NO autophagy
autophagy has important biological functions
autophagy activated during fasting or nutrient deprivation
recycling role
cellular garbage disposal
clearance of intracellular pathogens
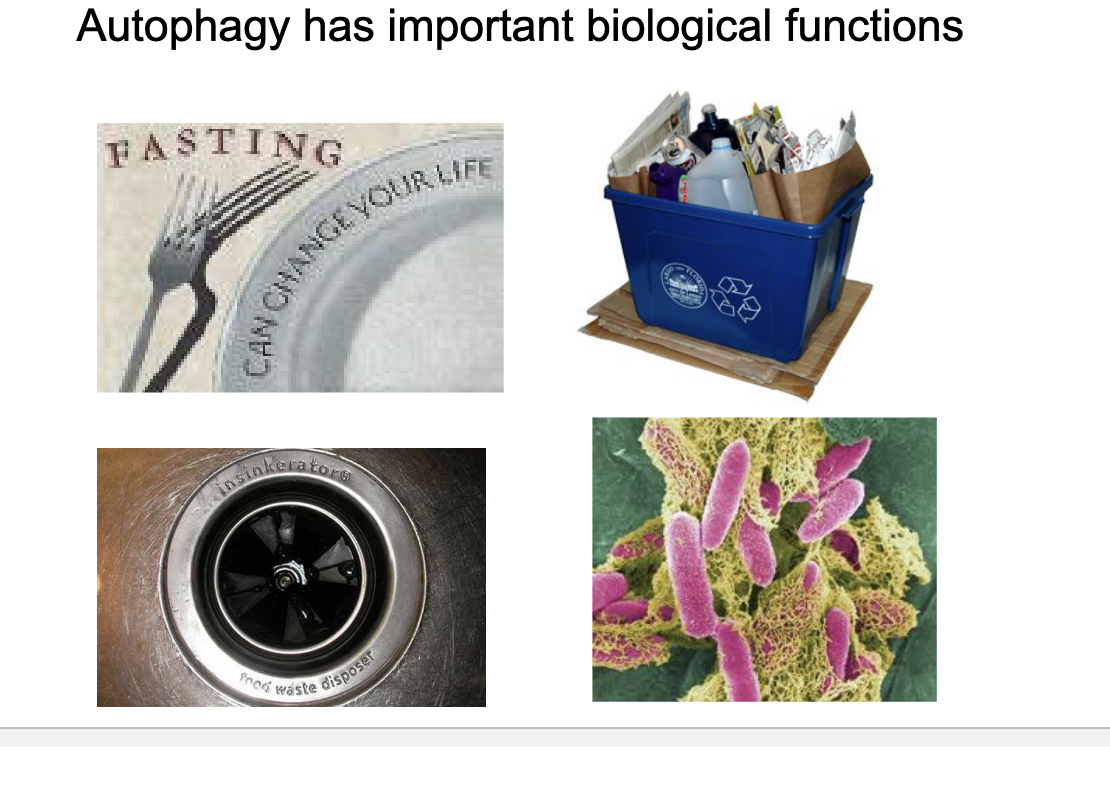
atg5
atg5 knockout pups do NOT survive during neonatal nutrient storage
atg5 is required for autophagosome formation → without it neonatals CANNOT adapt to early nutrient deprivation
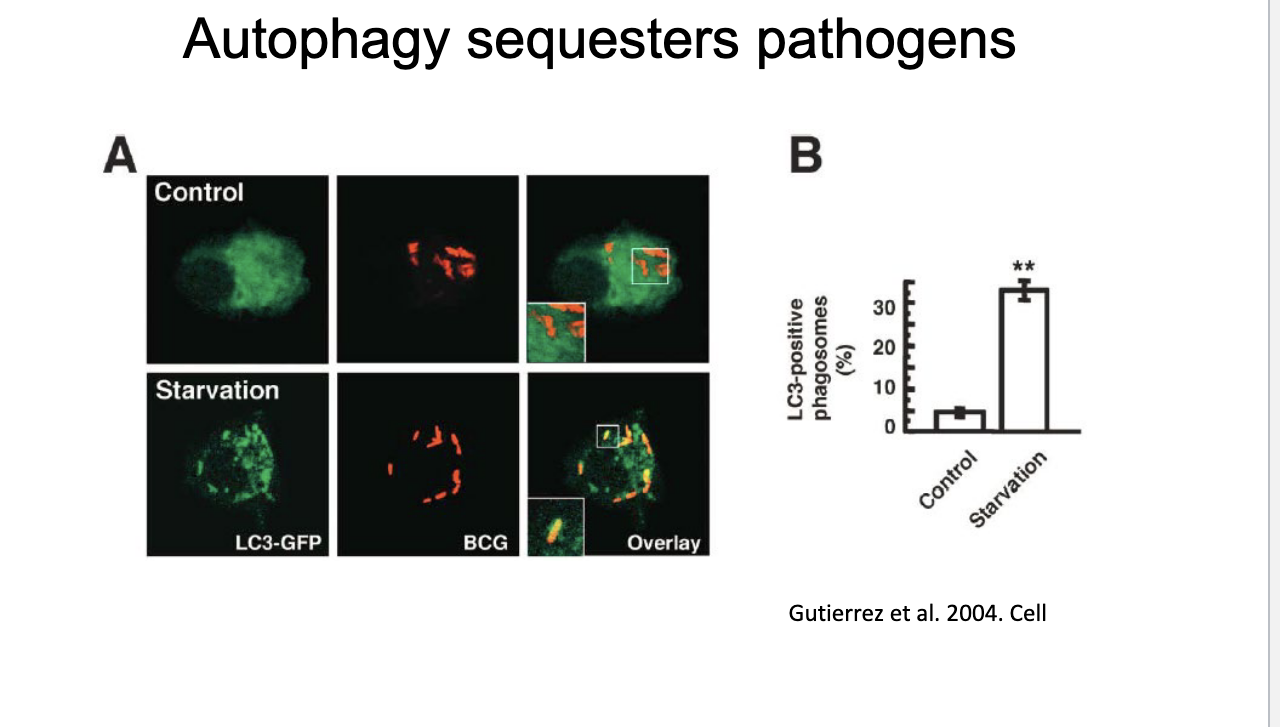
autophagy sequesters pathogens
in starvation → increased LC3
BCG = pathogen
yellow shows where LC3 surrounds BCG
limited autophagic response as theres not much yellow in the control but more in starvation → starvation enhances autophagic sequestrationa of bacteria
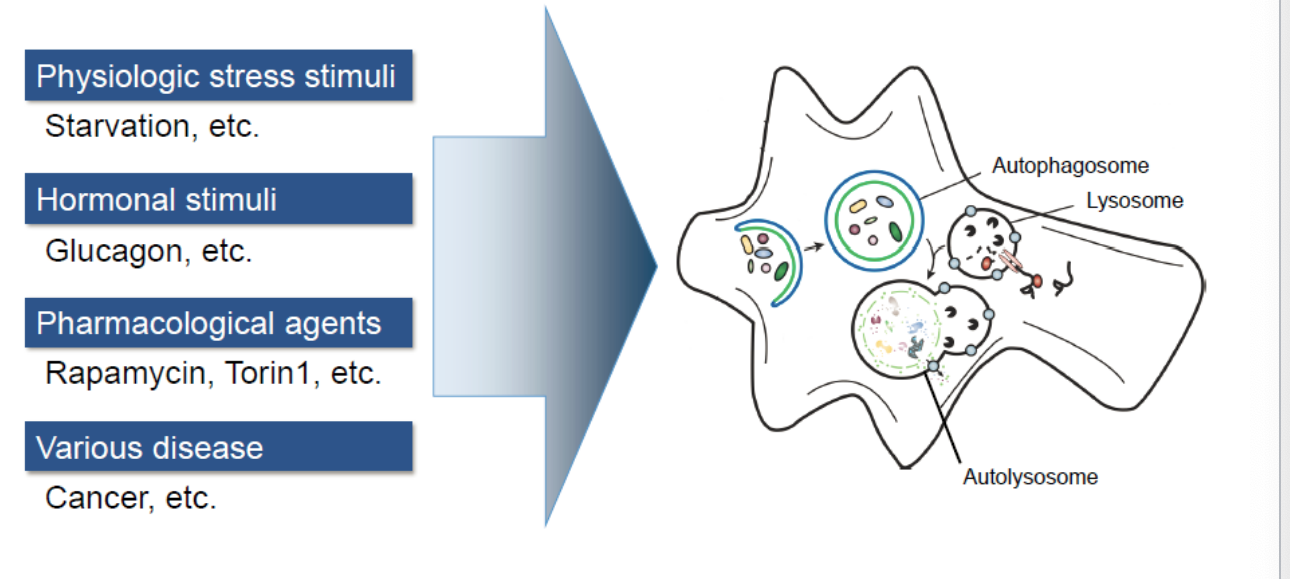
autophagy inducers
physiological stress stimuli
starvation
hormonal stimuli
glucagon
pharmacological agents
rapamycin, torin 1
various disease
cancer
the role of autophagy in human diseases
involves in diseases such as:
developmental defects
crohn’s disease
infection and immunity
neurodegenerative disease
cancer
heart disease
myopathies
ageing
metabolic disorders
pro-tumourigenic vs anti-tumourigenic
role of autophagy in cancer
pro-tumourigenic → PRO TUMOR
sustain the elevated metabolic needs of established tumors by degrading and recycling cellular components biosynthetic and energy generation
prevents cell death
anti-tumourigenic
buffering metabolic and oxidative stress by preventing toxic buildup of misfolded protein and damaged organelles
inhibit cell growth and proliferation
promotes non-apoptotic cell death
protects genomic integrity
chloroquine inhibits lysosome acidification and fusion with the autophagosome, blocking the final step → accumulation of undegraded material and blocked autophagy
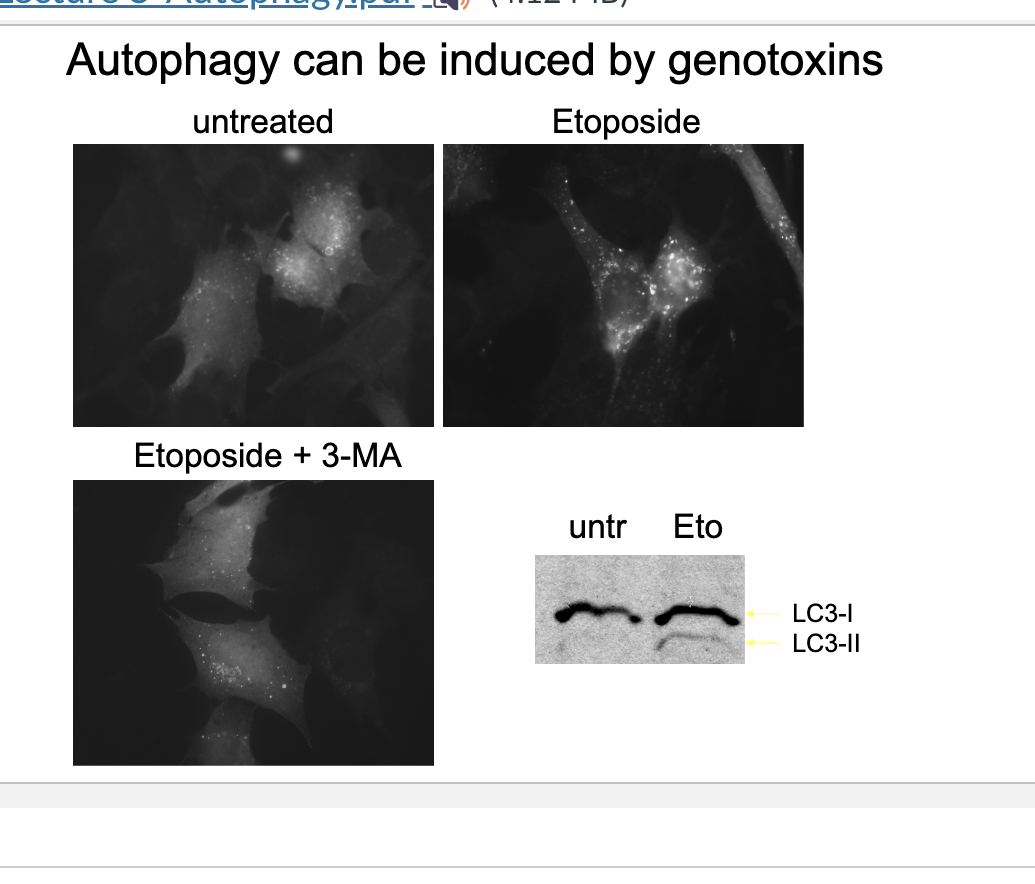
autophagy can be induced by genotoxins
Eto increases LC3-II (membrane bound form)→ confirming autophagy induction
Eto treated cells show many brigh LC3-puncta → autophagosomes forming in response to DNA damage
eto + 3-MA → fewer LC3 puncta → 3MA inhibits autophagy by blocking class III PI3K
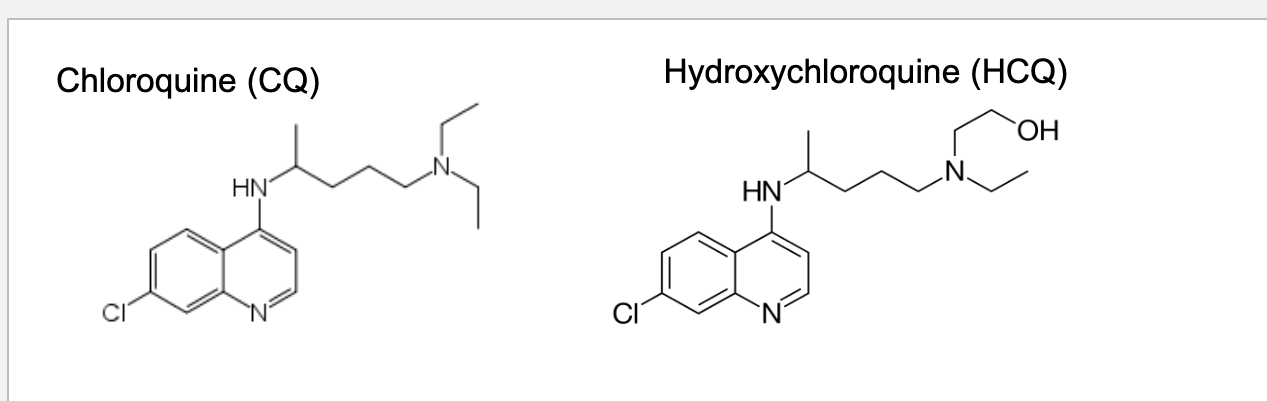
chloroquine (CQ) and hydroxychloroquine (HCQ)
discovered in 1934 at bayer
trialed for treating malaria by the US army during WWII → approved in 1947
deprotonated in the cytosol (physiological pH) → permeable to membrane → trapped in lysosomes where it is protonated at low pH
when protonated, CQ forms a toxic complex with heme (food for malaria parasites in RBCs → NO food b/c toxic so parasite death)
CQ and HCQ raise lysosomal pH (they are weak bases so when they are protonated they TAKE an H+ from lysosome → makes it more basic) which inhibits lysosomal activity → blocks autophagy which is good b/c disease can use autophagy to recycle nutrients and avoid cell death (NO autophagy → no nutrients → die)