USABO
1/866
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
867 Terms
Eukaryote
An organism whose cells contain complex structures enclosed within membranes. The defining membrane bound structure thats sets them apart from prokaryotic cells is the nucleas or nuclear envelope within which the genetic material is carried. All species of large complex species are eukaryotes, including animals, plants and funghi.
Euchromatin
This shows as pale areas in the nucleas under electro magnification. It is a less densly packed form of chromatin; 10% is even less condensed and in this form it can be actively transcribed to produce RNA.
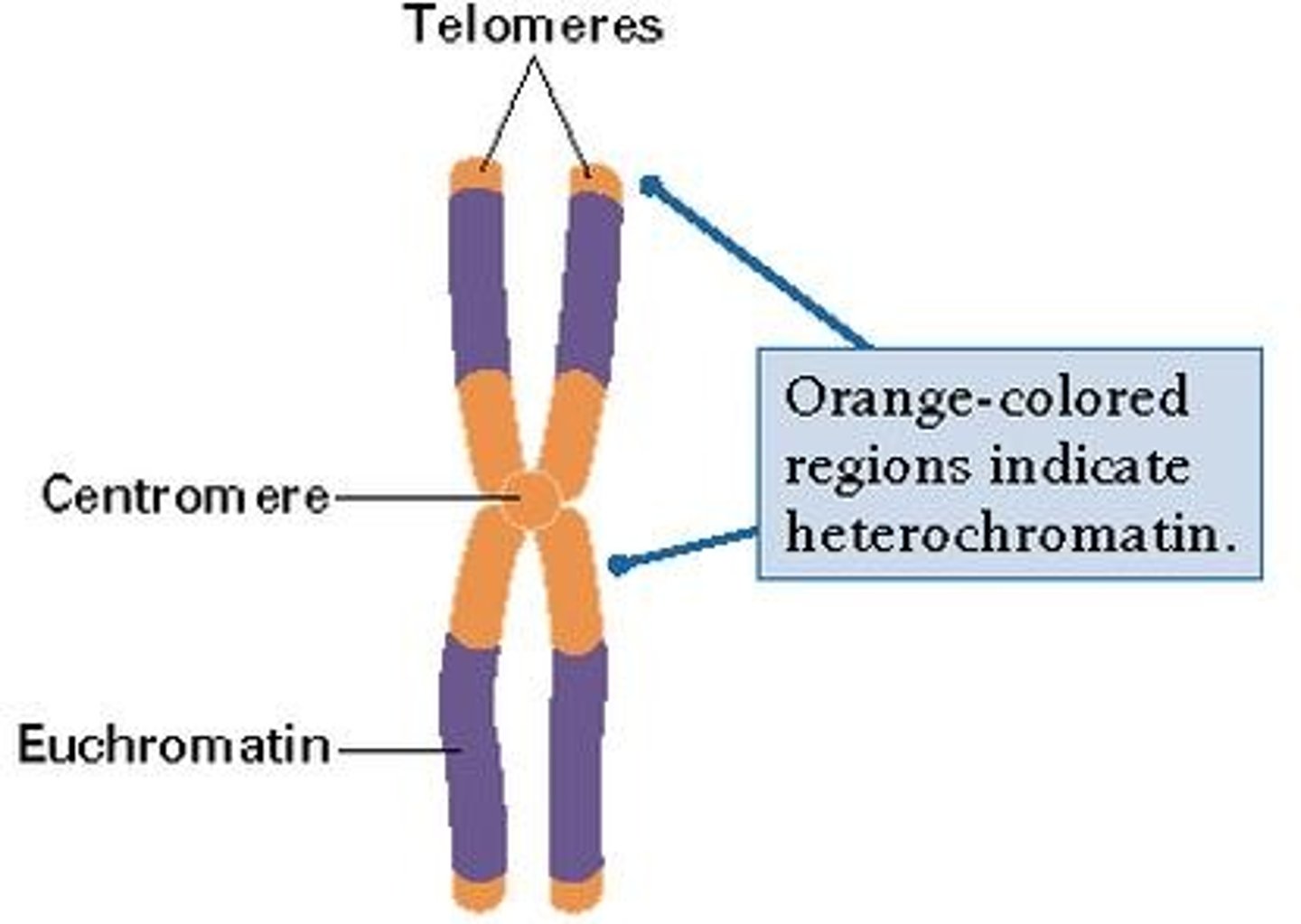
Heterochromatin
This shows as dark areas on the eukaryotic nucleas; it is densly packed chormatin (DNA and protein complex) which cannot be transcribed.
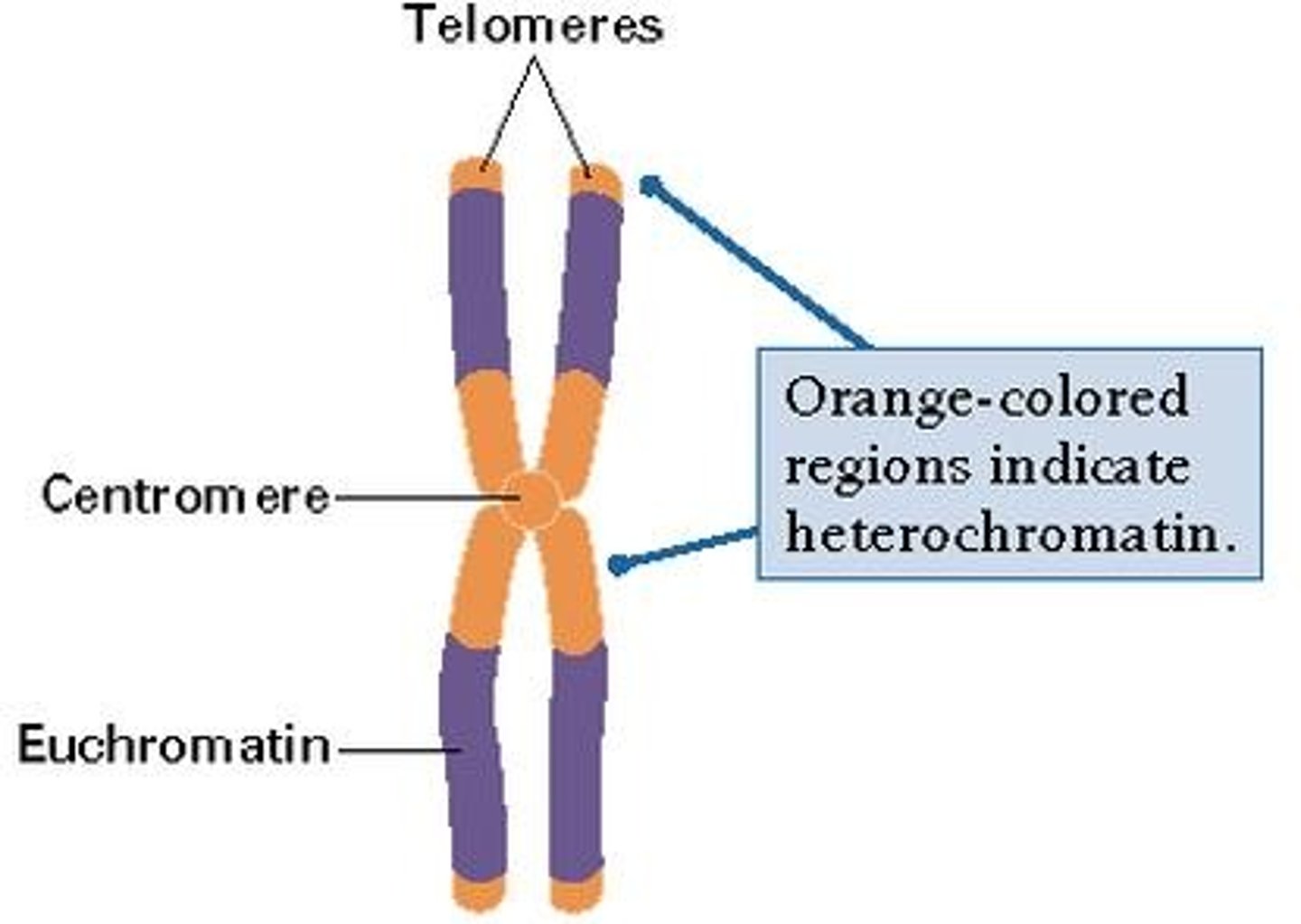
Histones
Special proteins around which DNA is wrapped.
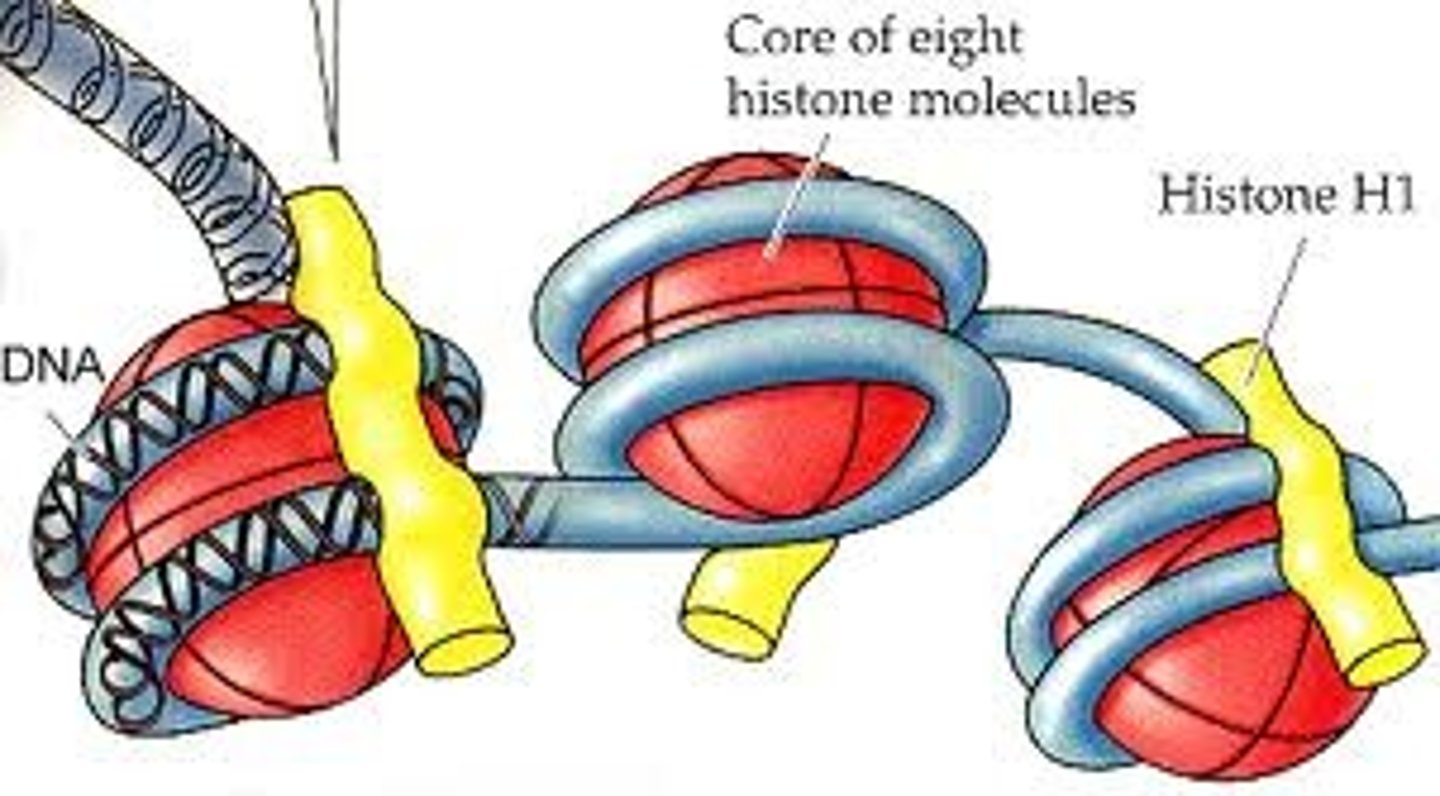
Lamins
Intermediate filament proteins which protect the structure of the nucleas, they polymerize to from a network of filaments that lie just within the nuclear membrane. The network of these filament proteins is called the nuclear lamina.
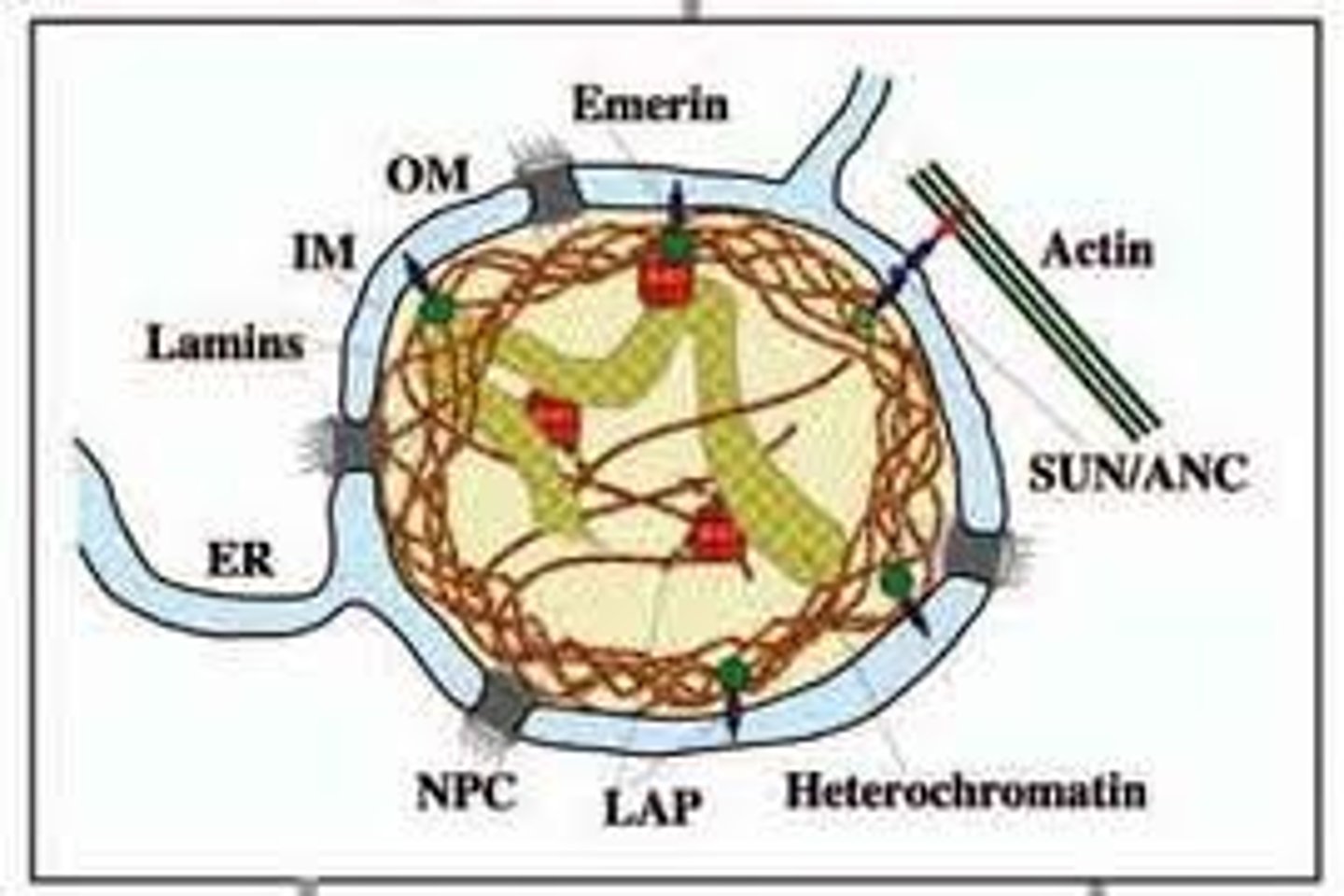
Nuclear Lamina
The network of intermediate nuclear filament proteins (Lamins) which is located just below the nuclear membrane and is linked to the membrane and chromatin.
Nuclear Pores
9nm (approx) gaps in the nuclear envelope that allow the passage of RNA and ribosomes out of the nucleas and the entry of selected small proteins and small water soluble molecules.
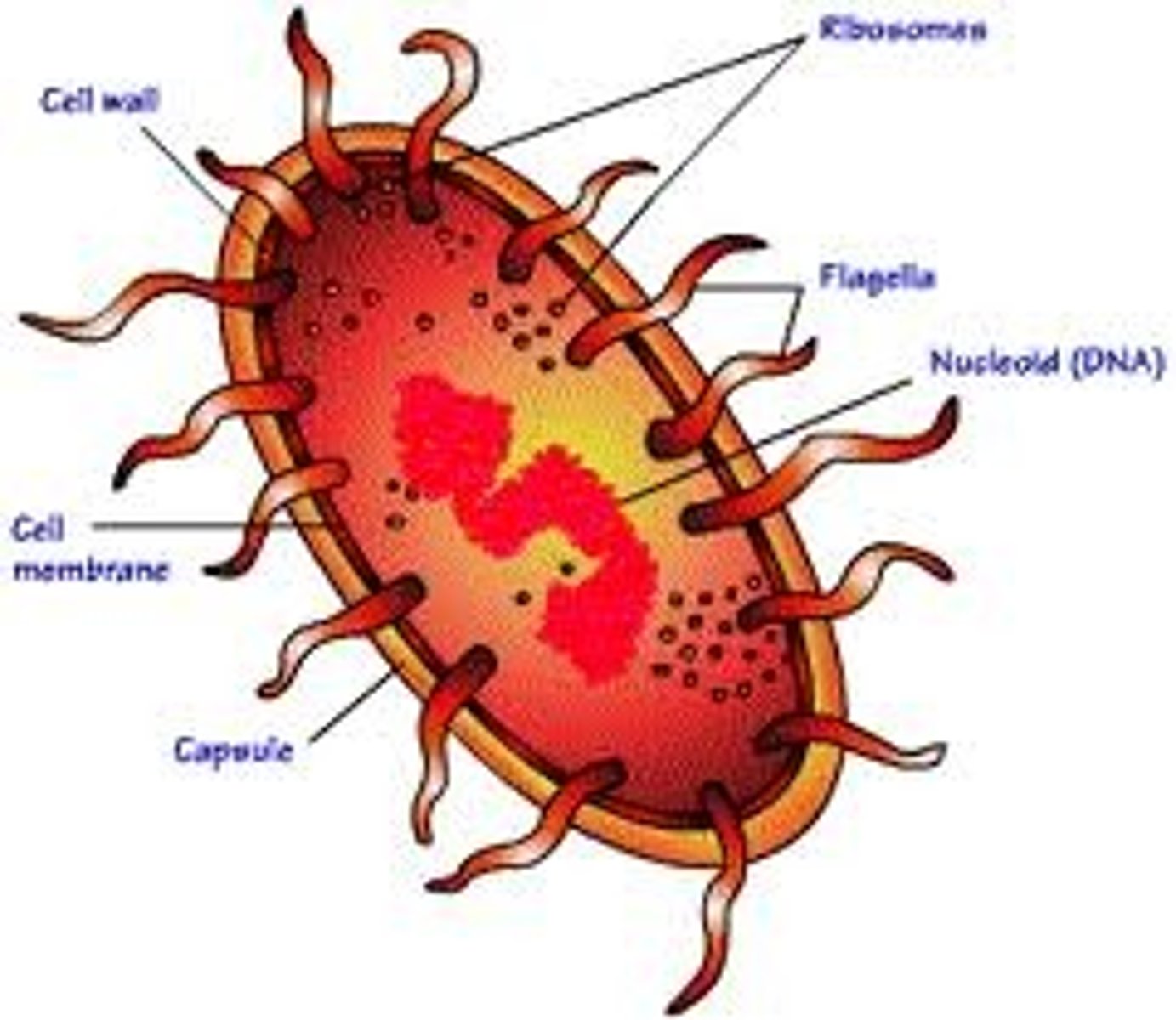
Prokarayotes
An organism of the kingdom of Monera, comprising the bacteria and cyanobacteria. Characterised by the abscence of a distinct, membrane bound nucleas or membrane bound organelles and by DNA that is not organised in to chromosomes. Also called moneran.
Protoctist
Any of various unicellular eukaryotic organisms and their multicellular, coenocytic or colocial descendants that belong to the kingdom of Protocista according to some taxonomic systems. The protoctists include protozoans, slime moulds, various algae and other groups. In many new classification systems, all proctists are considered protists.
Signal Sequence
Affectionately known as the 'address label' of a polypeptide. A short (3-60 amino acids long) peptide chain that directs the transport of a protein. These may also be called targeting signals, signal peptides, transit peptides, or localization signals.
The amino acid sequences of these direct proteins (which are synthesized in the cytosol) to certain organelles such as the nucleus, mitochondrial matrix, endoplasmic reticulum, chloroplast, apoplast and peroxisome. Some signal peptides are cleaved from the protein by signal peptidase after the proteins are transported.
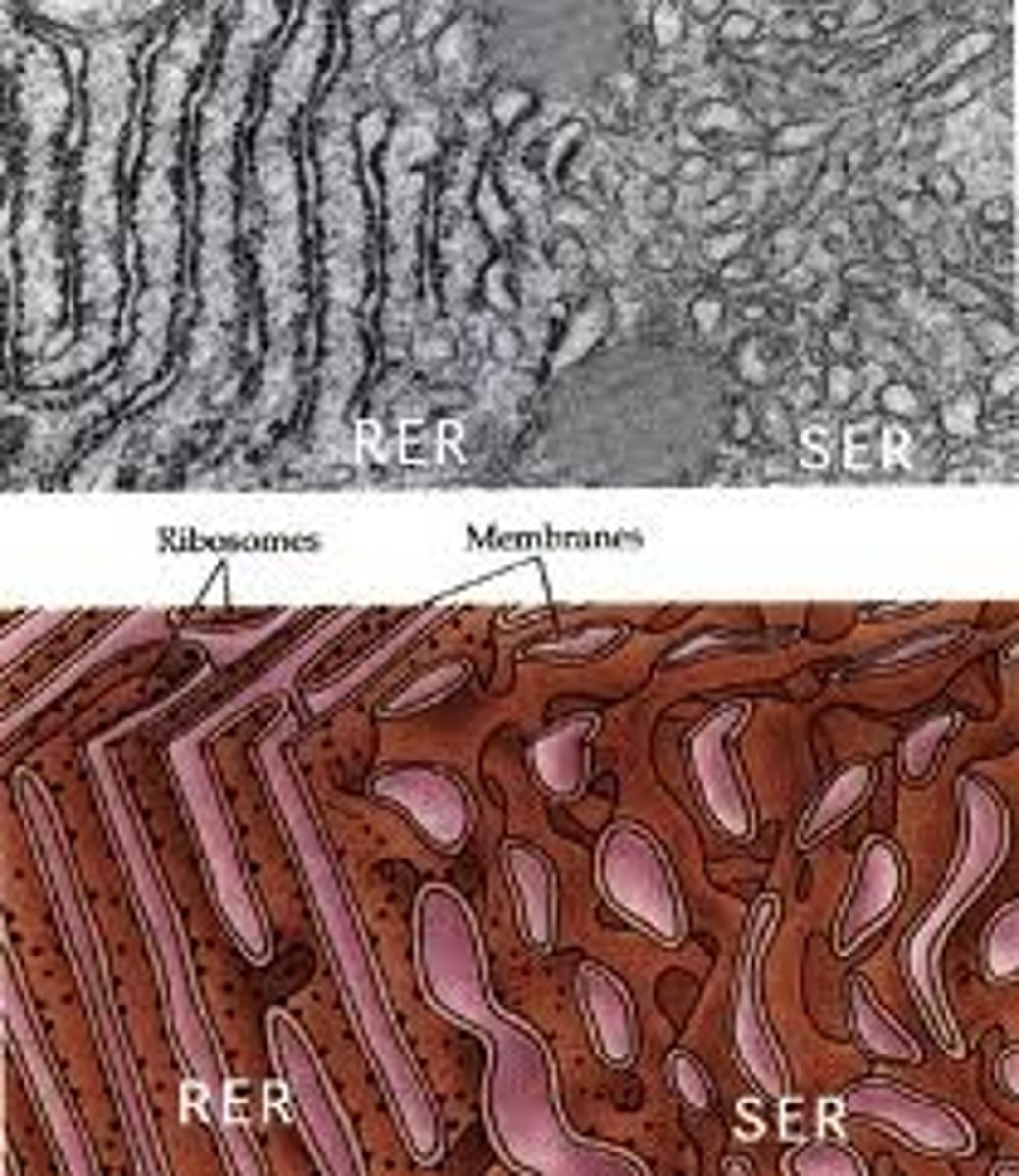
Endoplasmic Reticulum
The endoplasmic reticulum (ER) is a eukaryotic organelle that forms an interconnected network of tubules, vesicles, and cisternae within cells. Rough endoplasmic reticula synthesize proteins, while smooth endoplasmic reticula synthesize lipids and steroids, metabolize carbohydrates and steroids (but not lipids), and regulate calcium concentration, drug metabolism, and attachment of receptors on cell membrane proteins. Sarcoplasmic reticula solely regulate calcium levels.
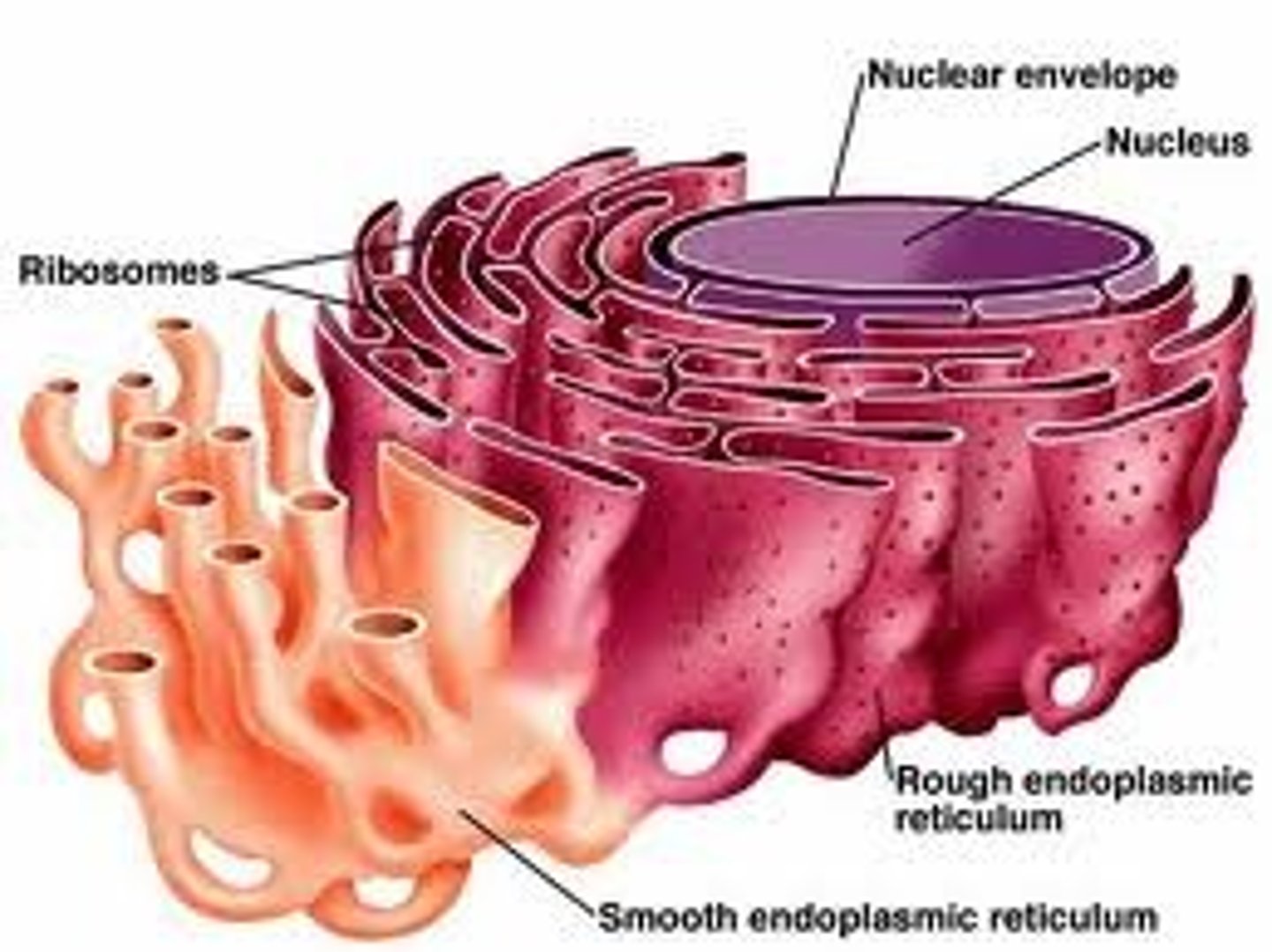
Smooth endoplasmic reticulum
Cell organelle responsible for attachment of receptors on cell membrane proteins, synthesizing lipids and steroids, metabolizing carbohydrates and steroids (but not lipids) and regulating calcium concentration and drug metabolism.
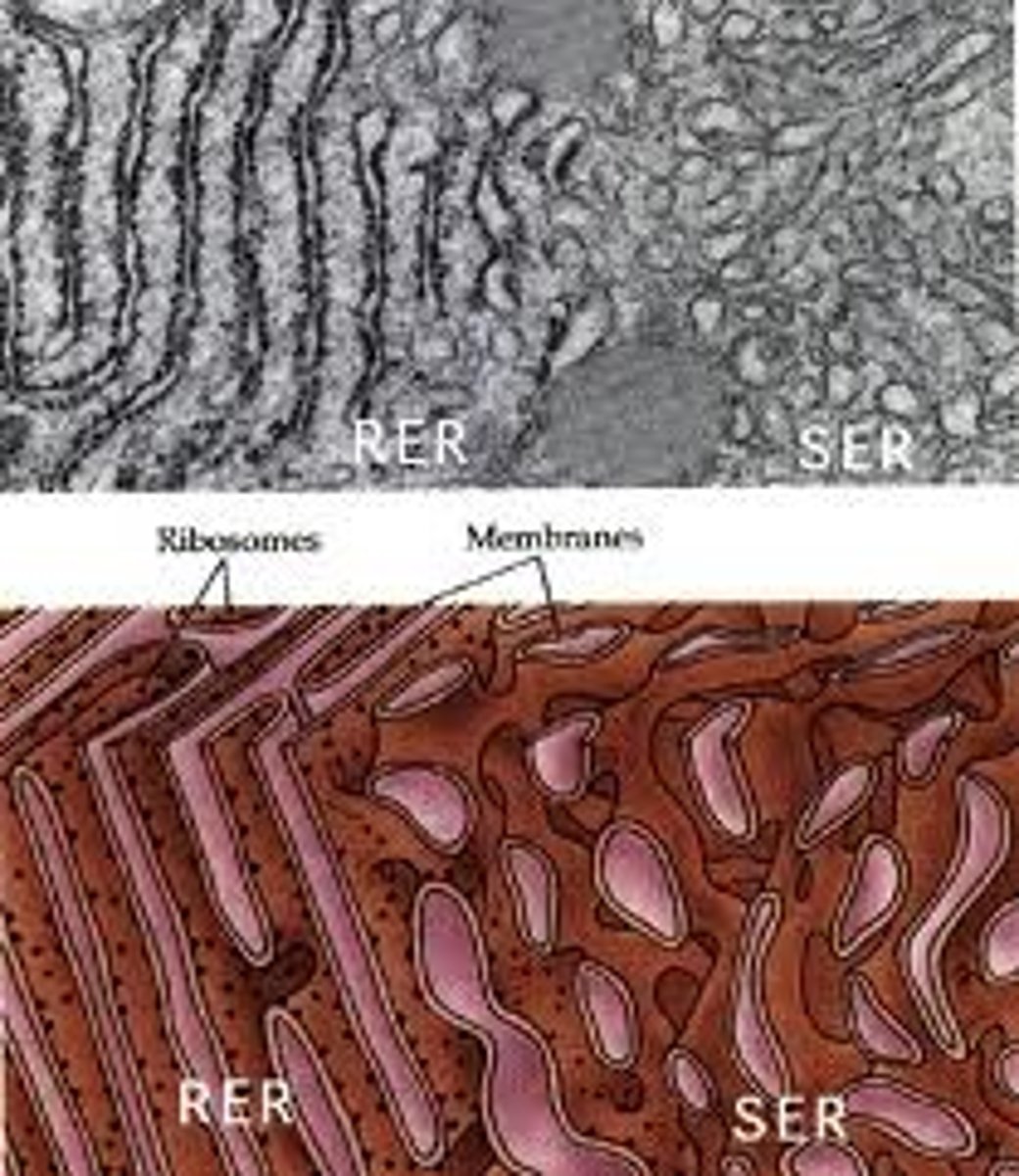
Rough endoplasmic reticulum
Cell organelle responsible for synthesizing proteins.
Catalytic site
In molecular biology this site is part of an enzyme where substrates bind and undergo a chemical reaction.The majority of enzymes are proteins but RNA enzymes called ribozymes also exist. The active site of an enzyme is usually found in a cleft or pocket that is lined by amino acid residues (or nucleotides in ribozymes) that participate in recognition of the substrate. Residues that directly participate in the catalytic reaction mechanism are called active site residues.
Vesicles
Small lipid-bounded spheres which transport proteins, glyco proteins and newly synthesized lipids (which are imbedded in the sphere itself) from the endoplasmic reticulum to the Golgi Apparatus or from the Golgi apparatus to another destination. They move short distances by the process of difussion, moving long distances requires the assistance of proteins associated with microtubules.
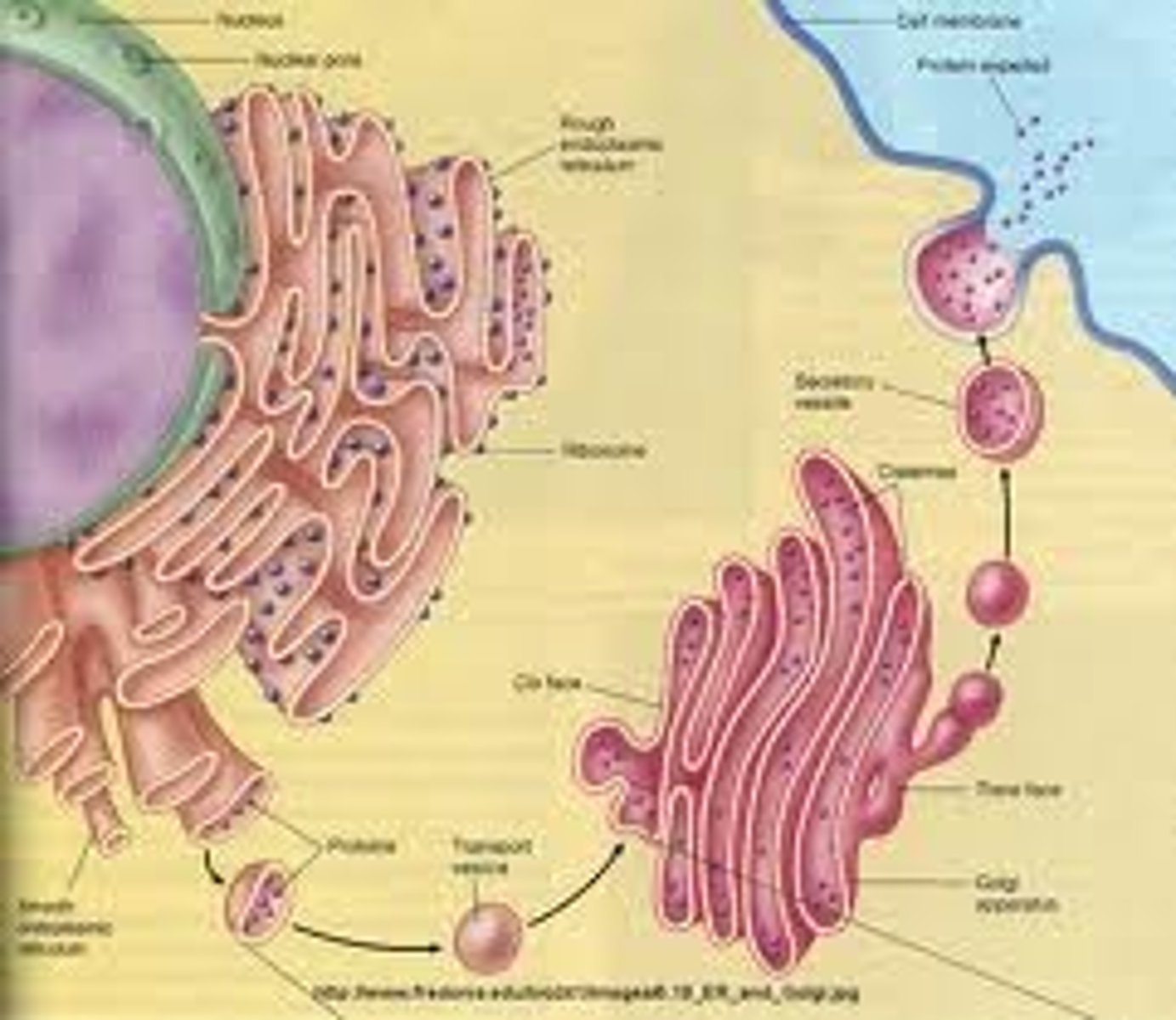
Golgi apparatus
This is an organelle found in all eukaryotic cells.It was identified in 1897 by the Italian physician Camillo Golgi, after whom it is named. It processes and packages proteins after their synthesis and before they make their way to their destination; it is particularly important in the processing of proteins for secretion. Its size varies in different types of cells depending on cell function; a hormone secreting cell will contain a far larger version of this organelle than a muscle cell for example. It also forms a part of the cellular endomembrane system.
Constitutive release
The constant release of small amounts of a substances from the cell membrane.
Regulated release
The release of substances from a cell membrane only when specific conditions exist. A good example is the release of gastrointestinal hormones and digestive enzymes in response to food.
Exocytosis
The process by which substances are exported from a cell.
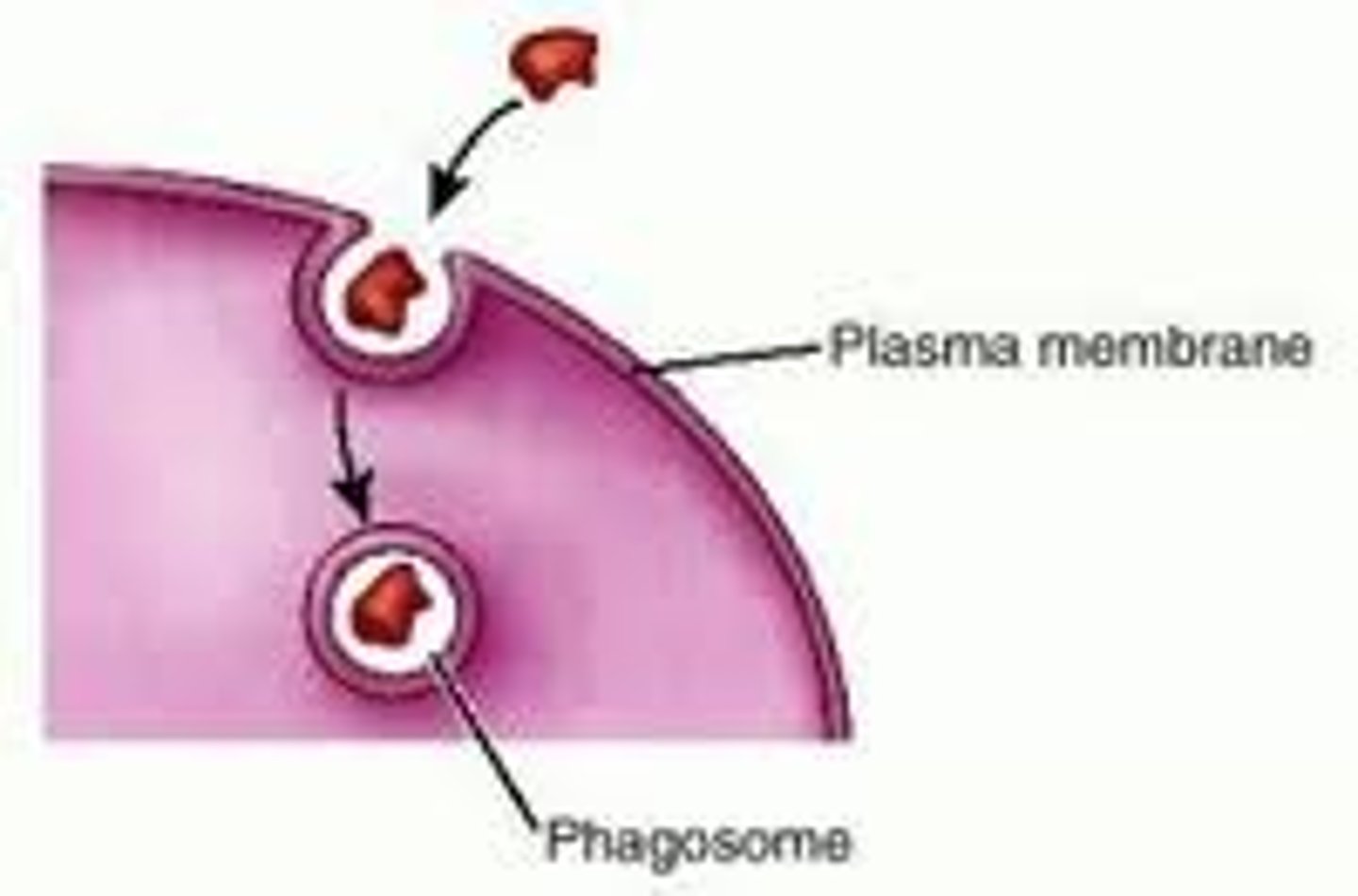
Phagocytosis/Endocytosis
The process by which substances or pathogens are taken in to a cell by engulfment by a vesicular structure surrounded by cell membrane.
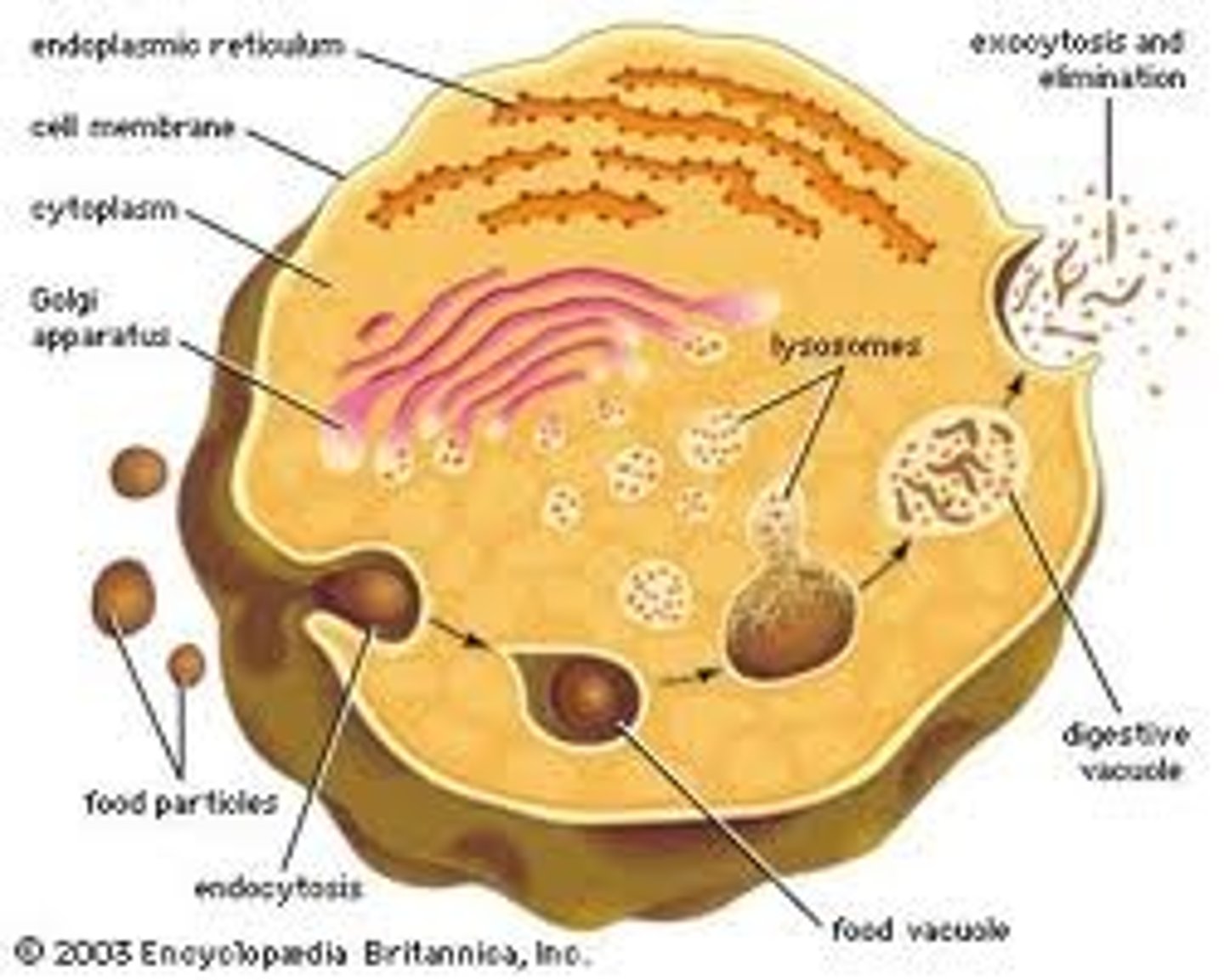
Lysosomes
Small organelles which contain digestive enzymes with an internal pH of around 5. They are responsible for breaking down large molecules taken in to the cell by phagocytosis and also for the breaking down of old organelles.
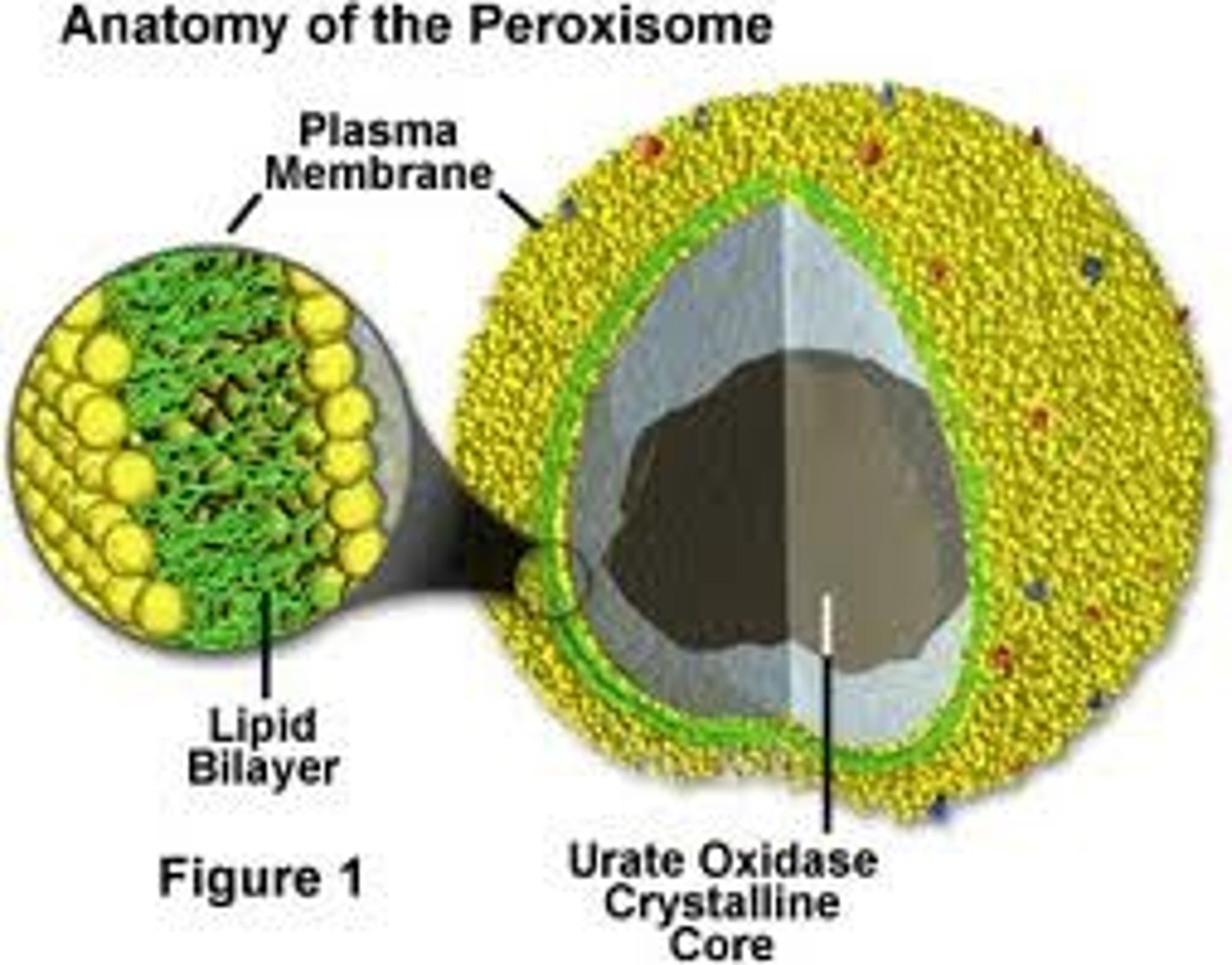
Peroxisomes
Organelles that are plentiful in liver cells and adipocytes, responsible for breaking down fatty acids and amino acids in to hydrogen peroxide (among other things) via the action of an enzyme known as catalayse.
MItochondrian
Sausage shaped organelles with a double membrane. The inner membrane folds in to cristae. This organelle plays a fundamental role in the production of ATP in eukarayote cells and they are abundant in cells which require high amounts of energy such as muscle cells.
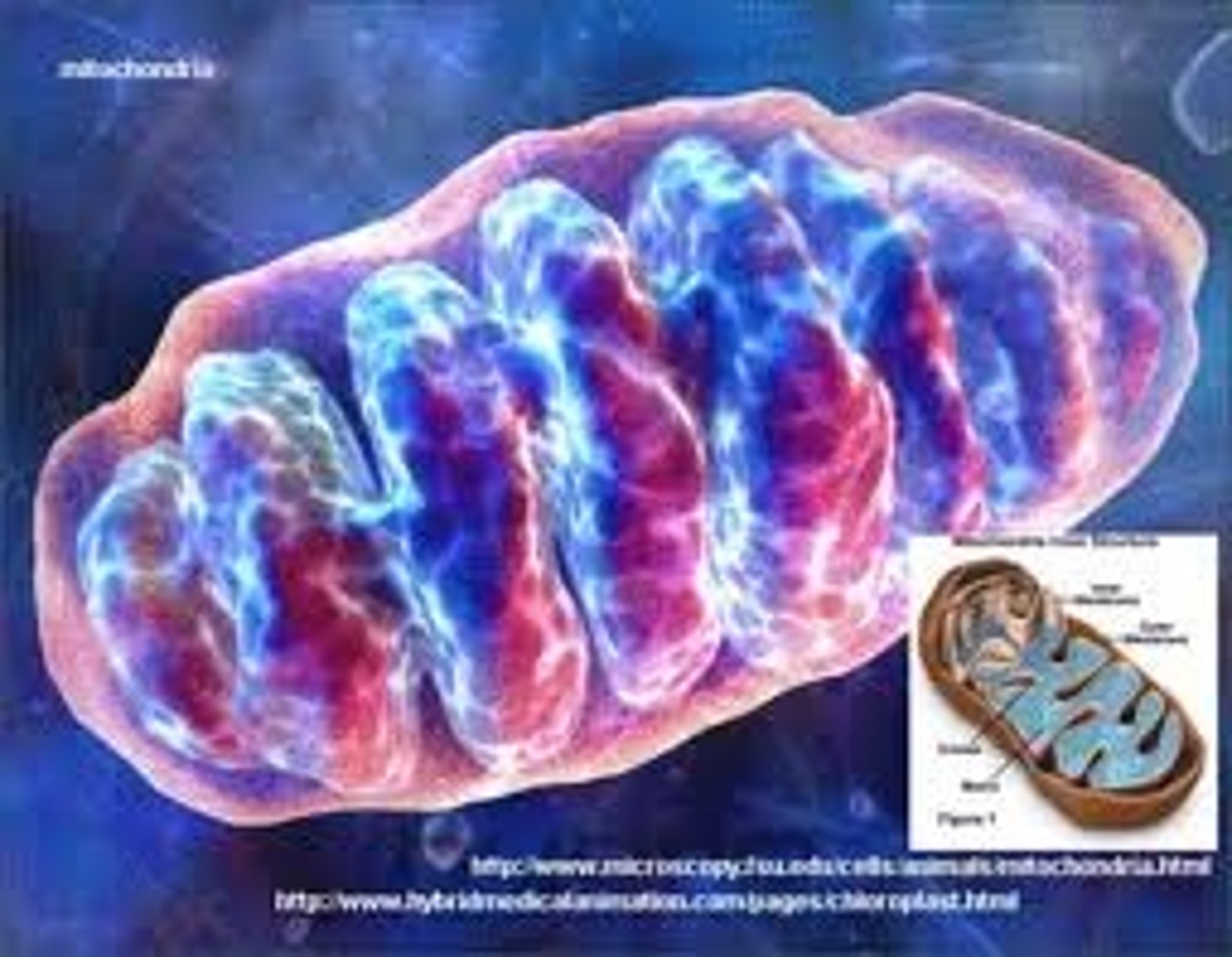
Cristae
Internal compartments formed by the inner membrane of a mitochondrion. They are studded with proteins, including ATP synthase and a variety of cytochromes. The maximum surface for chemical reactions to occur is within the mitochondria. This allows cellular respiration (aerobic respiration since the mitochondrion requires oxygen) to occur.
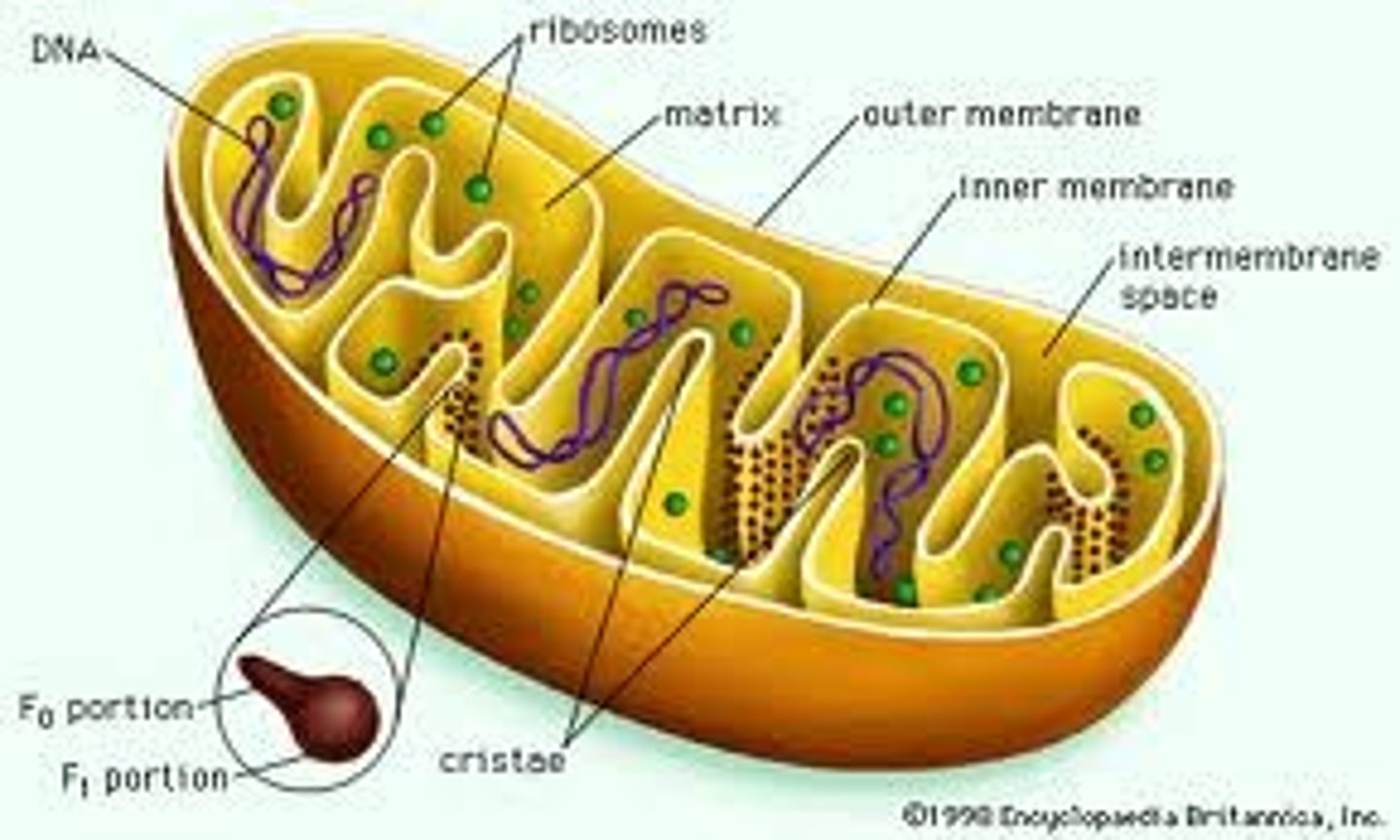
Mitochondrial DNA
The DNA located in mitochondria
It can be regarded as the smallest chromosome, and was the first significant part of the human genome to be sequenced. In most species, including humans, mtDNA is inherited solely from the mother. The DNA sequence of mtDNA has been determined from a large number of organisms and individuals (including some organisms that are extinct), and the comparison of those DNA sequences represents a mainstay of phylogenetics, in that it allows biologists to elucidate the evolutionary relationships among species. It also permits an examination of the relatedness of populations, and so has become important in anthropology and field biology.
Mitochondrial Matrix
This matrix contains soluble enzymes that catalyze the oxidation of pyruvate and other small organic molecules.
It also contains the mitochondria's DNA and ribosomes. The word "matrix" stems from the fact that this space is viscous, compared to the relatively aqueous cytoplasm.
Cell Wall
An extracellular structure in plants which is rigid and surrounds the cell membrane giving it shape and support, like playtex for plants lol! It is primarily composed of cellulose which is a polysaccharide.
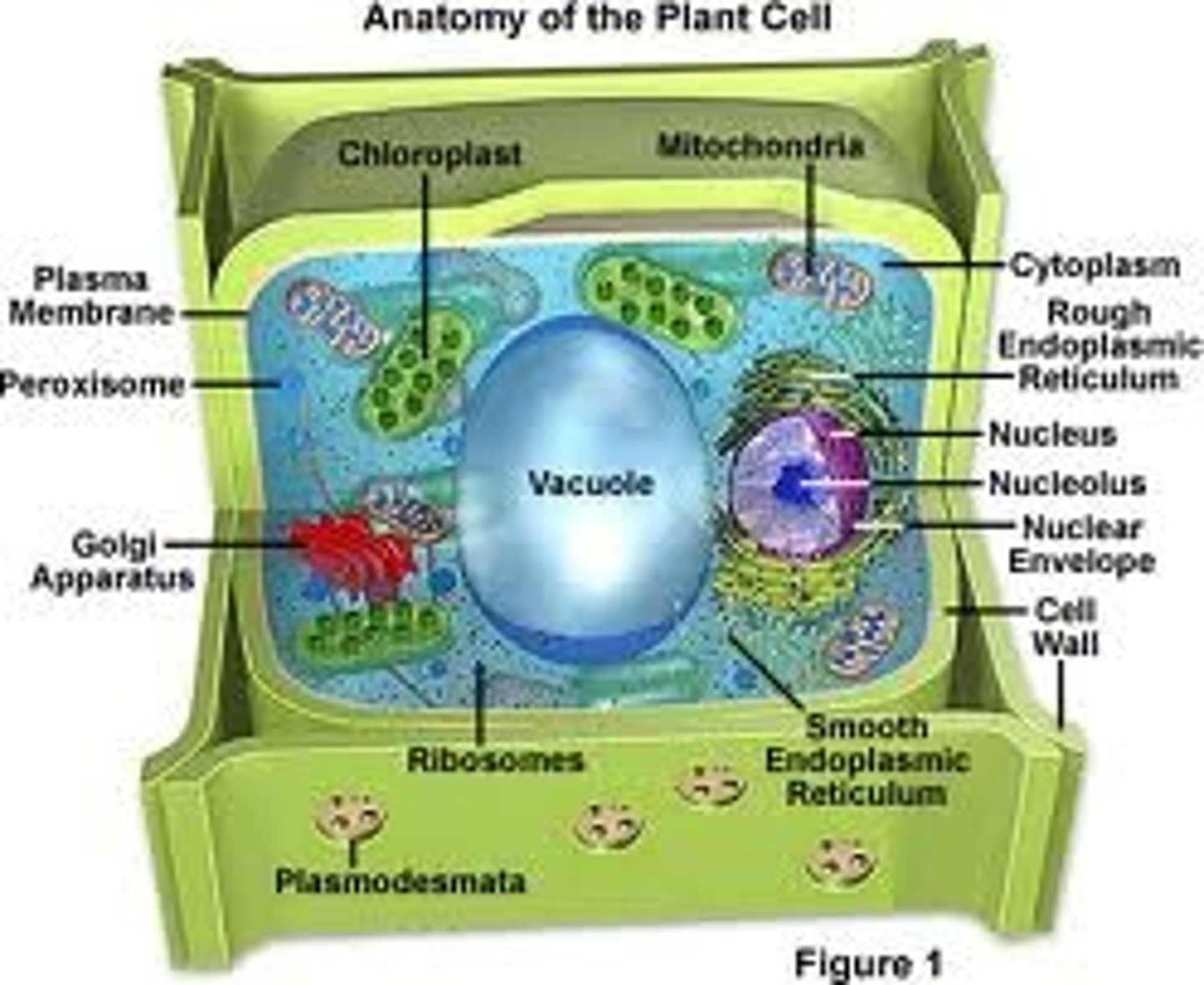
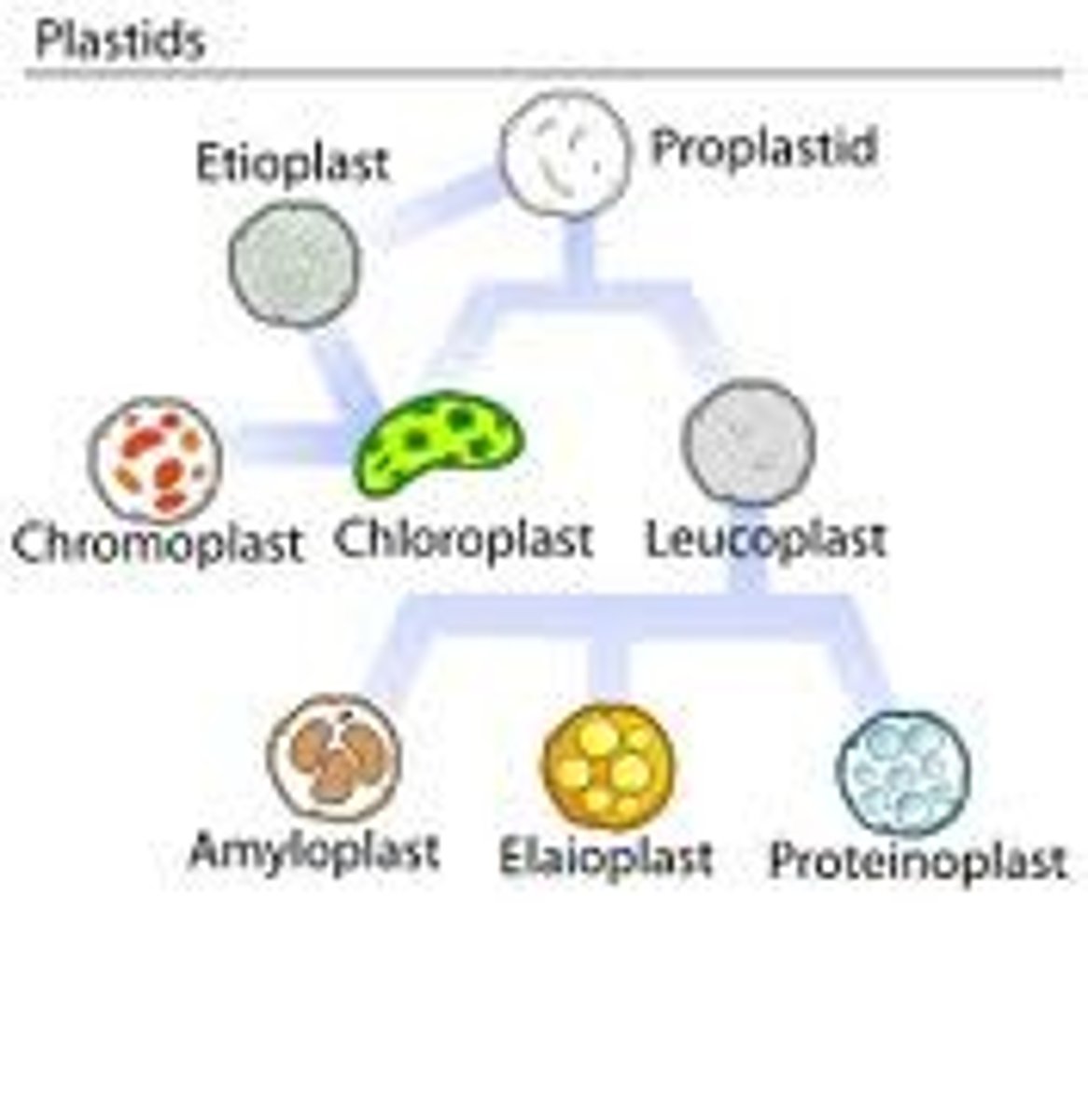
Plastids
These are major organelles found in the cells of plants and algae. They are the site of manufacture and storage of important chemical compounds used by the cell, often containing pigments used in photosynthesis. The types of pigments present can change or determine the cell's color.These organelles are responsible for photosynthesis, storage of products like starch and for synthesis. All types are derived from proplastids (formerly "eoplasts", eo-: dawn, early), which are present in the meristematic regions of the plant. Proplastids and young chloroplasts commonly divide, but more mature chloroplasts also have this capacity.
Chloroplasts
These plant organelles have their own DNA like mitochondria. They are normally larger than mitochondria though and they also have a three membrane system.
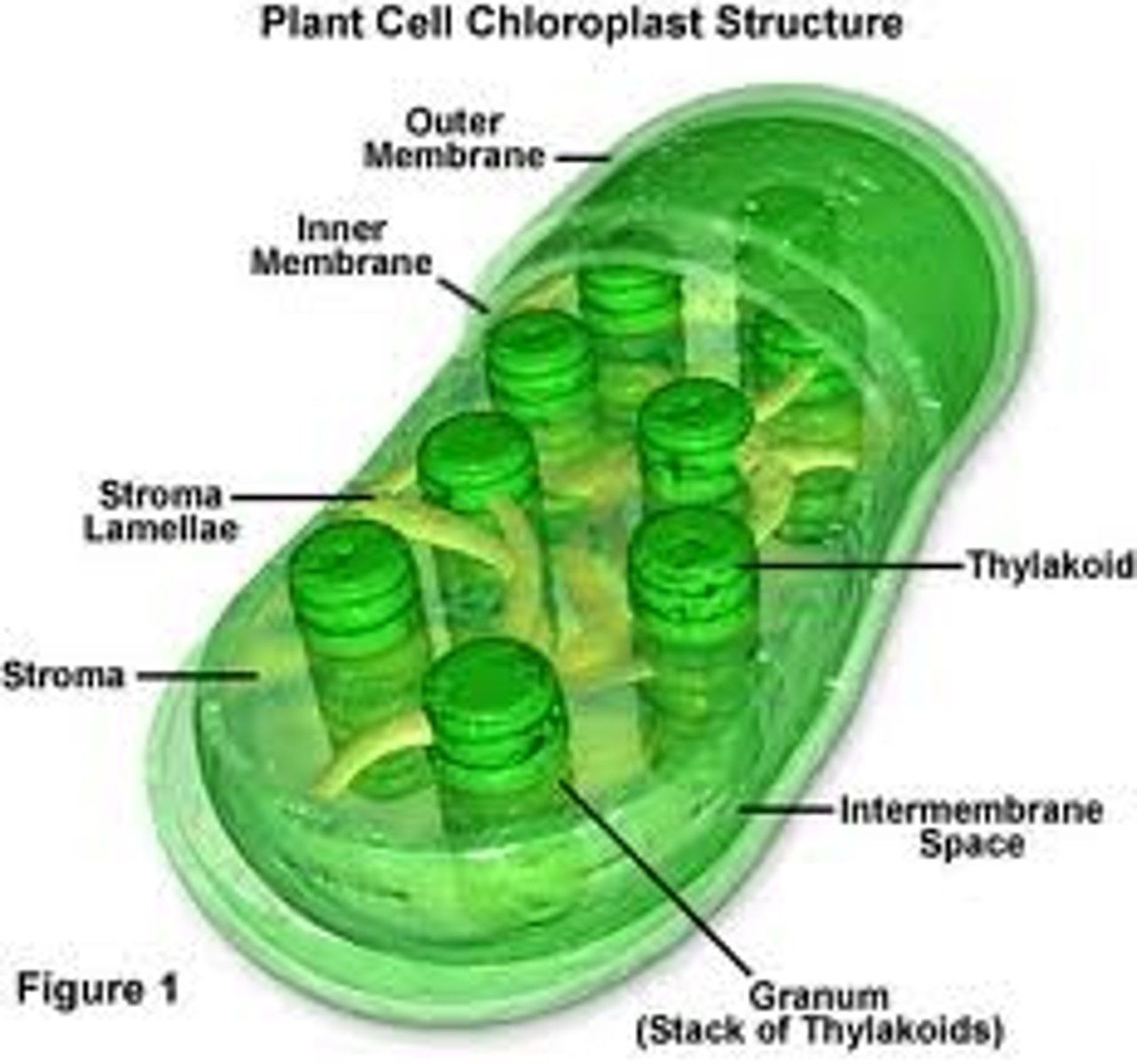
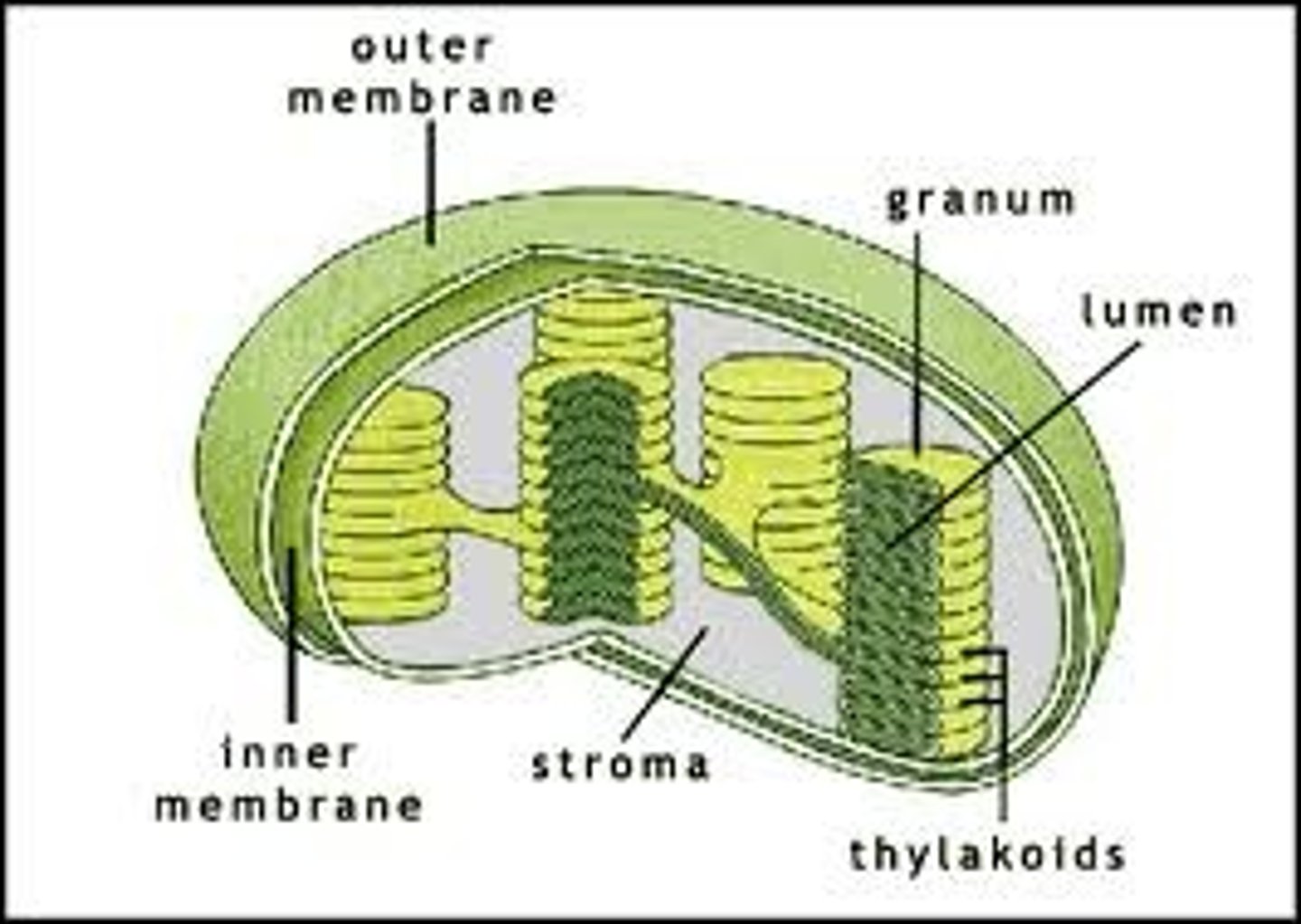
Thylakoid
A thylakoid is a membrane-bound compartment inside chloroplasts and cyanobacteria. They are the site of the light-dependent reactions of photosynthesis. Thylakoids consist of a thylakoid membrane surrounding a thylakoid lumen. Chloroplast thylakoids frequently form stacks of disks referred to as grana (singular: granum). Grana are connected by intergrana or stroma thylakoids, which join granum stacks together as a single functional compartment.
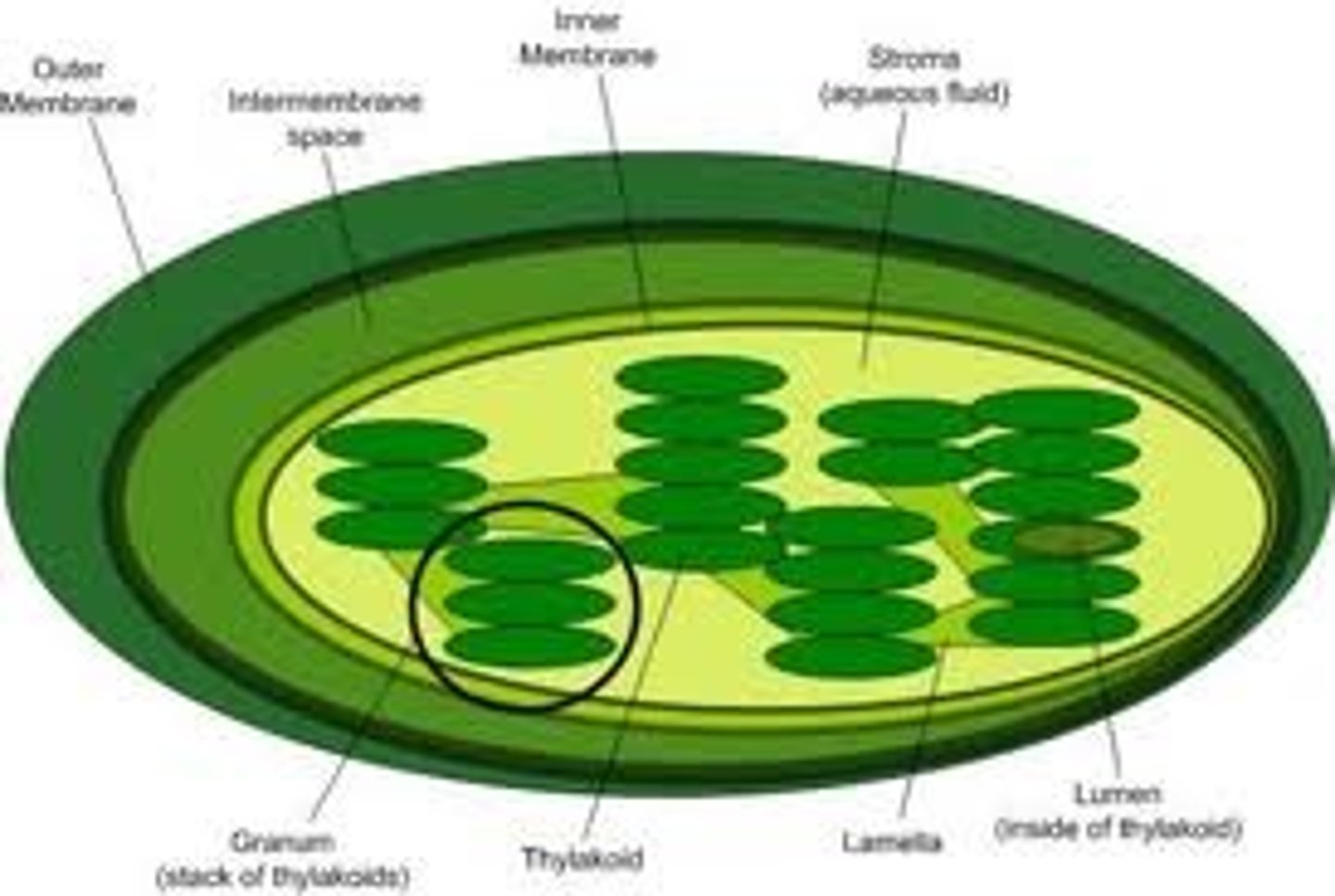
Stroma
Stroma (fluid), the fluid in between grana, where carbohydrate formation reactions occur in the chloroplasts of plant cells photosynthesizing
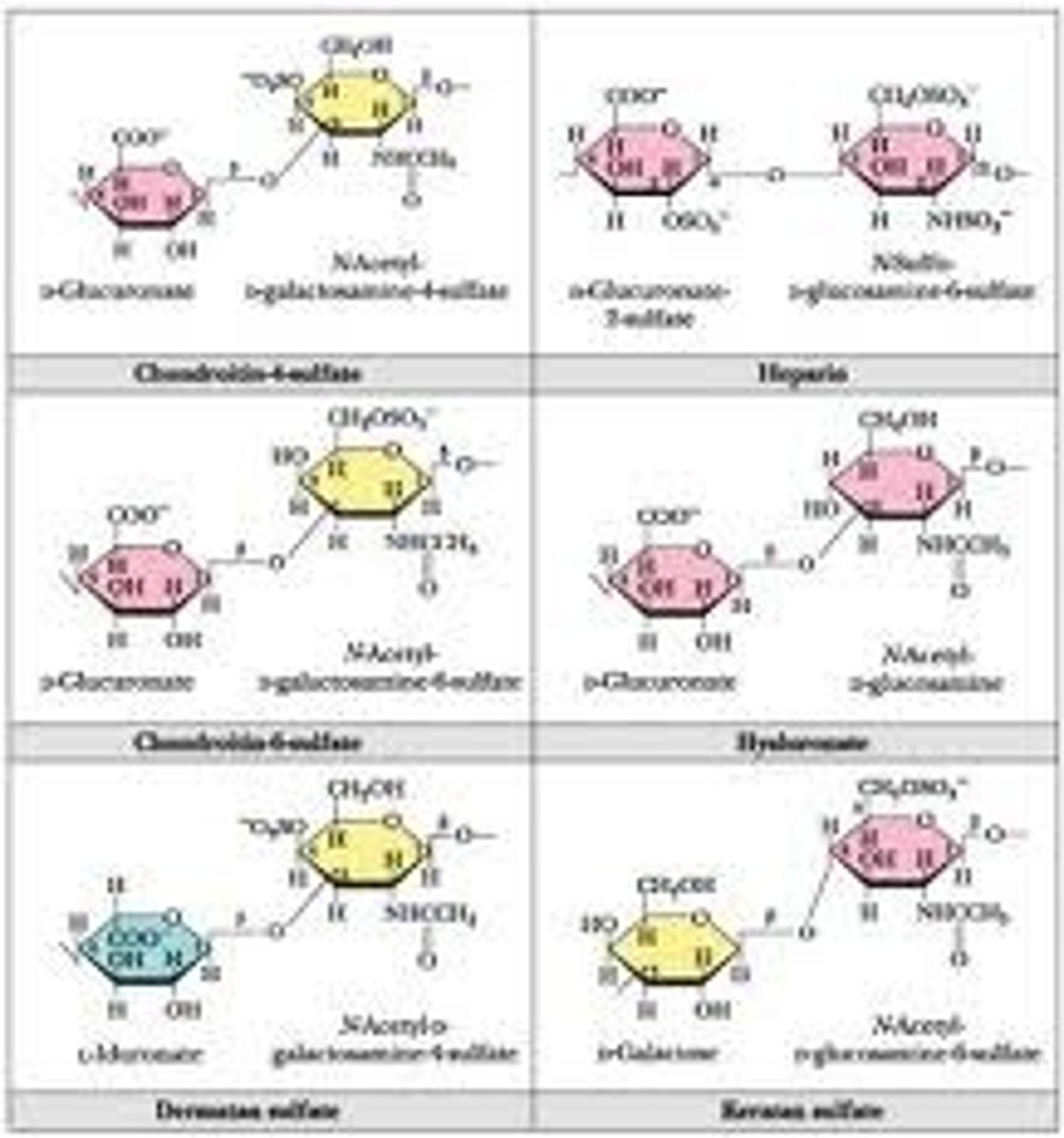
Glycosaminoglycans (GAG's)
Also known as mucopolysaccharides these are long unbranched polysaccharides consisting of a repeating disaccharide unit. The repeating unit consists of a hexose (six-carbon sugar) or a hexuronic acid, linked to a hexosamine (six-carbon sugar containing nitrogen). These are the major component of the 'gel' found in the extracellular matrix of tissue. They are negatively charged and thus attract ions, especially sodium which aids diffusion of water in to the tissue, giving tissue it's compression resistance.
Extracellular Matrix
This matrix is secreted by cells and laid down externally and it's properties vary enormously depending on it's chemical composition and which tissue is being examined. In some cells it acts as cement or scaffolding. In plants it can be associated with individual cells.
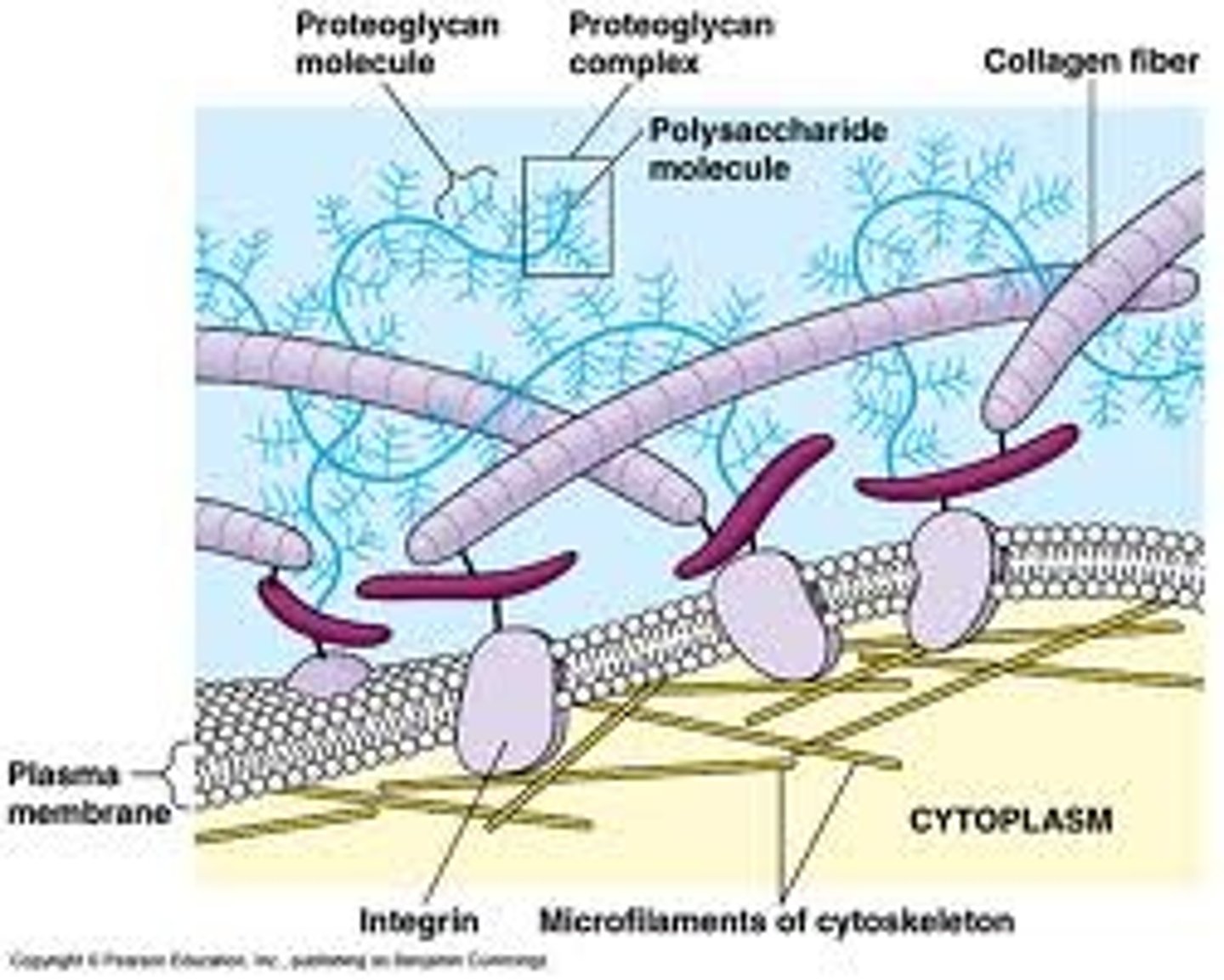
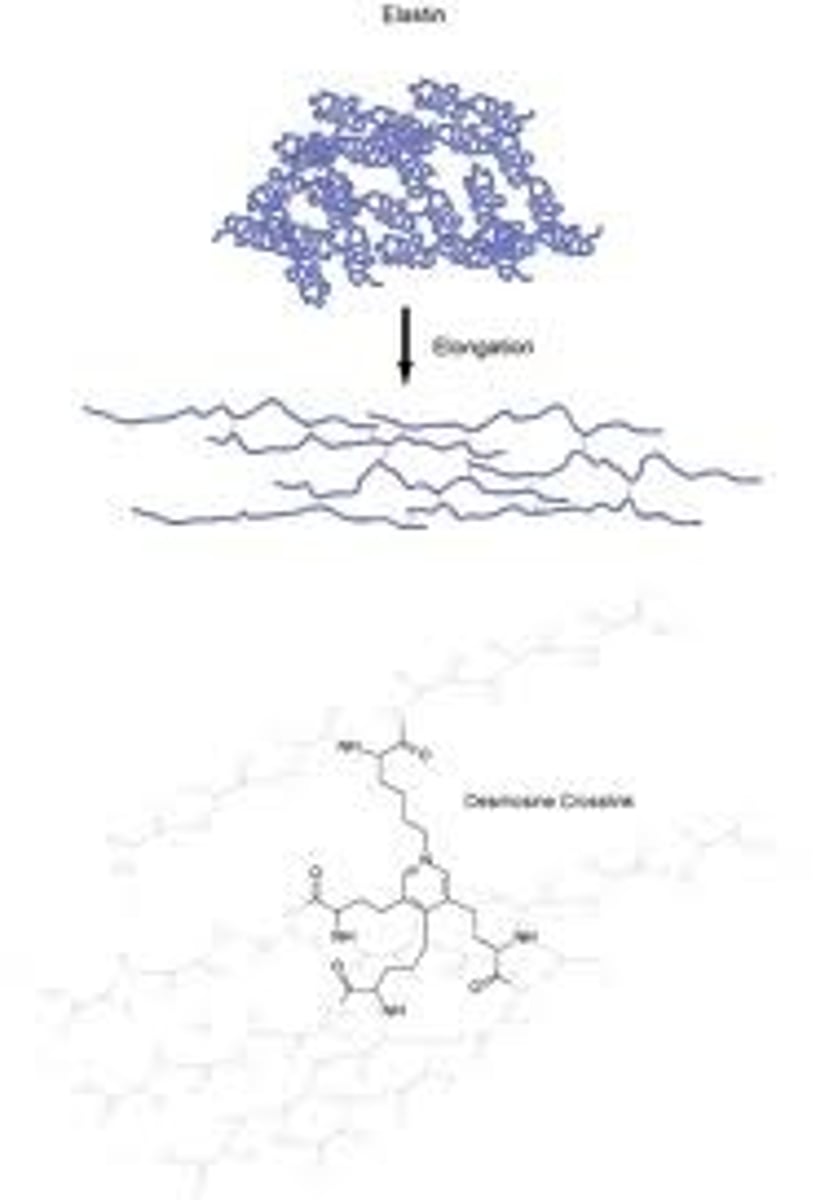
Elastin
An flexible protein found in the extra cellular matrix of blood vessels.
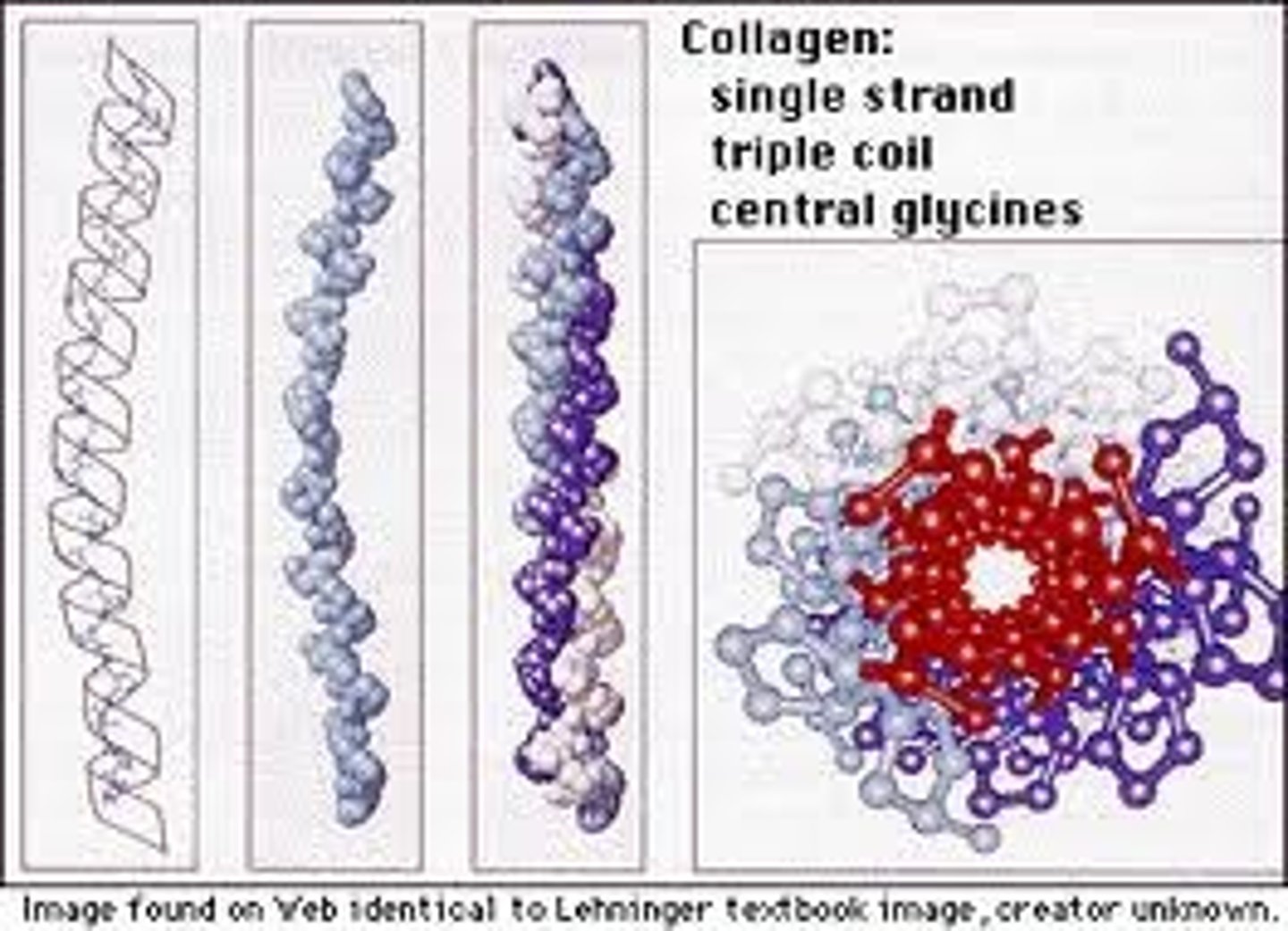
Collagen
A strong protein which can provide strength and/or flexibility found in animal tissue.
Connective Tissue
The name often given to tissue that contain a large proportion of extracellular matrix. In this tissues the cells that are secreting the materials are often quite far from each other.
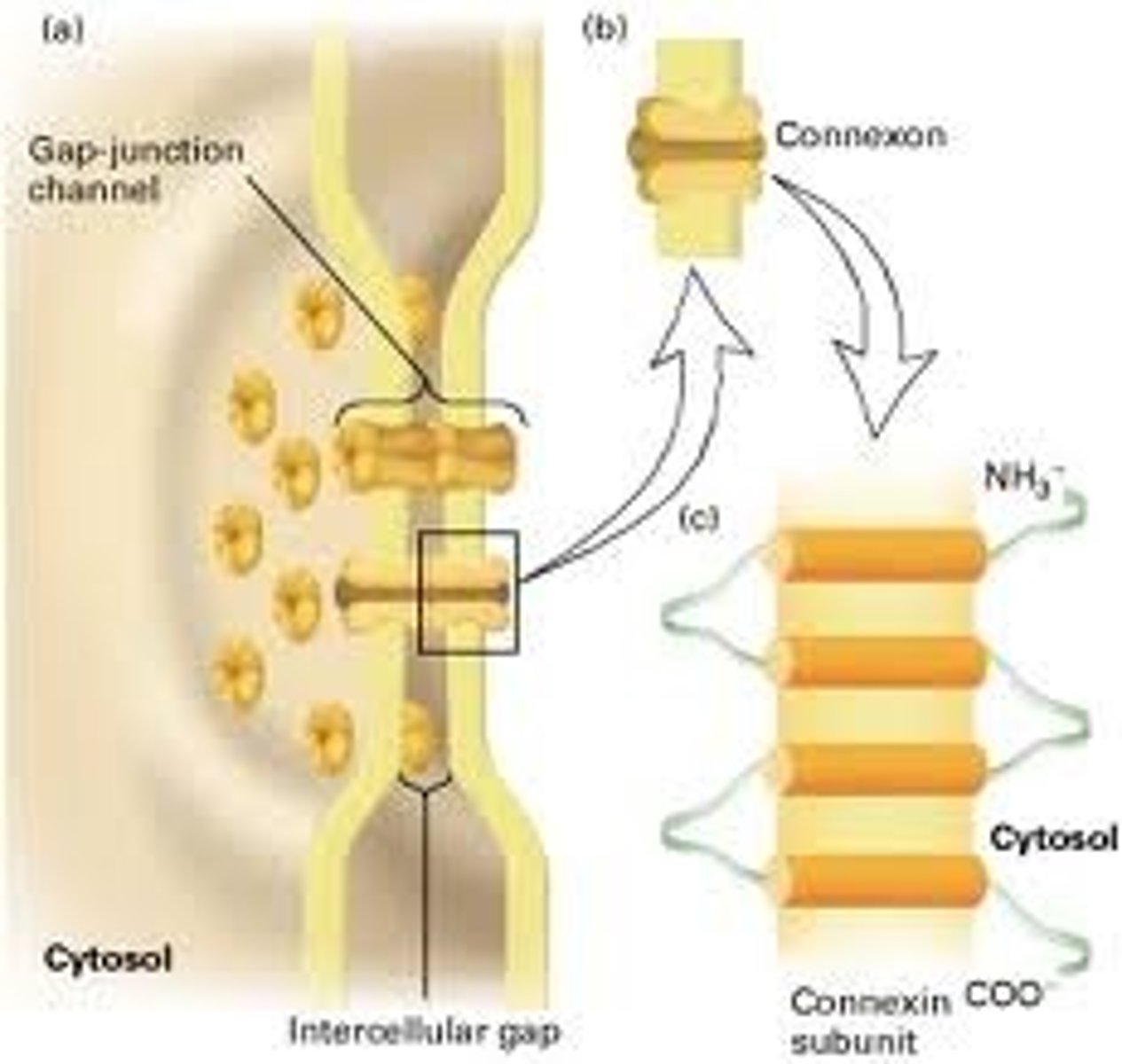
Gap junction
A specialised type of cell junction, an example of which is the smooth muscle of the intestine. The gap's allow for effective transmission of molecules and electrical activity between the cells.
Tight junctions
These cell junctions are linked very closely and prevent movement of membrane proteins, in the skin for example or in the role mainting the polarity of the cells of the intestine.
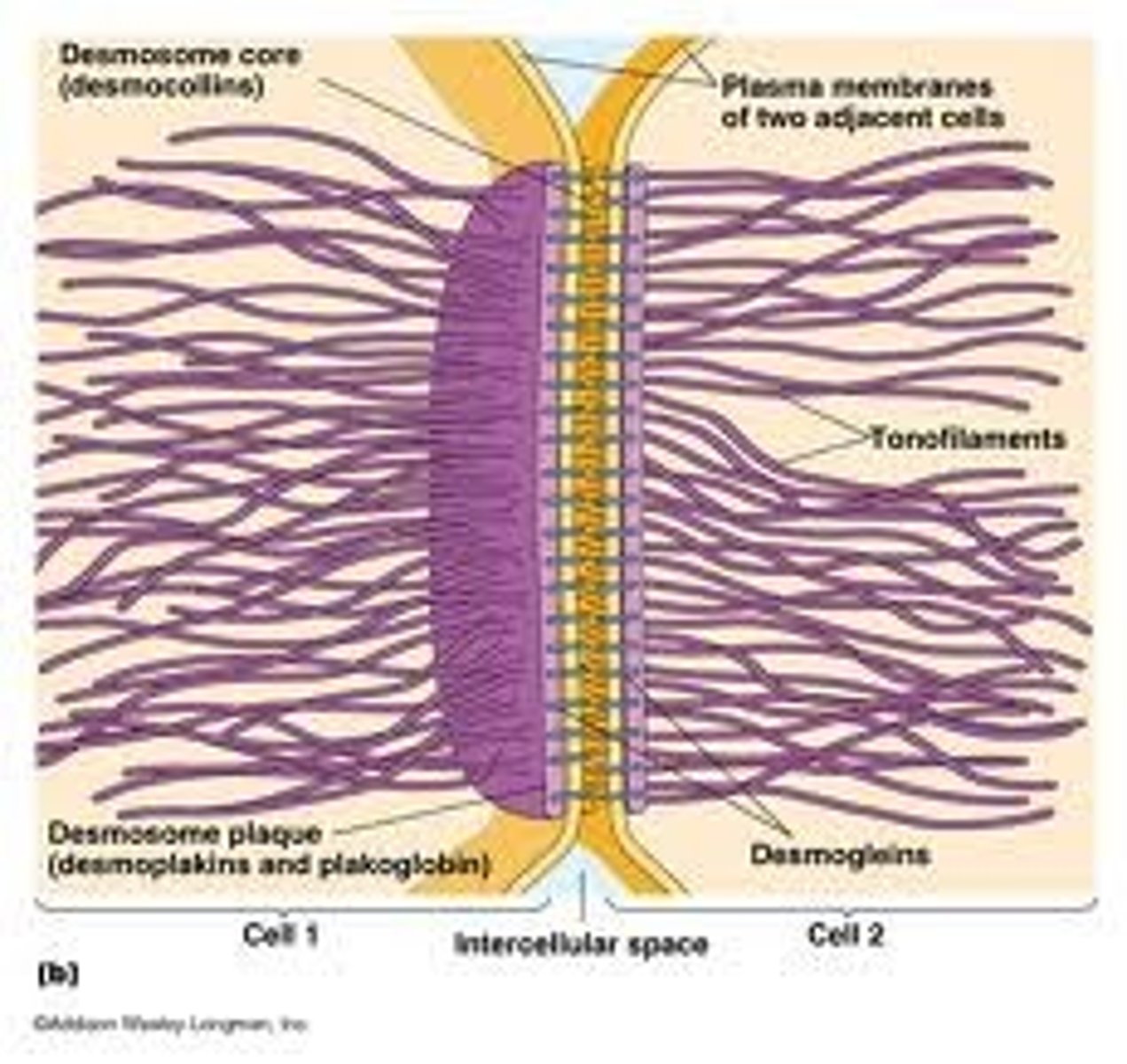
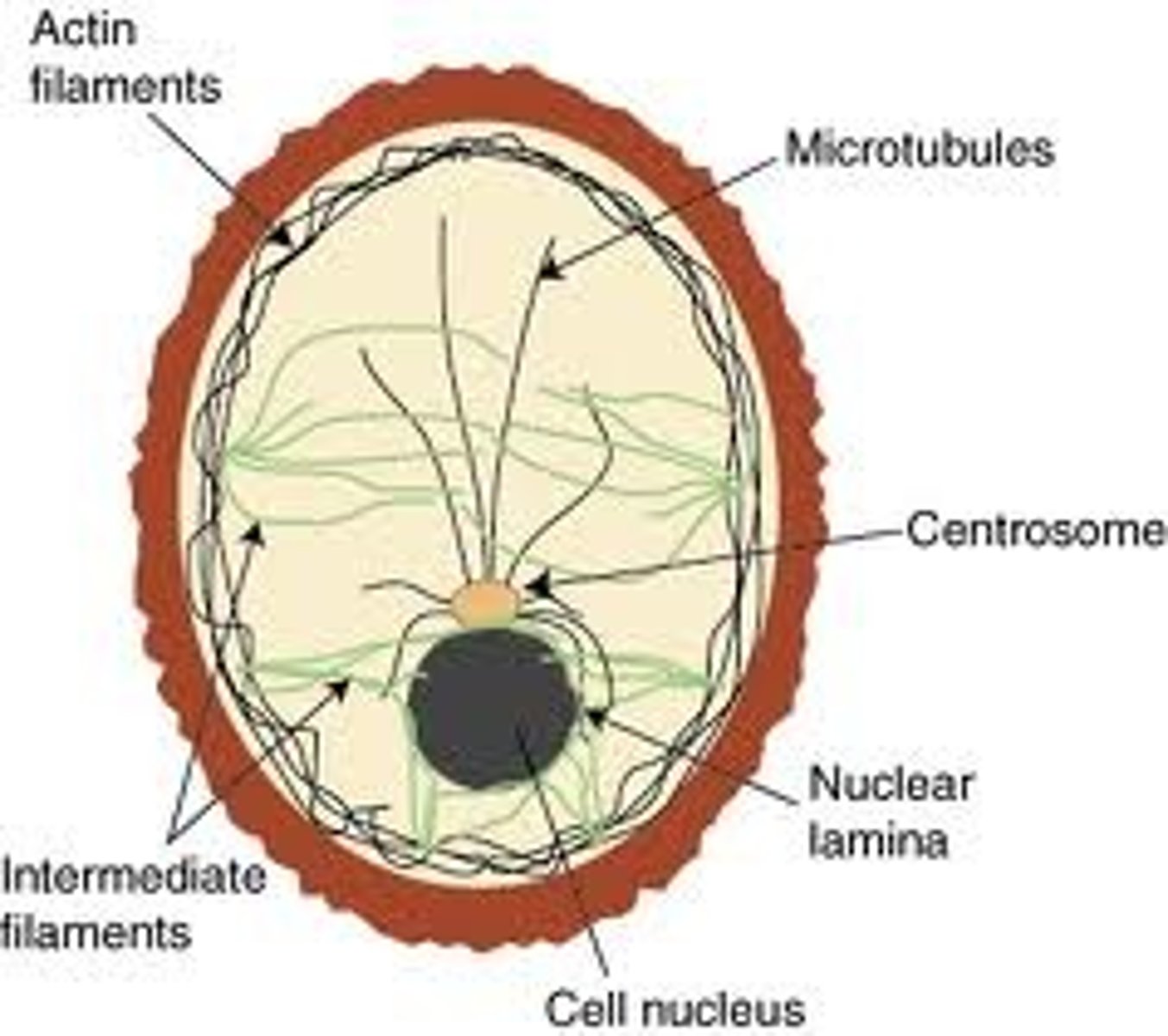
Cytoskeleton
A system of specialised long filament like proteins found in the cytosol of eukaryote cells which forms the constantly changing 'scaffolding'. They have many roles such as movement of motile cells, transport of organelles around the cell and intracellular movement of chromosomes during mitosis.
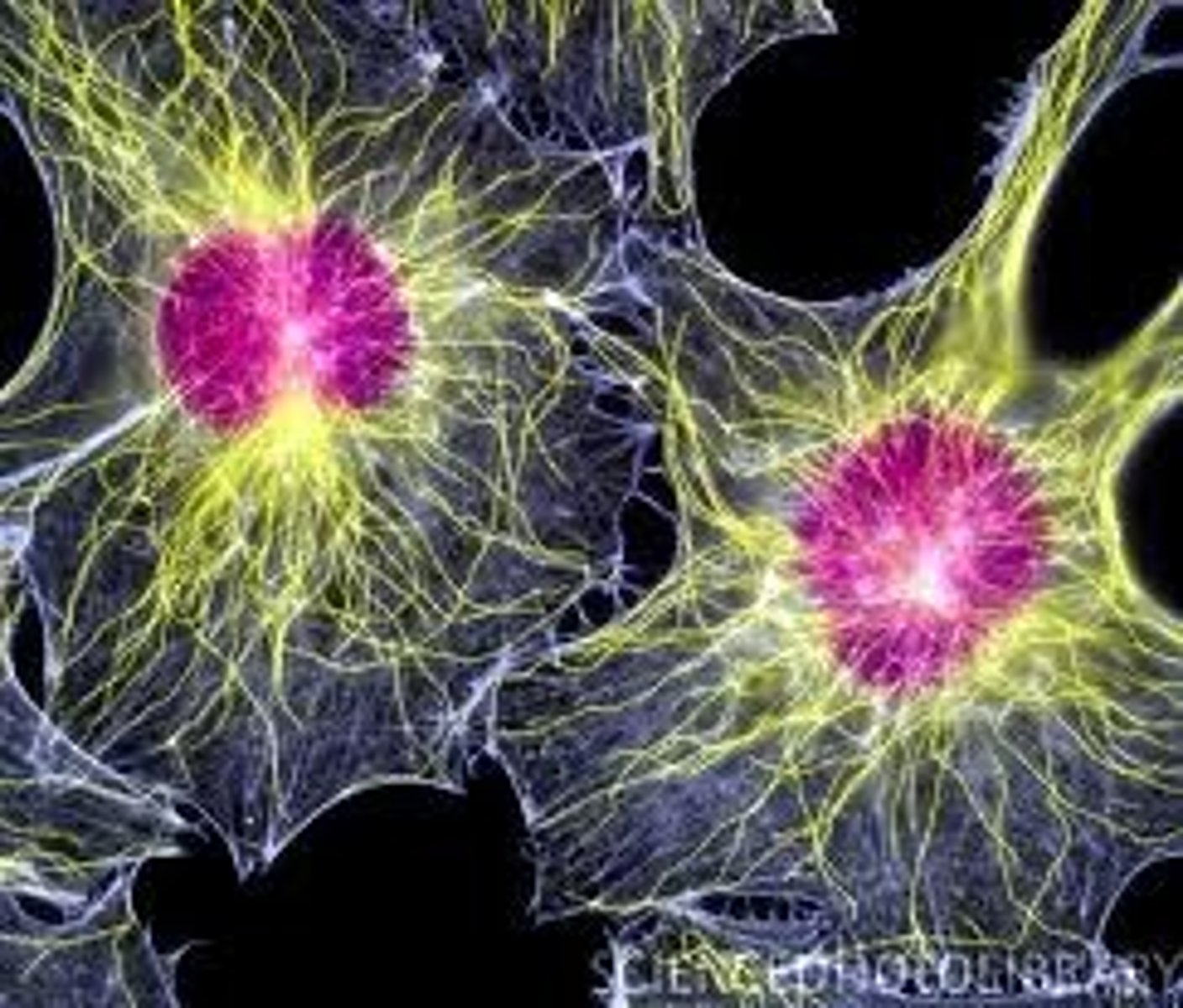
Microfilaments
Also known as actin filaments,one of three protein sub units that make up the eukaryote cytoskeleton. Found in highest concentration around the edges of the cell just below the cell membrane, they tend to form bundles. Actin polymers have the ability to disassemble and re-assemble meaning they are particulary useful for cell locomotion and in the microvilli of absorptive epithelial cells.
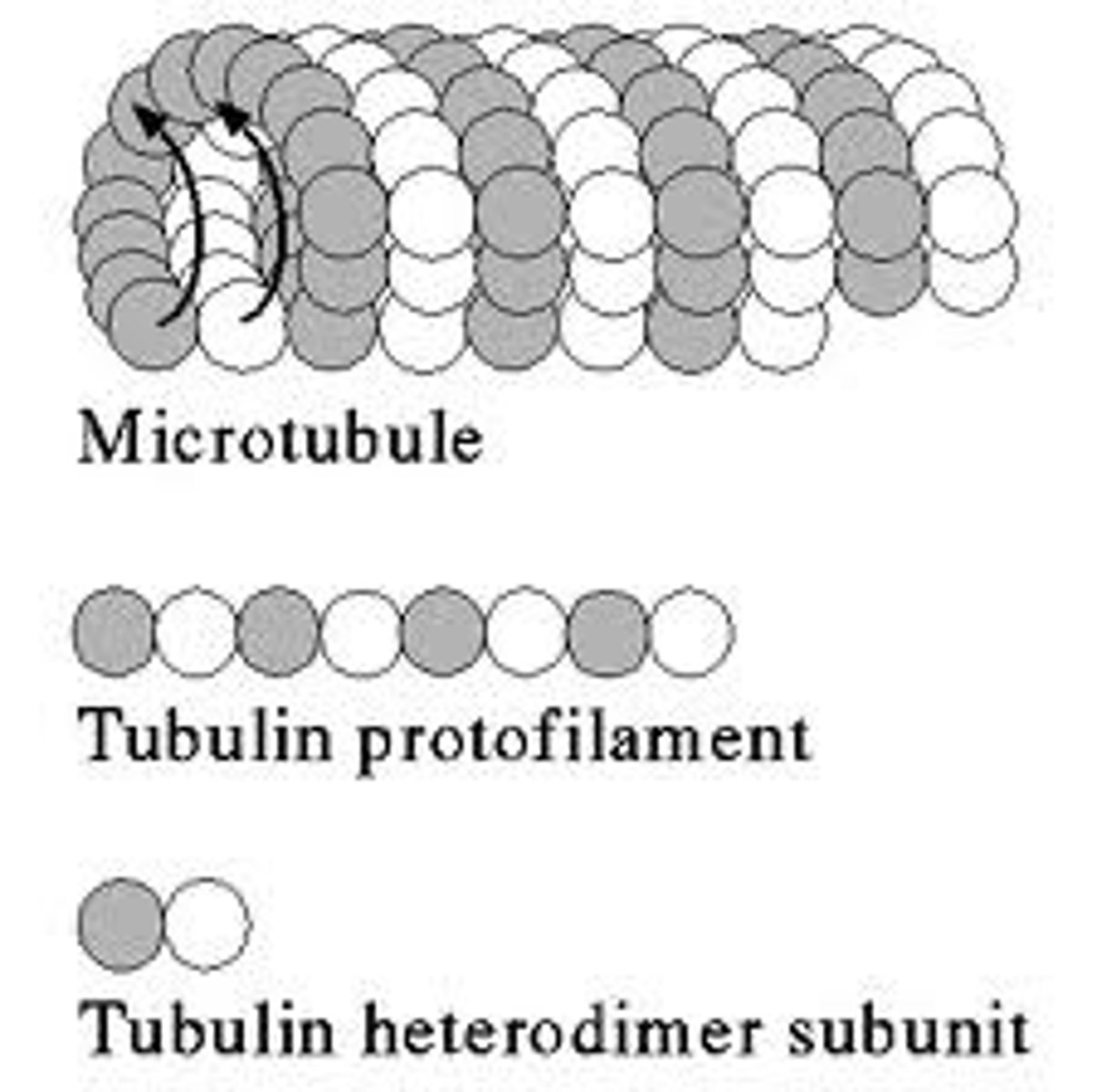
Tubulin
The protein of which the microtubules of the eukaryote cytoskeleton are formed.
Microtubules
Hollow tubes composed of thirteen parallel filaments of polymerized tubulin, measuring about 25 nm in external diameter. Part of the cytoskeleton of ALL eukaryote cells radiating from the centrosome in the nucleas towards the edges of the cell. They are very unstable and are constantly disassembling and reassembling so most do not reach the cell cortex. These tubules play a crucial role in cell organisation, movement of organelles and the reorganization of chormosomes in to daughter cells during mitosis.
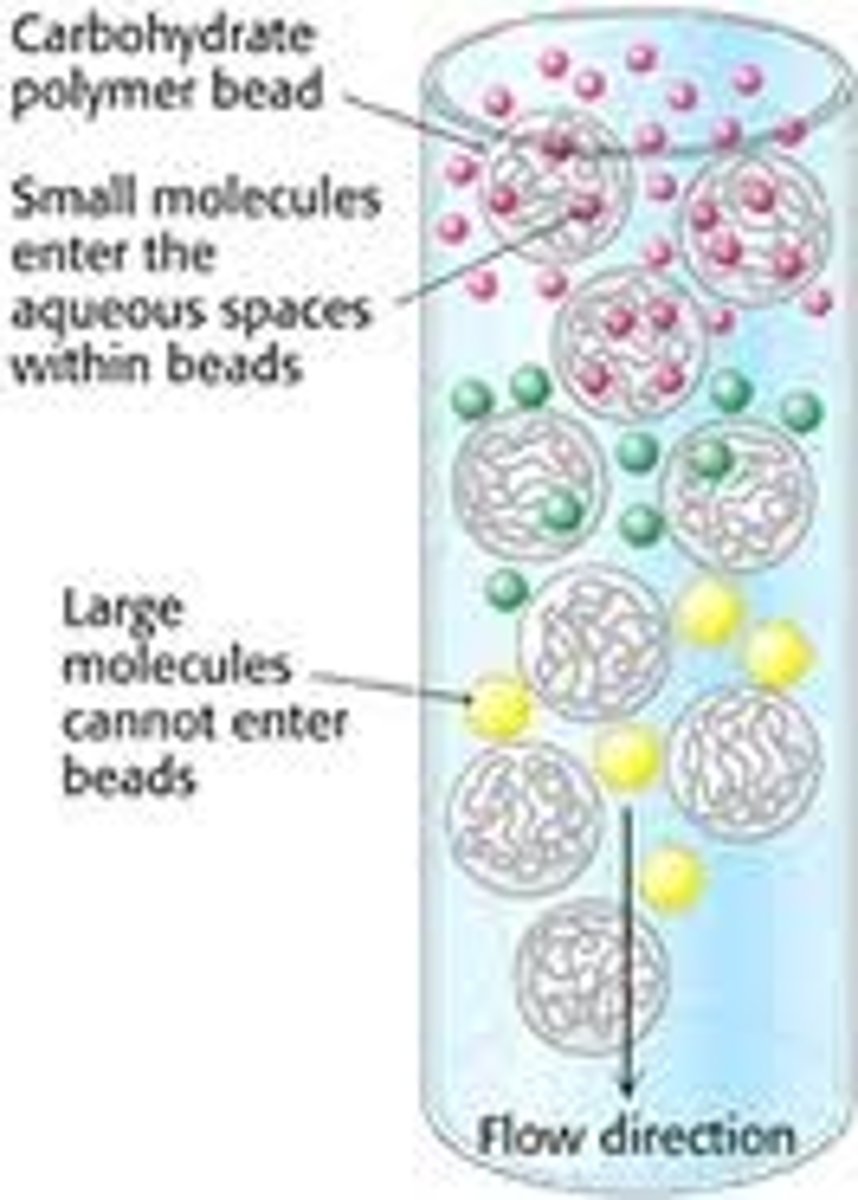
Gel Filtration
This form of filtration chromatography seprarates proteins, peptides, and oligonucleotides on the basis of size. Molecules move through a bed of porous beads, diffusing into the beads to greater or lesser degrees. Smaller molecules diffuse further into the pores of the beads and therefore move through the bed more slowly, while larger molecules enter less or not at all and thus move through the bed more quickly. Both molecular weight and three-dimensional shape contribute to the degree of retention. This technique may be used for analysis of molecular size, for separations of components in a mixture, or for salt removal or buffer exchange from a preparation of macromolecules.
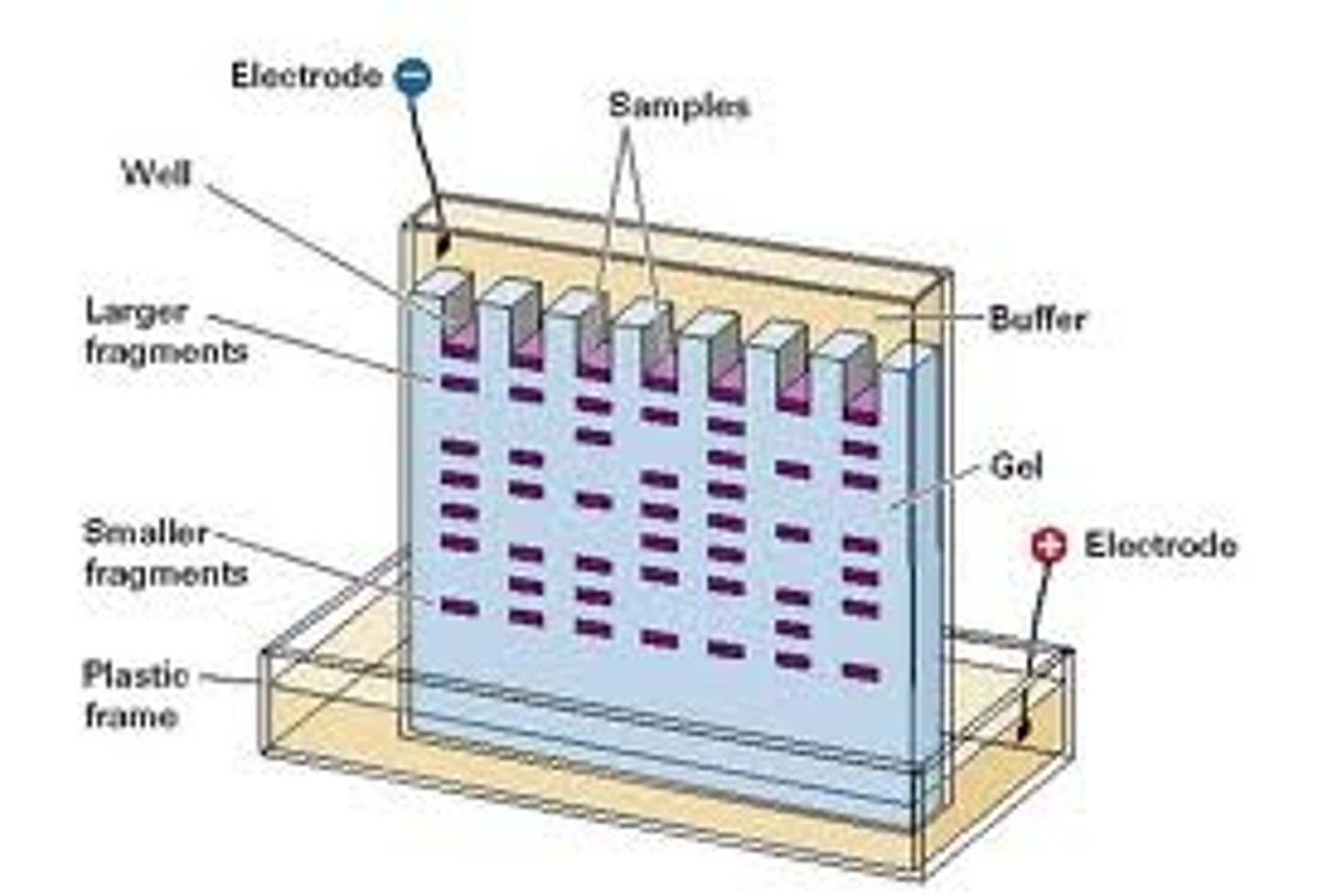
Gel Electrophoresis
In simple terms: This is a procedure which enables the sorting of molecules based on size and charge. Using an electric field, molecules (such as DNA) can be made to move through a gel made of agar. The molecules being sorted are dispensed into a well in the gel material. The gel is placed in an electrophoresis chamber, which is then connected to a power source. When the electric current is applied, the larger molecules move more slowly through the gel while the smaller molecules move faster. The different sized molecules form distinct bands on the gel.
SDS-PAGE
SDS-PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis, is a technique widely used in biochemistry, forensics, genetics and molecular biology to separate proteins according to their electrophoretic mobility (a function of length of polypeptide chain or molecular weight). SDS gel electrophoresis of samples that have identical charge per unit mass due to binding of SDS results in fractionation by size. This method can be used to separate all types, even those that are not water soluble.
Two Dimensional PAGE
Also known as 2-D electrophoresis, begins with 1-D electrophoresis but then separates the molecules by a second property in a direction 90 degrees from the first. In 1-D electrophoresis, proteins (or other molecules) are separated in one dimension, so that all the proteins/molecules will lie along a lane but that the molecules are spread out across a 2-D gel. Because it is unlikely that two molecules will be similar in two distinct properties, molecules are more effectively separated in 2-D electrophoresis than in 1-D electrophoresis.
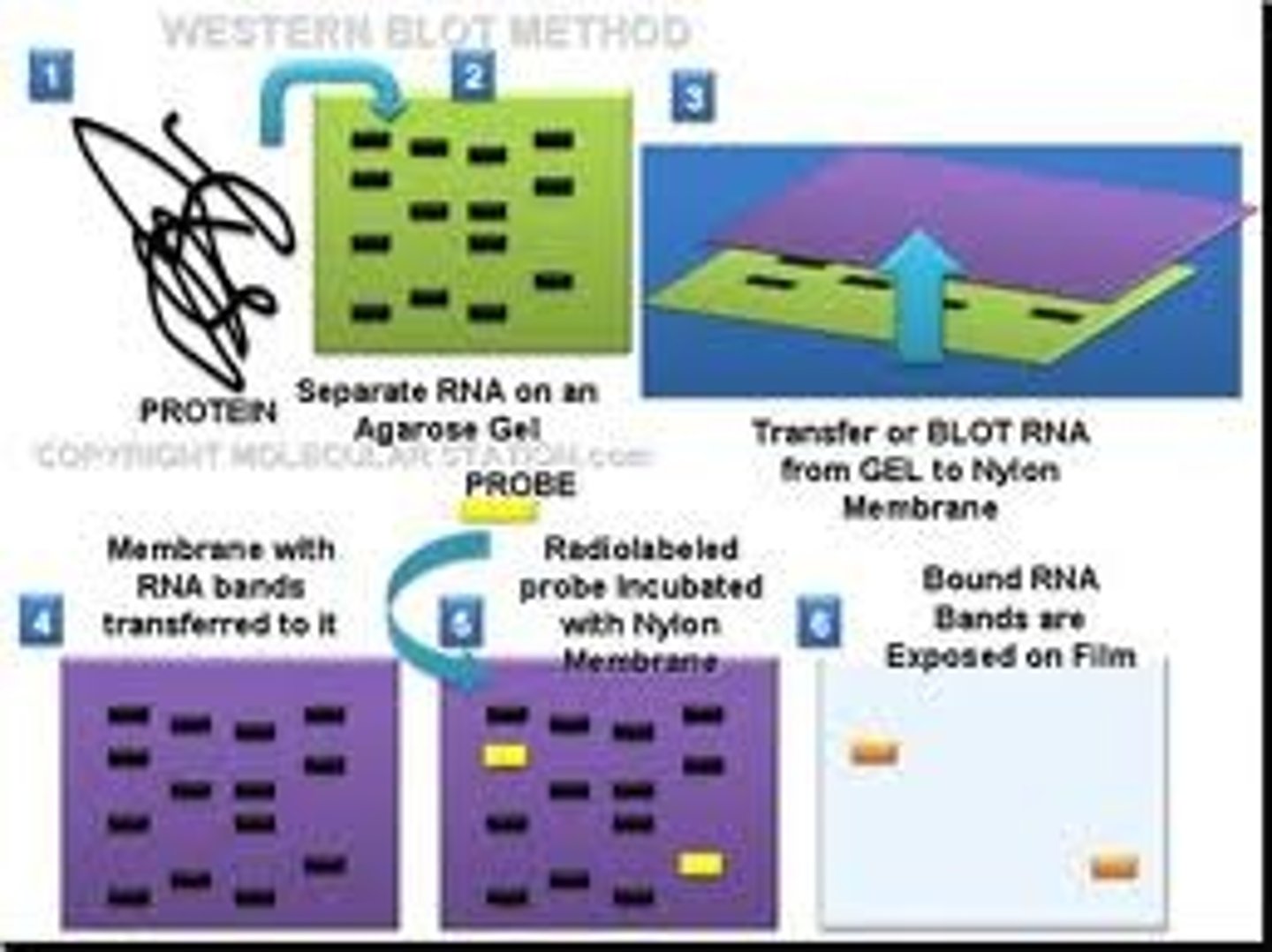
Western Blotting
This protein seperation technique (sometimes called the protein immunoblot) is a widely used analytical technique used to detect specific proteins in the given sample of tissue homogenate or extract. It uses gel electrophoresis to separate native or denatured proteins by the length of the polypeptide (denaturing conditions) or by the 3-D structure of the protein (native/ non-denaturing conditions). The proteins are then transferred to a membrane (typically nitrocellulose or PVDF), where they are probed (detected) using antibodies specific to the target protein.
There are now many reagent companies that specialize in providing antibodies (both monoclonal and polyclonal antibodies) against tens of thousands of different proteins. Commercial antibodies can be expensive, although the unbound antibody can be reused between experiments. This method is used in the fields of molecular biology, biochemistry, immunogenetics and other molecular biology disciplines.
Tertiary Structure
In biochemistry and molecular biology, this structure of a protein or any other macromolecule is its three-dimensional structure, as defined by the atomic coordinates.[6] Proteins and nucleic acids are capable of diverse functions ranging from molecular recognition to catalysis. Such functions require a precise three-dimensional tertiary structure. While such structures are diverse and seemingly complex, they are composed of recurring, easily recognizable tertiary structure motifs that serve as molecular building blocks. Tertiary structure is considered to be largely determined by the biomolecule's primary structure, or the sequence of amino acids or nucleotides of which it is composed. Efforts to predict tertiary structure from the primary structure are known generally as structure prediction.
![<p>In biochemistry and molecular biology, this structure of a protein or any other macromolecule is its three-dimensional structure, as defined by the atomic coordinates.[6] Proteins and nucleic acids are capable of diverse functions ranging from molecular recognition to catalysis. Such functions require a precise three-dimensional tertiary structure. While such structures are diverse and seemingly complex, they are composed of recurring, easily recognizable tertiary structure motifs that serve as molecular building blocks. Tertiary structure is considered to be largely determined by the biomolecule's primary structure, or the sequence of amino acids or nucleotides of which it is composed. Efforts to predict tertiary structure from the primary structure are known generally as structure prediction.</p>](https://knowt-user-attachments.s3.amazonaws.com/b4ccf142-5adb-42f5-8059-0ceb2d04eafe.jpg)
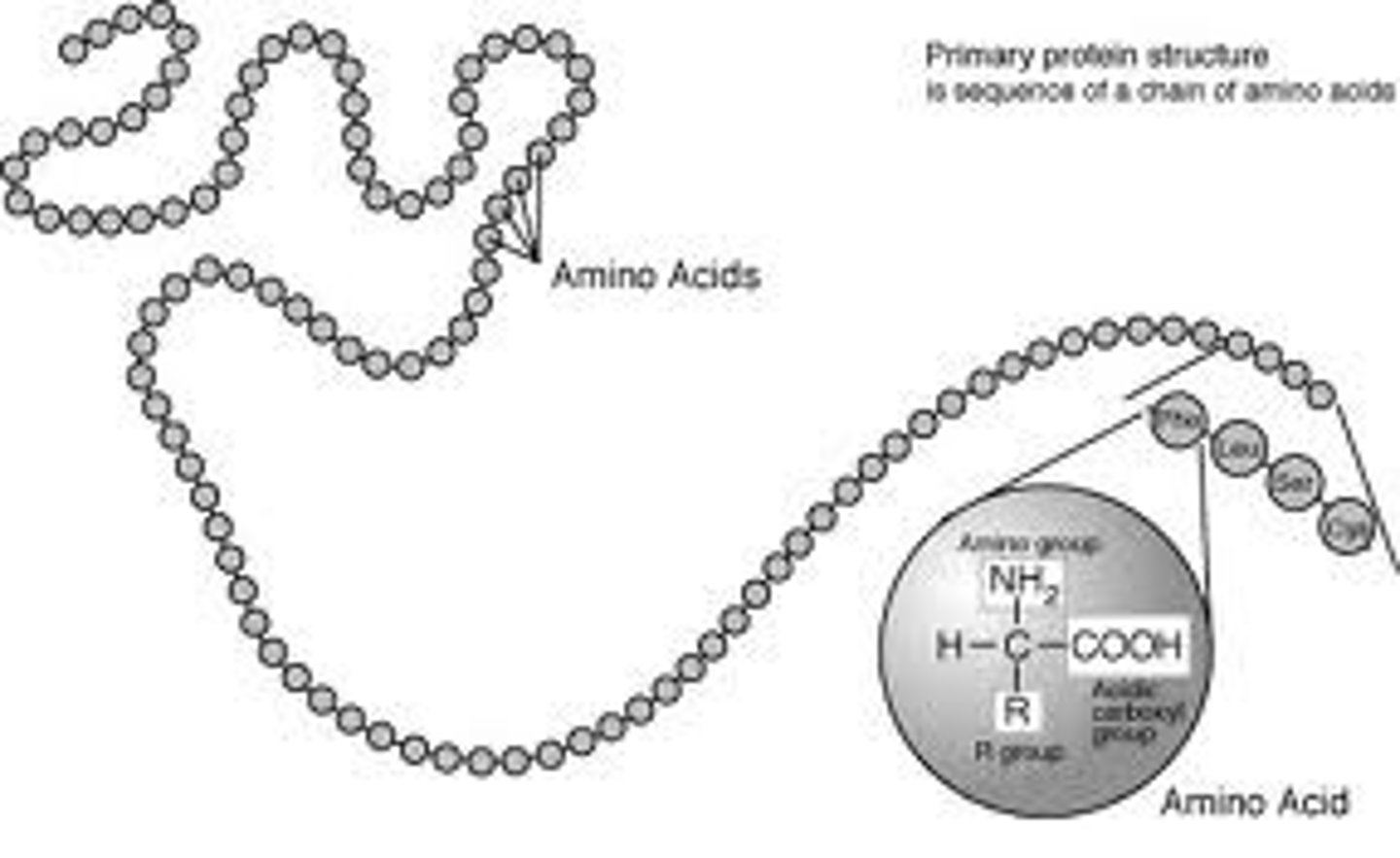
Primary Structure
This is the name given to the sequence of amino acid monomer units, or residues of which a compound is composed.
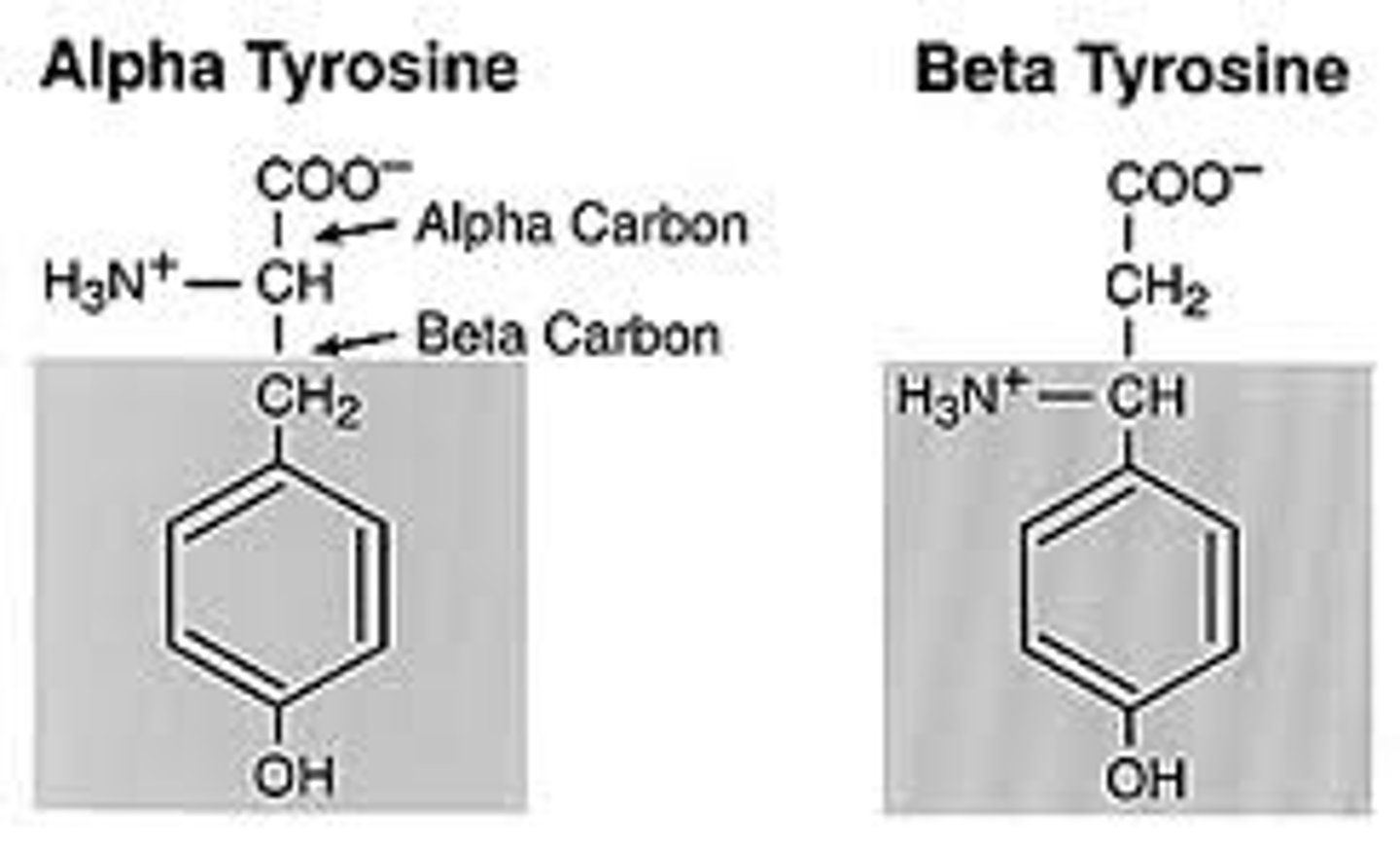
Alpha carbon
The carbon bonded to the carboxyl group in an amino acid.
R Group
This is the group which varies in proteins and can be any of twenty amino acids, the polarity of this Group dictates how a protein will behave in certain pH conditions. This explains why enzymes require a certain pH to function.
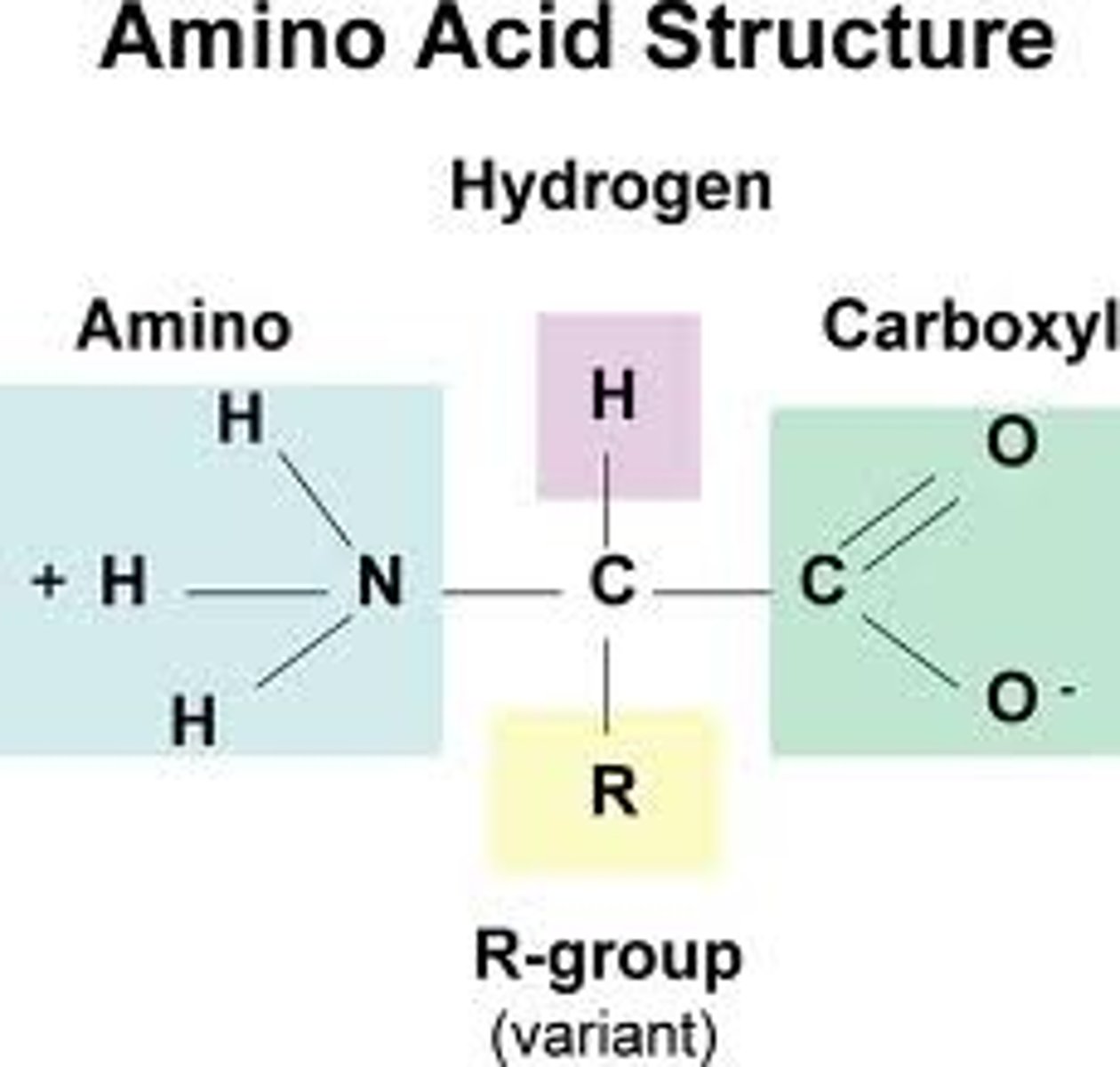
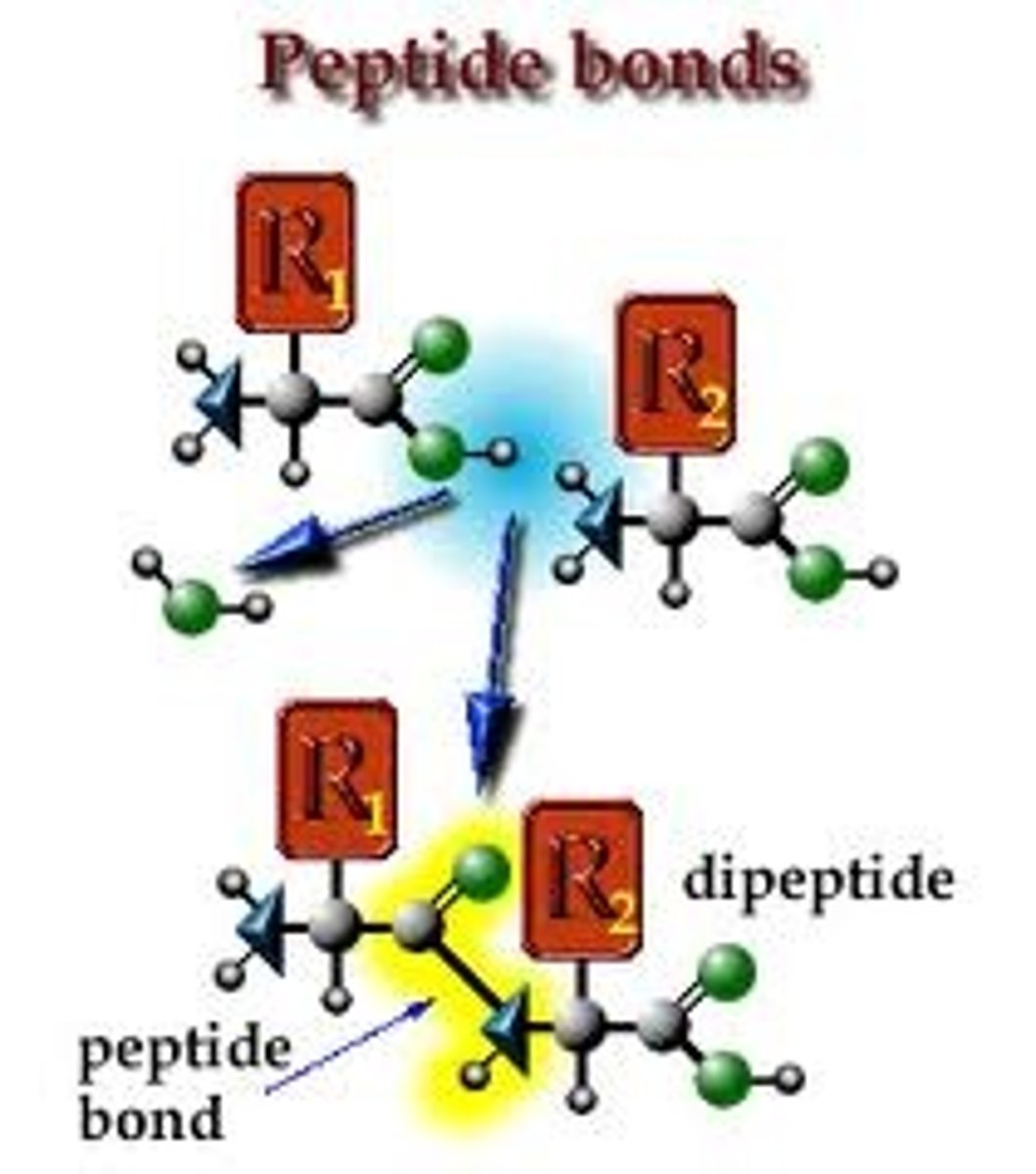
Peptide Bond
This bond occurs when the amino group from one protein joins with the carboxyl group of another, forming a dipeptide.
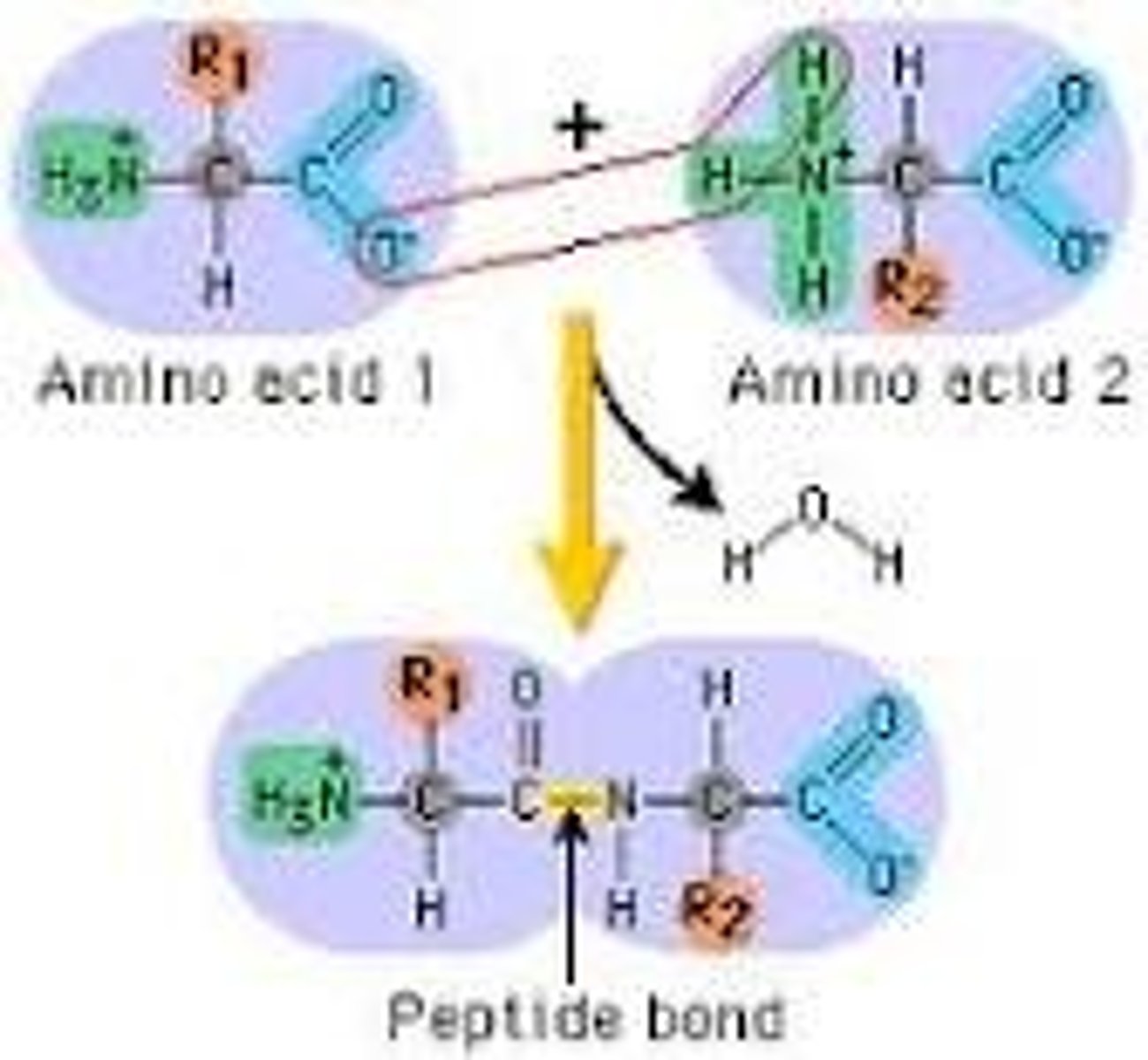
Dipeptide
A protein formed by two amino acids linked by a peptide bond,
Amino Terminus
The name by which the -NH₂ free end of a peptide is known.
Carboxyl Terminus
The name by which the -COOH (carboxyl) free end of a peptide is known.
Peptide
Small chains of amino acids.
Mr
Shorthand for molecular mass.
Hydrophobic residues/amino acid
Amino acids which are non polar and are repelled by water example are Alanine, Valine, Leucine, Isoleucine, Proline, Methionine, Phenylalanine, Tryptophan and Cystine. Hydrophbicity is also affected by pH levels in some cases.
Hydrophillic residues/amino acid
Amino acids which are polar and are attracted to water examples are Glutamine, Serine,Theronine, Histodine, Lysine. Hydrophbicity is also affected by pH levels in some cases.
Polar
Pertaining to a compound exhibiting polarity or dipole moment, that is a compound bearing a partial positive charge on one side and a partial negative charge on the other.
Non Polar
Molecule which has no separation of charge, so no positive or negative poles are formed.
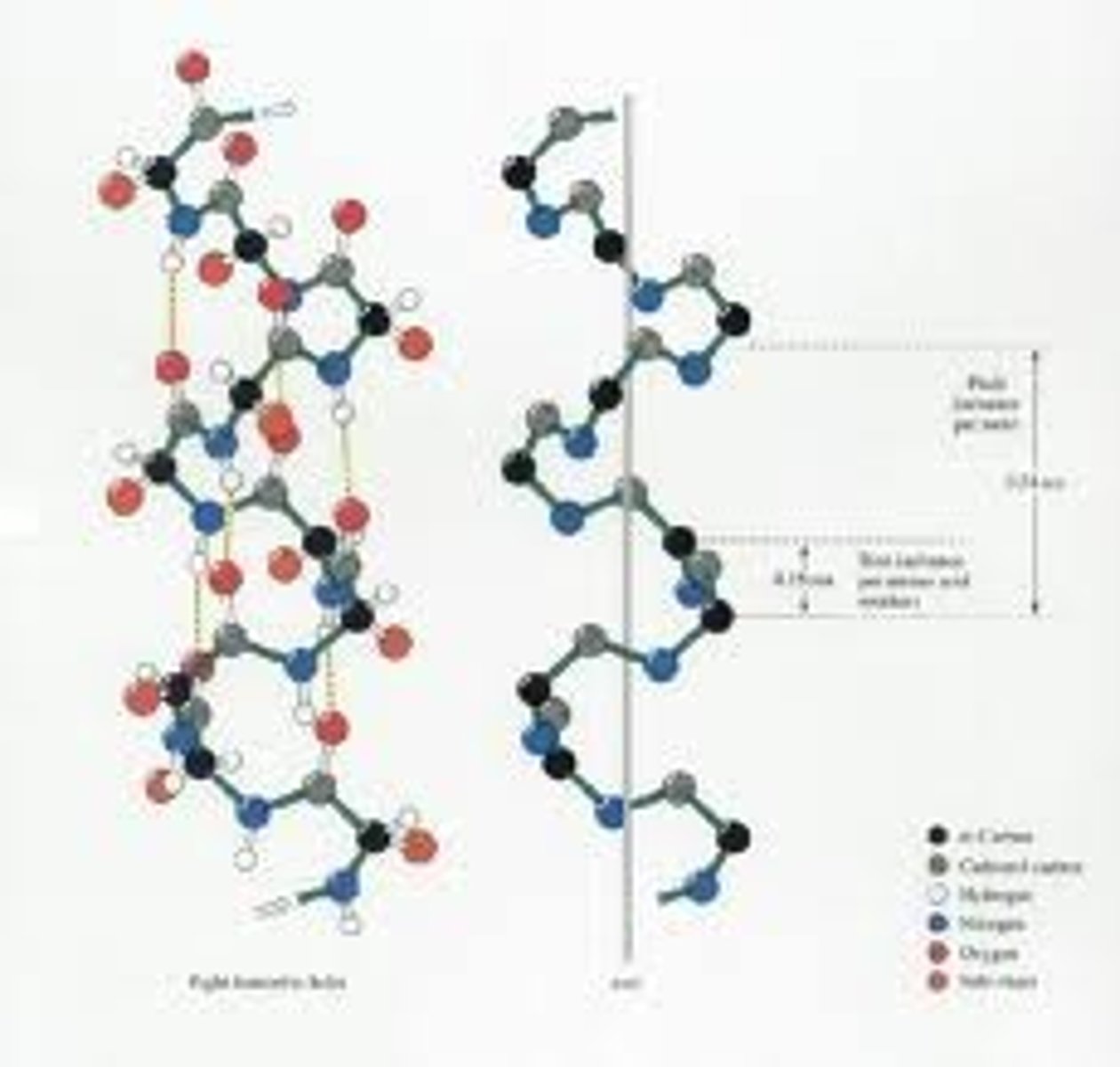
α Helix
A common motif in the secondary structure of proteins, the alpha helix (α-helix) is a right-handed coiled or spiral conformation, in which every backbone N-H group donates a hydrogen bond to the backbone C=O group of the amino acid four residues earlier (i+4 \rightarrow i hydrogen bonding). This secondary structure is also sometimes called a classic Pauling-Corey-Branson alpha helix . Among types of local structure in proteins, the α-helix is the most regular and the most predictable from sequence, as well as the most prevalent.
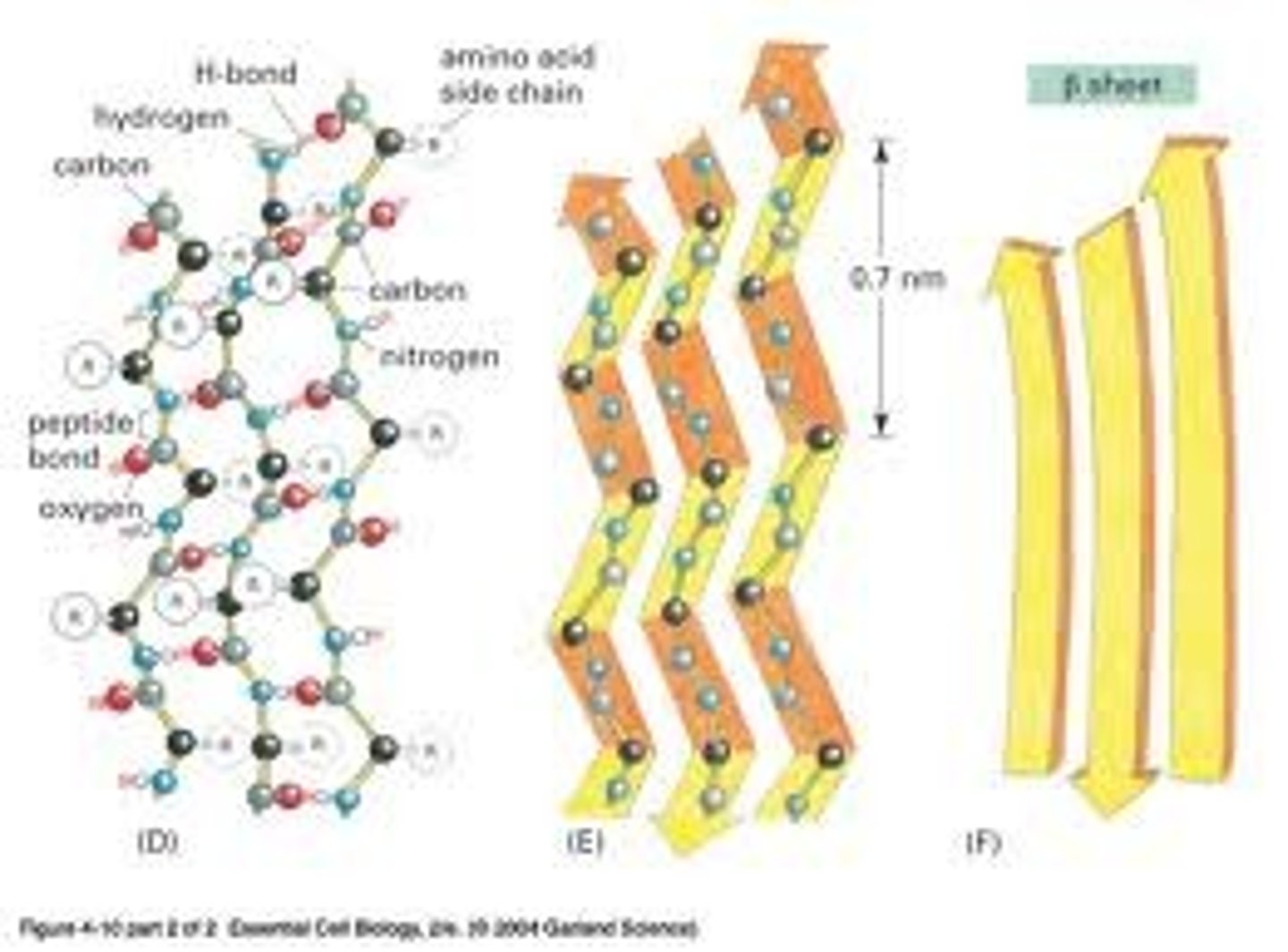
β sheet
The β sheet (also β-pleated sheet) is the second form of regular secondary structure in proteins, only somewhat less common than alpha helix. Beta sheets consist of beta strands connected laterally by at least two or three backbone hydrogen bonds, forming a generally twisted, pleated sheet. A beta strand (also β strand) is a stretch of polypeptide chain typically 3 to 10 amino acids long with backbone in an almost fully extended conformation. The higher-level association of β sheets has been implicated in formation of the protein aggregates and fibrils observed in many human diseases, notably the amyloidoses such as Alzheimer's disease.
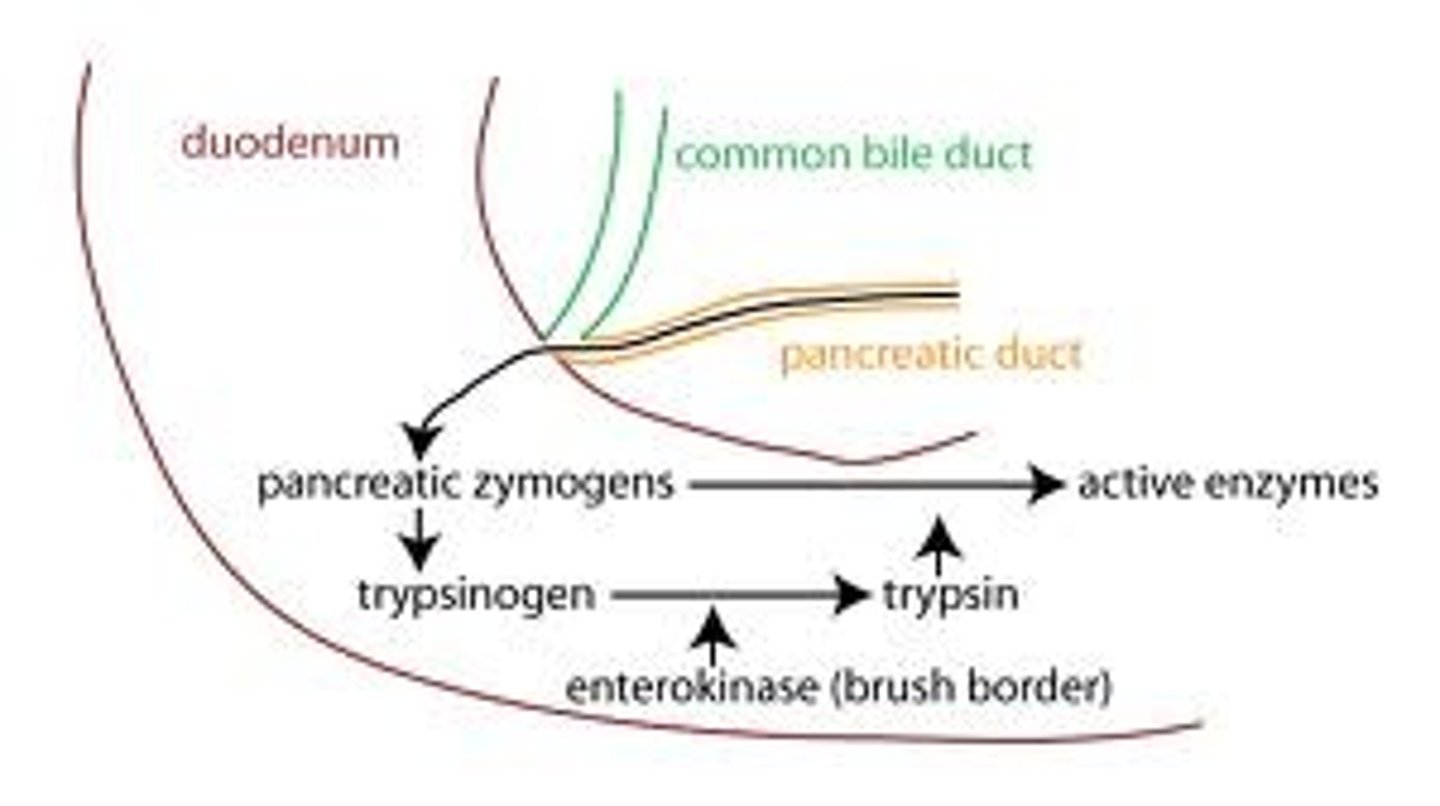
Zymogens
A zymogen (or proenzyme) is an inactive enzyme precursor. A zymogen requires a biochemical change (such as a hydrolysis reaction revealing the active site, or changing the configuration to reveal the active site) for it to become an active enzyme. The biochemical change usually occurs in a lysosome where a specific part of the precursor enzyme is cleaved in order to activate it. The amino acid chain that is released upon activation is called the activation peptide.
The pancreas secretes zymogens partly to prevent the enzymes from digesting proteins in the cells in which they are synthesised. Fungi also secrete digestive enzymes into the environment as zymogens. The external environment has a different pH than inside the fungal cell and this changes the zymogen's structure into an active enzyme.
Proteolyisis
This process is the directed degradation (digestion) of proteins which fail to fold correctly by cellular enzymes called proteases or by intramolecular digestion.
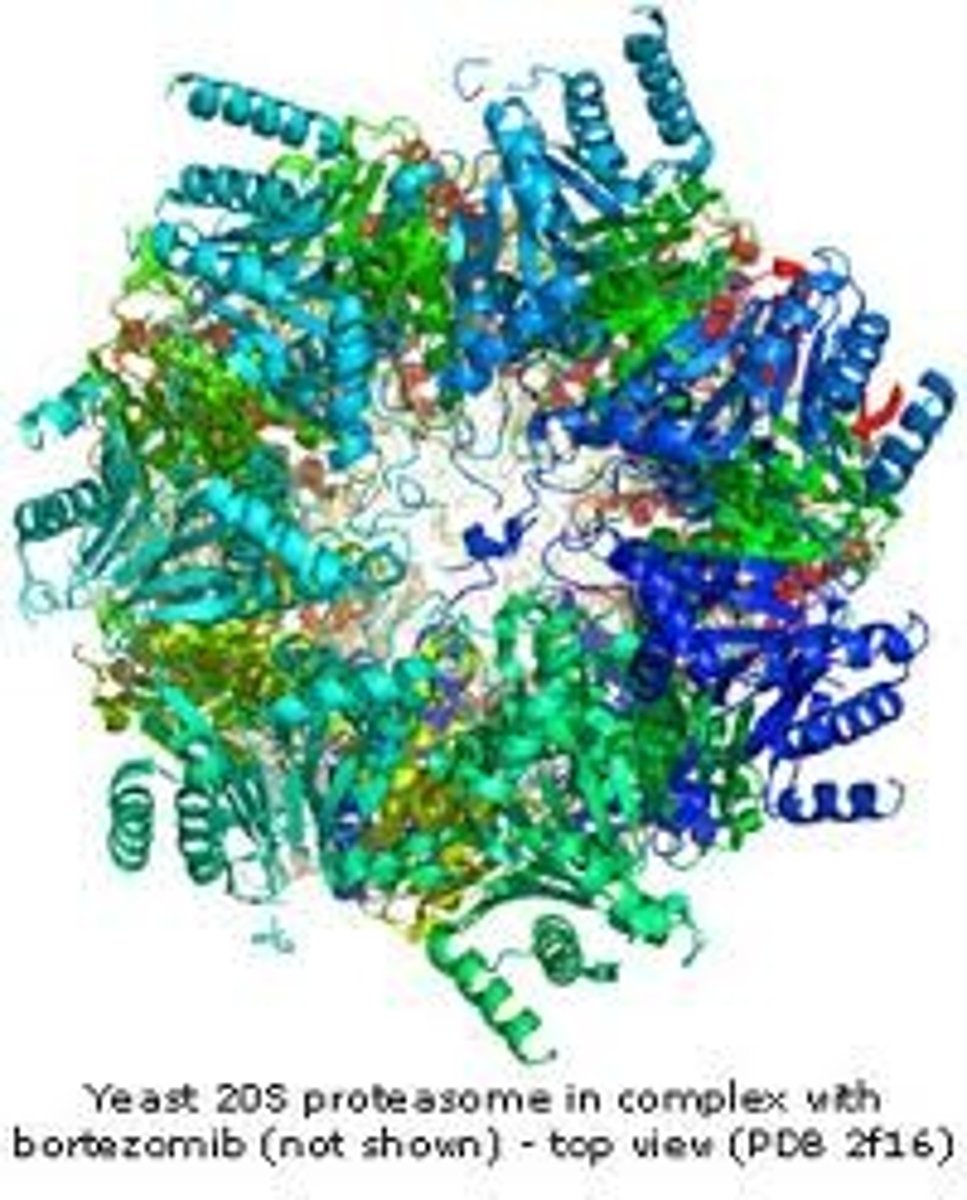
Proteasomes
Proteasomes are very large protein complexes inside all eukaryotes and archaea, and in some bacteria. In eukaryotes, they are located in the nucleus and the cytoplasm.The main function of the proteasome is to degrade unneeded or damaged proteins by proteolysis, a chemical reaction that breaks peptide bonds. Enzymes that carry out such reactions are called proteases. Proteasomes are part of a major mechanism by which cells regulate the concentration of particular proteins and degrade misfolded proteins. The degradation process yields peptides of about seven to eight amino acids long, which can then be further degraded into amino acids and used in synthesizing new proteins. Proteins to be destroyed are labelled by ubiquitin.
Ubiquitin
A small regulatory protein that has been found in almost all tissues (ubiquitously) of eukaryotic organisms. Among other functions, it directs protein recycling.It can be attached to proteins and label them for destruction. This protein tag directs proteins to the proteasome, which is a large protein complex in the cell that degrades and recycles unneeded proteins. This discovery won the Nobel Prize for chemistry in 2004.
The tags can also direct proteins to other locations in the cell, where they control other protein and cell mechanisms.
Post-translation modification
This is the chemical modification of a protein after its translation. It is one of the later steps in protein biosynthesis, and thus gene expression, for many proteins.A protein (also called a polypeptide) is a chain of amino acids. During protein synthesis, 20 different amino acids can be incorporated to become a protein. After translation, the posttranslational modification of amino acids extends the range of functions of the protein by attaching it to other biochemical functional groups (such as acetate, phosphate, various lipids and carbohydrates), changing the chemical nature of an amino acid (e.g. citrullination), or making structural changes (e.g. formation of disulfide bridges).Also, enzymes may remove amino acids from the amino end of the protein, or cut the peptide chain in the middle. For instance, the peptide hormone insulin is cut twice after disulfide bonds are formed, and a propeptide is removed from the middle of the chain; the resulting protein consists of two polypeptide chains connected by disulfide bonds. Also, most nascent polypeptides start with the amino acid methionine because the "start" codon on mRNA also codes for this amino acid. This amino acid is usually taken off during post-translational modification.Other modifications, like phosphorylation, are part of common mechanisms for controlling the behavior of a protein, for instance activating or inactivating an enzyme.
Enzyme Substrate Complex
A non-covalent complex composed of a substrate bound to the active site of the enzyme
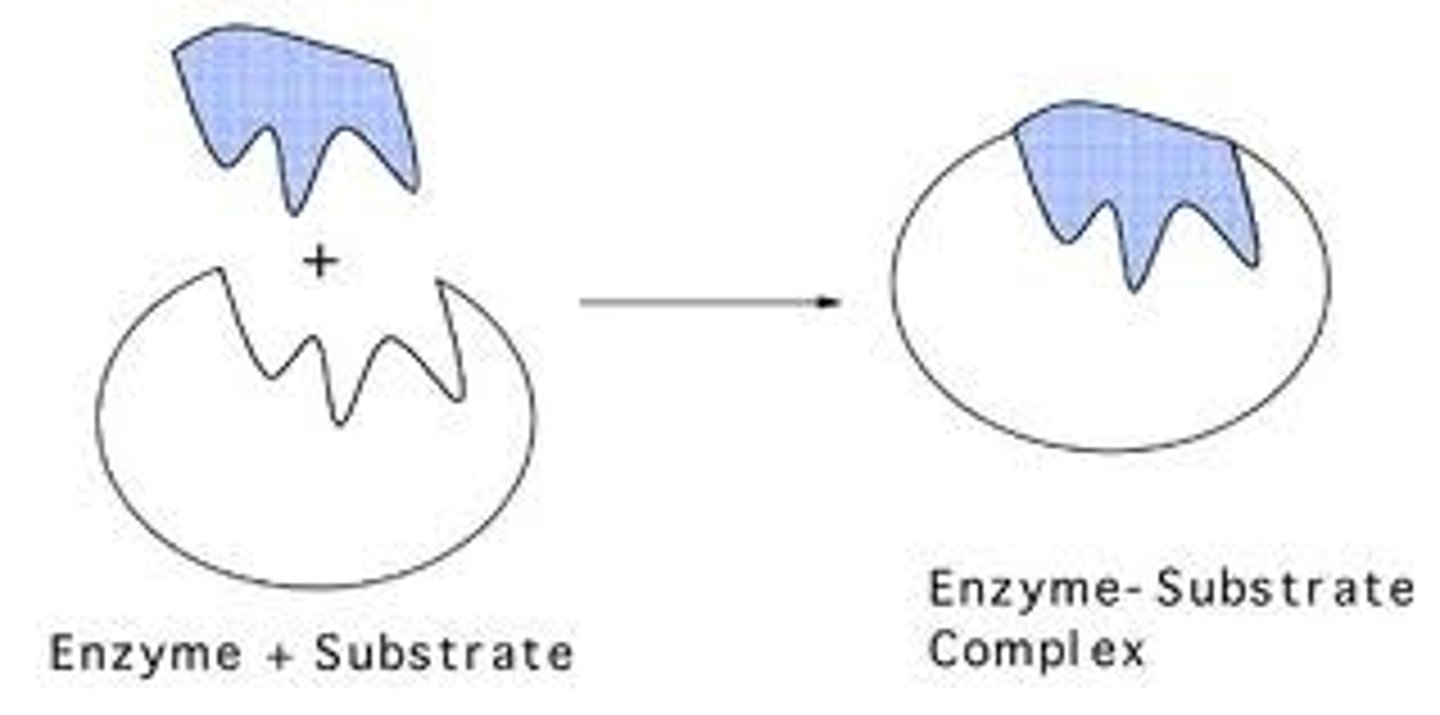
Substrate
In biochemistry, a substrate is a molecule upon which an enzyme acts. Enzymes catalyze chemical reactions involving the substrate(s). In the case of a single substrate, the substrate binds with the enzyme active site, and an enzyme-substrate complex is formed. The substrate is transformed into one or more products, which are then released from the active site. The active site is now free to accept another substrate molecule. In the case of more than one substrate, these may bind in a particular order to the active site, before reacting together to produce products.
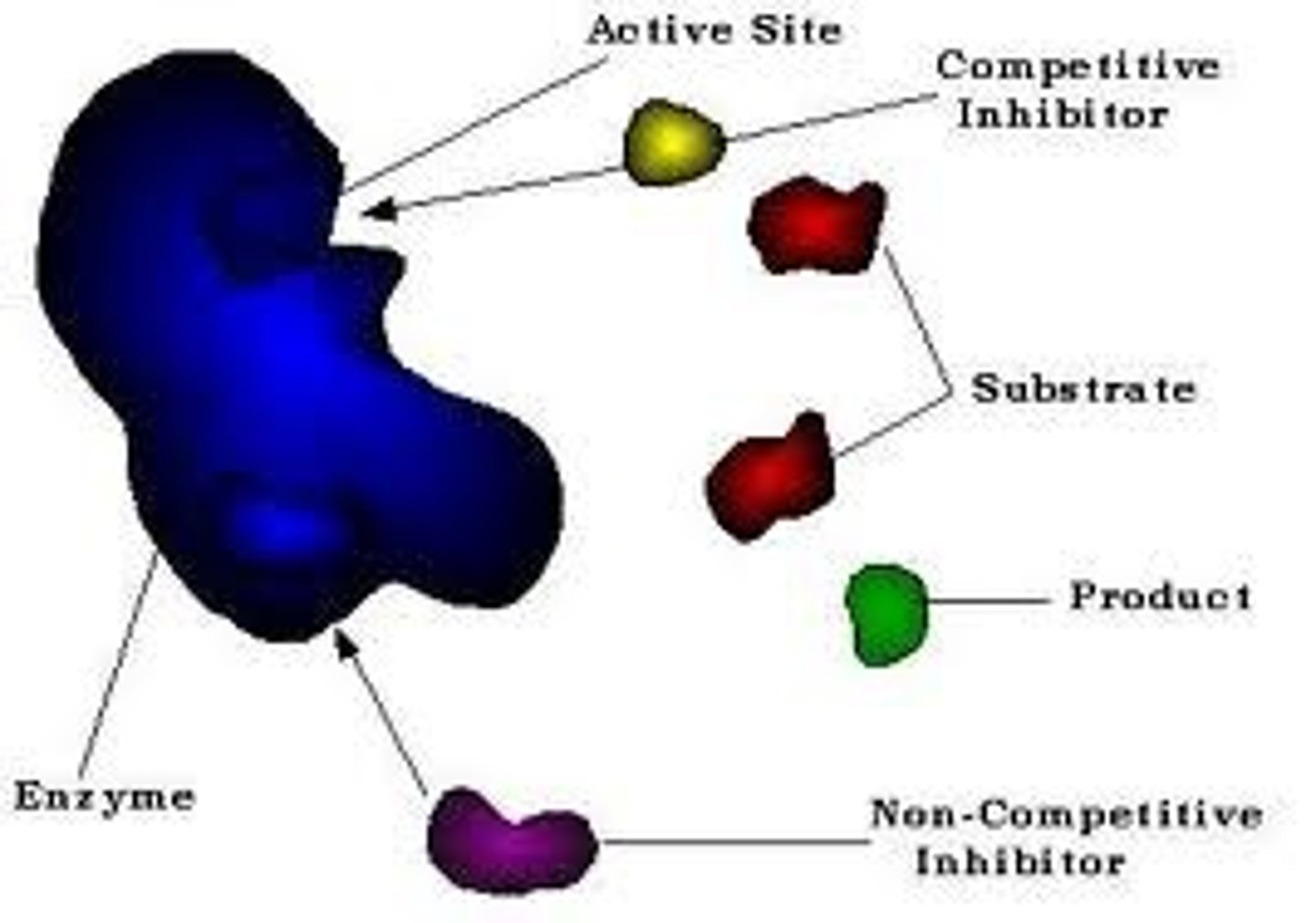
Substrate specificity
A characteristic feature of enzyme activity in relation to the kind of substrate on which the enzyme or catalytic molecule reacts. An ezyme can only react upon the substrate with which it locks.
Enzyme Assay
A technique used to measure the rate of activity of an enzyme by measuring the products expected of the enzyme activity, for example CO₂.
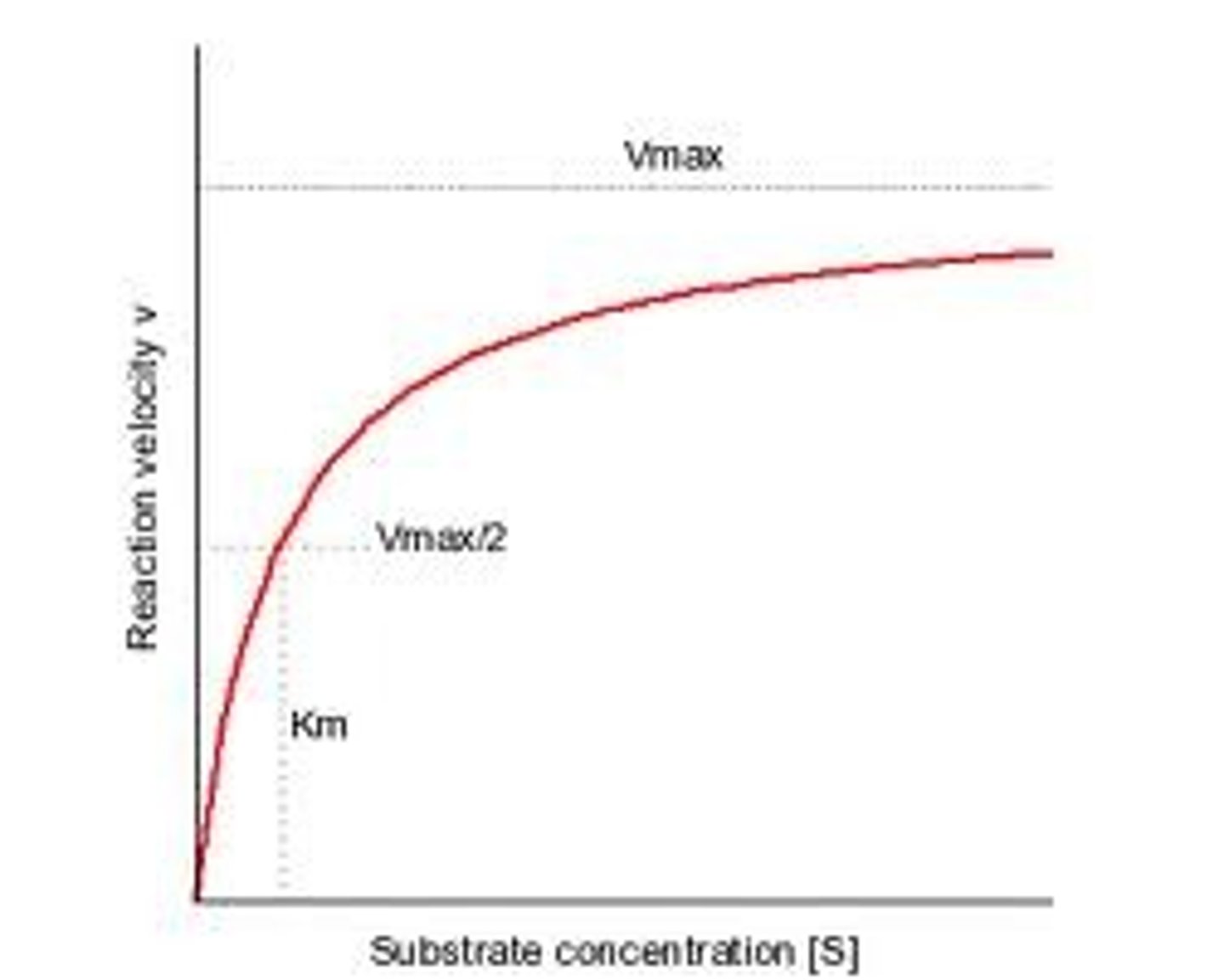
Km
The point on a rectangular hyperbola which is half the value of the Vmax. It indicates the affinity of an enzyme and substrate a high value means low affinity and a low value means high affinity (the enzyme substrate locks more securely and takes longer to seperate and release products).
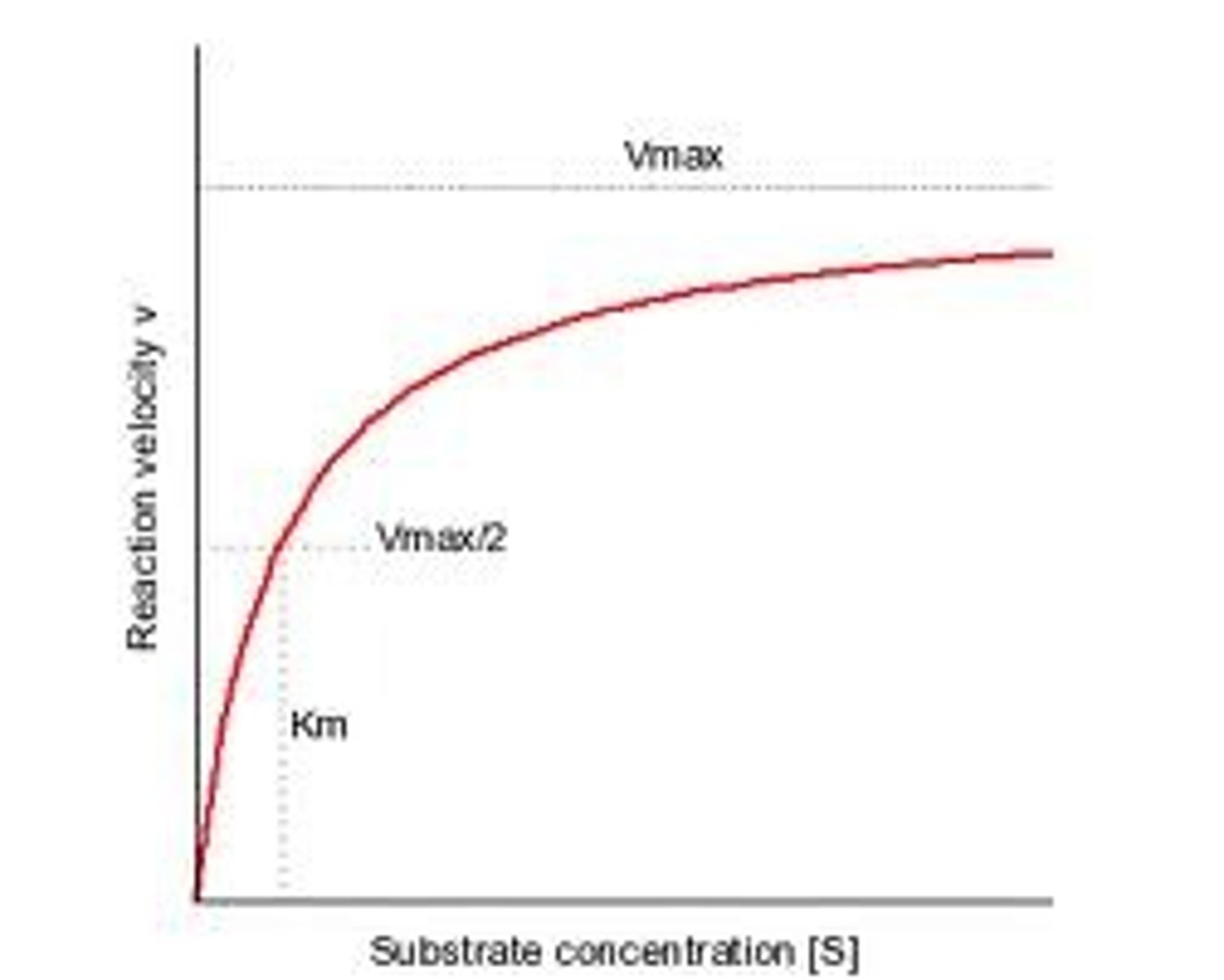
Vmax
The point on a hyperbolic plot/during an enzyme assay at which the maximum rate of substrate to product conversion is reached and the line begins to level out. This is often used to indicate the maximum rate of enzyme activity, however it is only approximate as the plot line never completely levels out.
Michaelis-Menton equation
This equation can be used if a range of [S] values is known, to plot a line.
E + S ↔ ES → E + P
Hofstee-Eadie plot
A plot used to obtain a more accurate indication of Km and Vmax. Simplified - v/[S] so the figures used to plot the original hyperbolic rectangle are used dviding the enzyme byt hte substrate. These new figures are then plotted on along the horizontal axis and a best fit line drawn along them. The point at which the line crosses the vertical axis is the Vmax, the point at which it crosses the horizontal axis is the Km.
![<p>A plot used to obtain a more accurate indication of Km and Vmax. Simplified - v/[S] so the figures used to plot the original hyperbolic rectangle are used dviding the enzyme byt hte substrate. These new figures are then plotted on along the horizontal axis and a best fit line drawn along them. The point at which the line crosses the vertical axis is the Vmax, the point at which it crosses the horizontal axis is the Km.</p>](https://knowt-user-attachments.s3.amazonaws.com/799ee21b-22cb-430a-a88e-755a13001d9d.jpg)
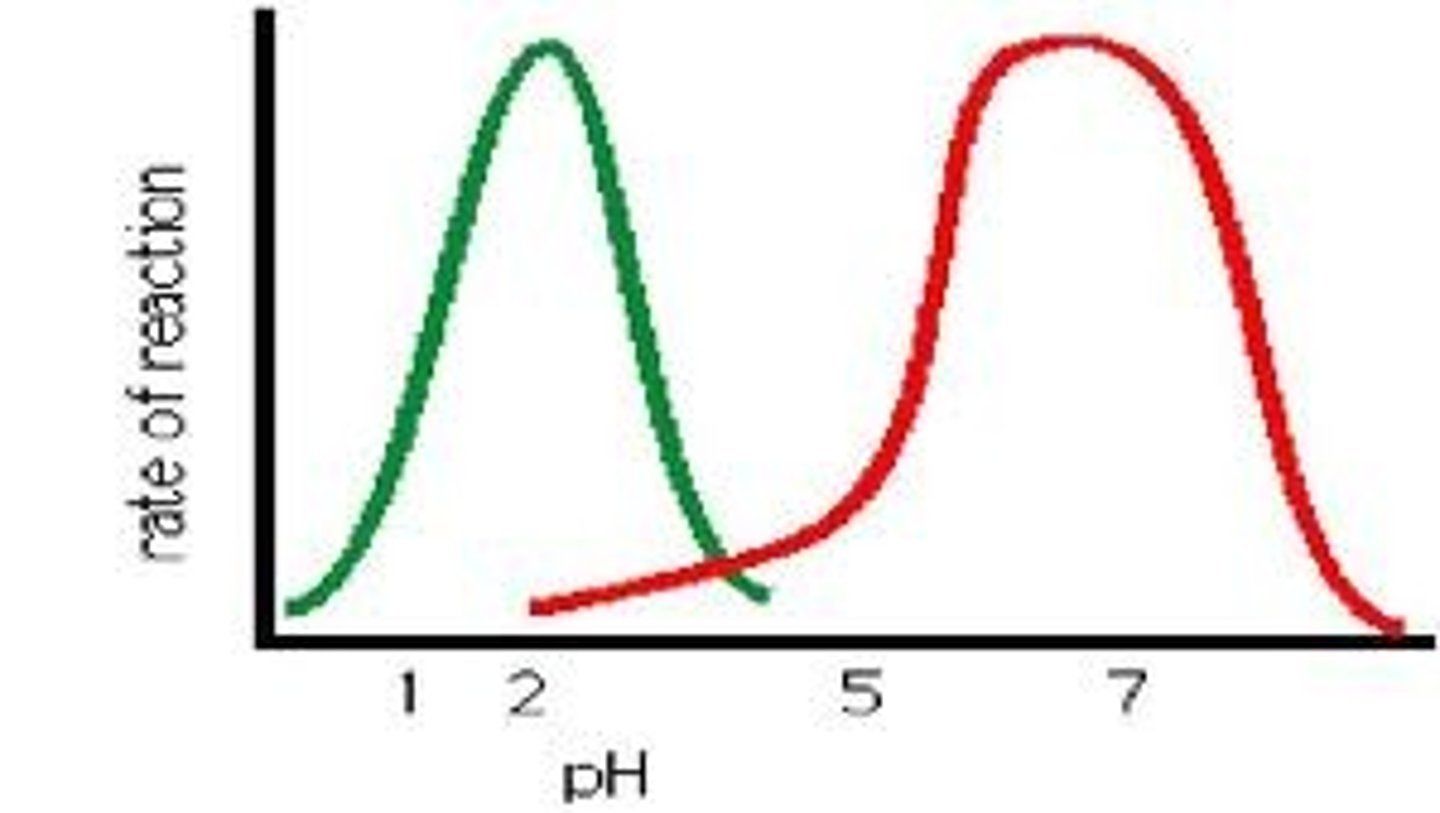
pH Optima
The optimum pH in which an enzyme is most active, this is normally related to the normal environment of the enzyme, for example Pepsin has a pH optima of around 2, ideal for the acidic environment of the vertebrate stomach.
Cofactors
Other conditions required by enzymes in order for them to perform their roles. For example metal ions (Mg²⁺ + K⁺) or coenzymes such as NDP, ATP or ADP. They cannot be made in the body of mammals and must be derived from vitamins in the diet.
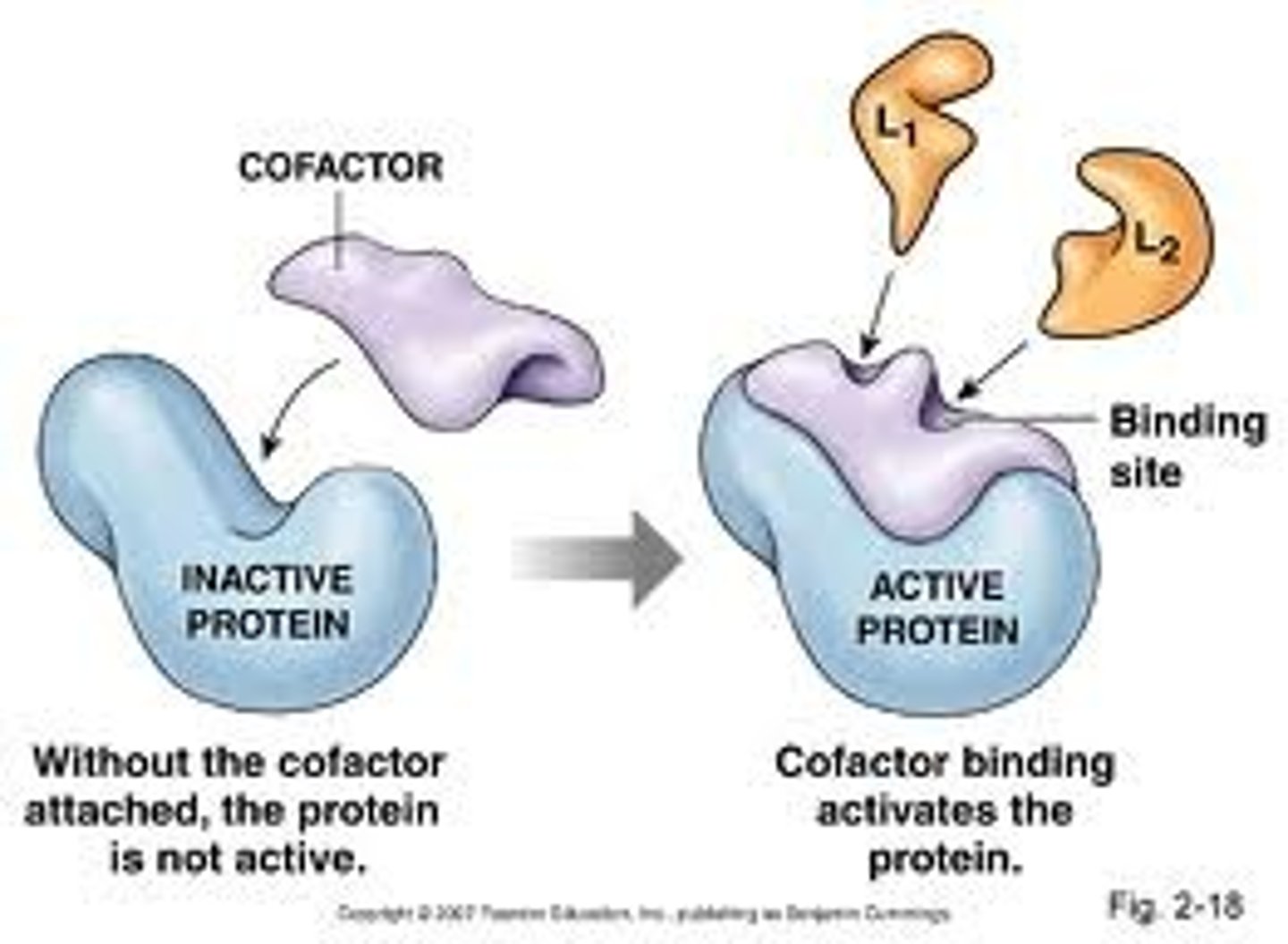
Enzyme Inhibition
An important control mechanism in metabolism, this is also the route of effect used by many drugs and also of many toxins.
Irreversible Inhibitors
Usually not of biological origins, these act by forming strong covalent bonds with the enzyme, poisoning them. The bond is so strong it is irreversible and example of this would be heavy metal toxicity.
Reversible Inhibitors
This inhibition can be relieved by the removal of the inhibitor from the enzyme solution due to weak reversible interactions they form with the enzyme.
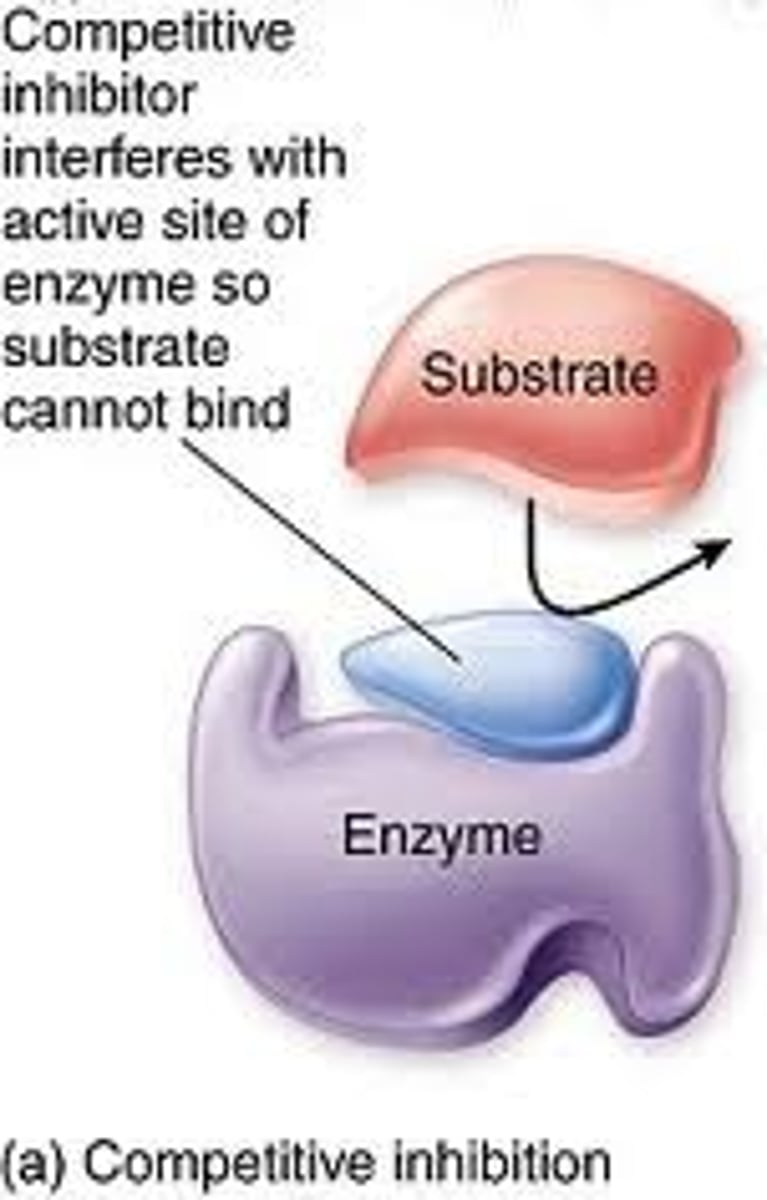
Competitive Inhibitor
This form of inhibitor is very similar in structure to the substrate and can thus form an enzyme inhibitor complex which prevents the enzyme substrate complex from forming. It is theorised that the higher concentration of substrate to that of inhibitor the lower the rate of inhibitor interaction thus allowing Vmax to be obtained.
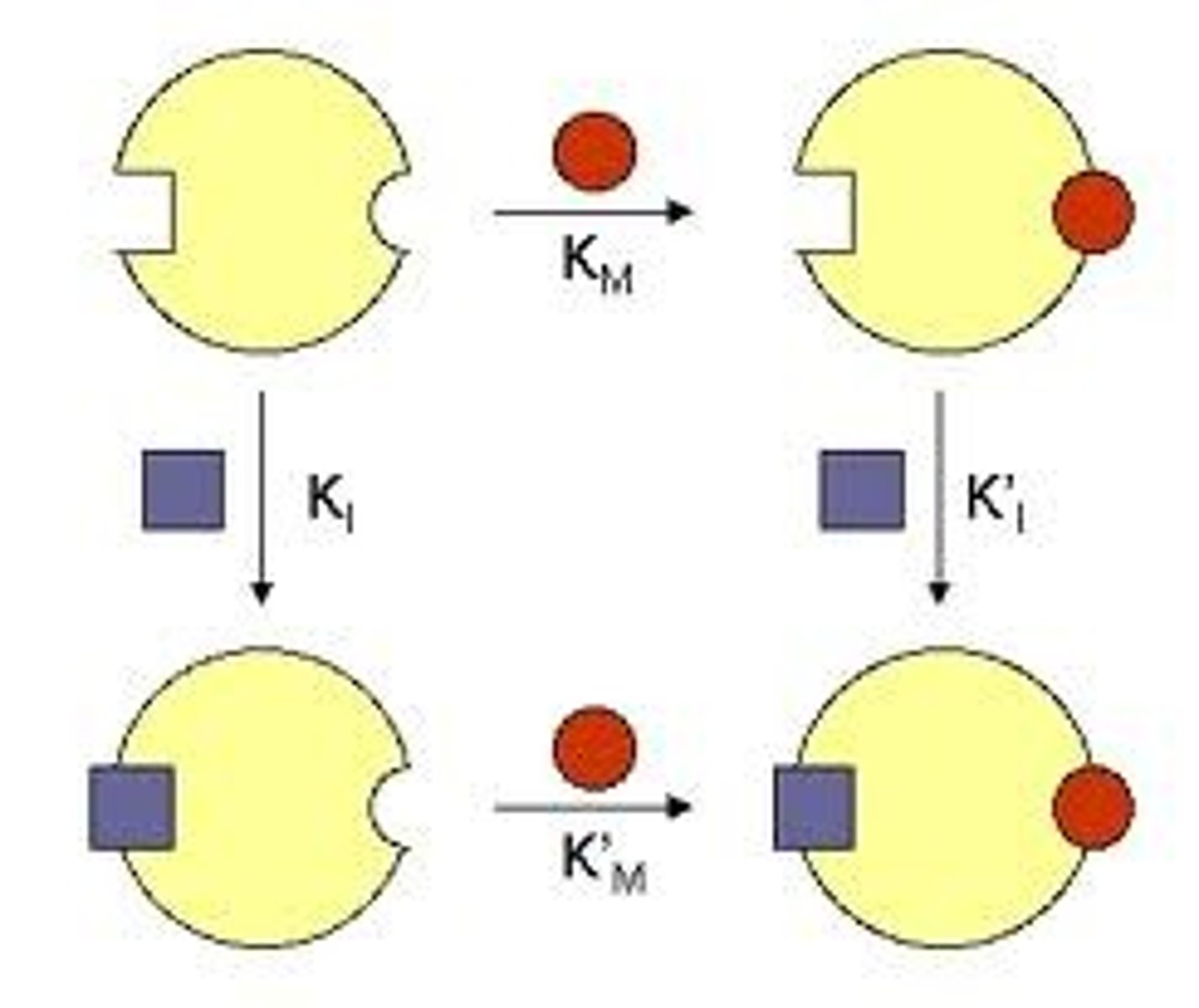
Non Competitive Inhibitor
These inhibitors do not appear to have the same structure as substrate, it therefor binds at a different site on the enzyme and the ES complex can still form. It does however hinder the catylitic action of the enzyme and the end product is never produced.
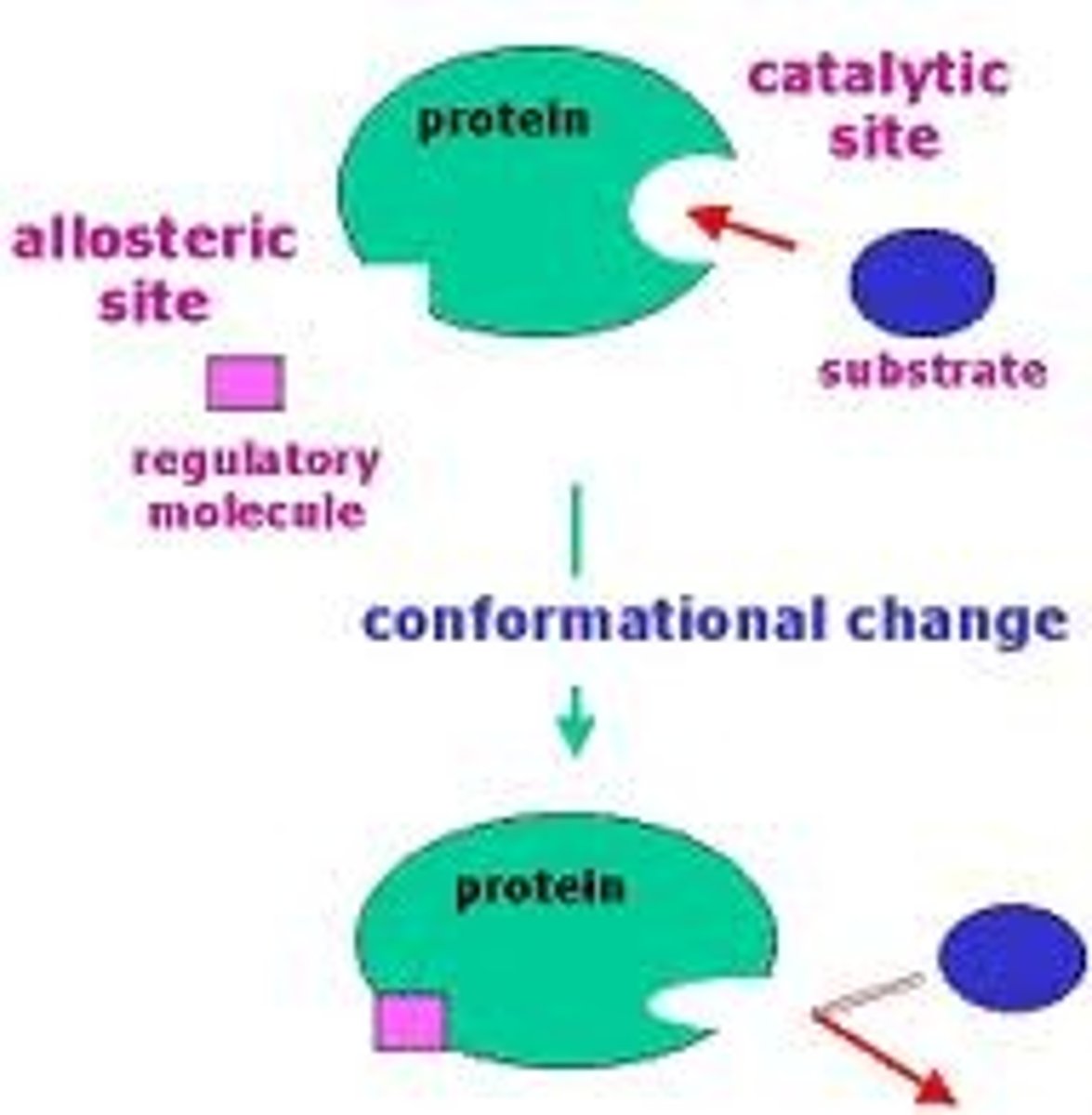
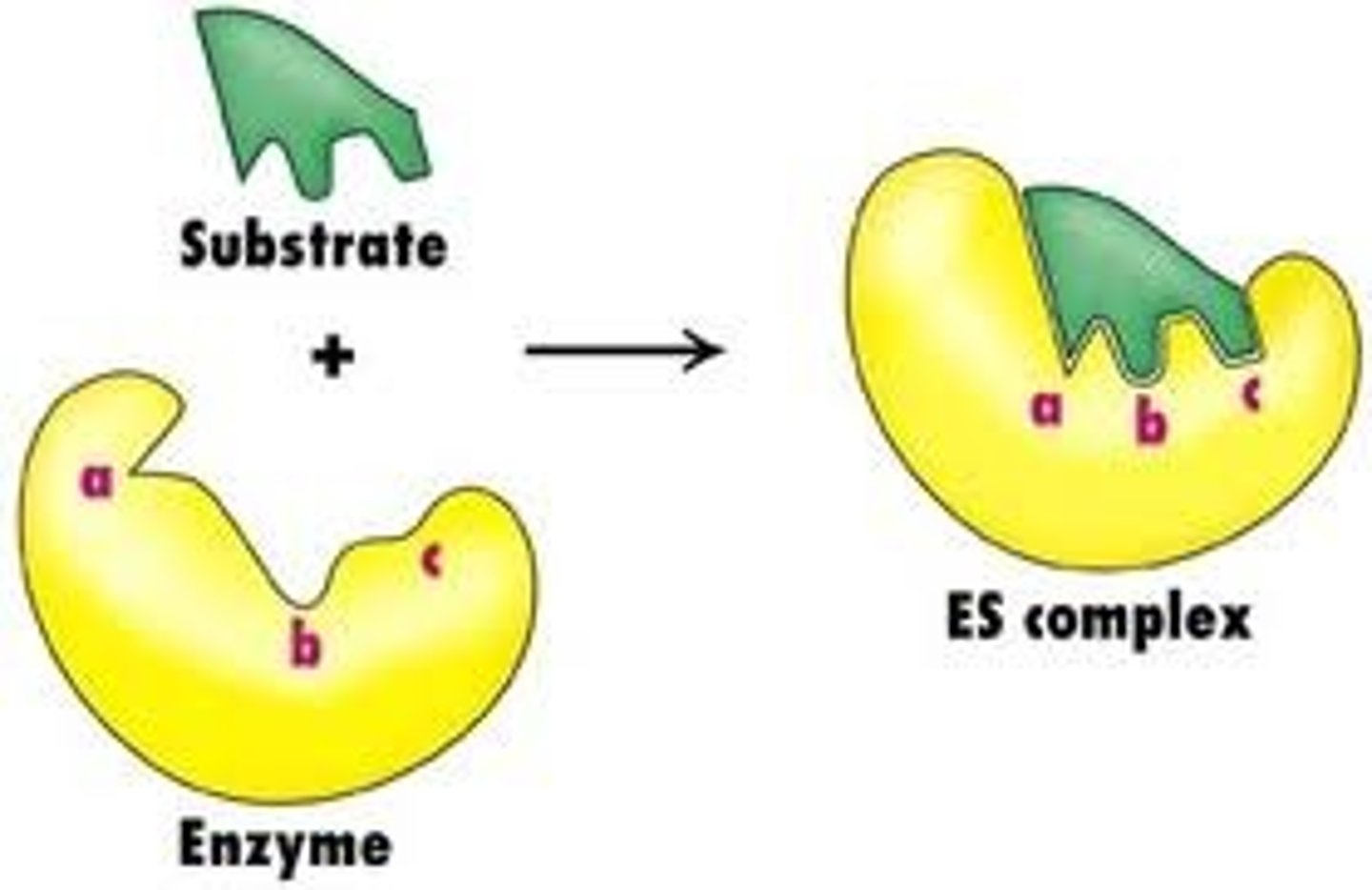
Induced Fit
The term used to describe the effect of a substrate on the active site of an enzyme on binding. This suggests that the active site and substrate are not an exact fit but the actual binding of the substrate induces a change in the structure of the active site.
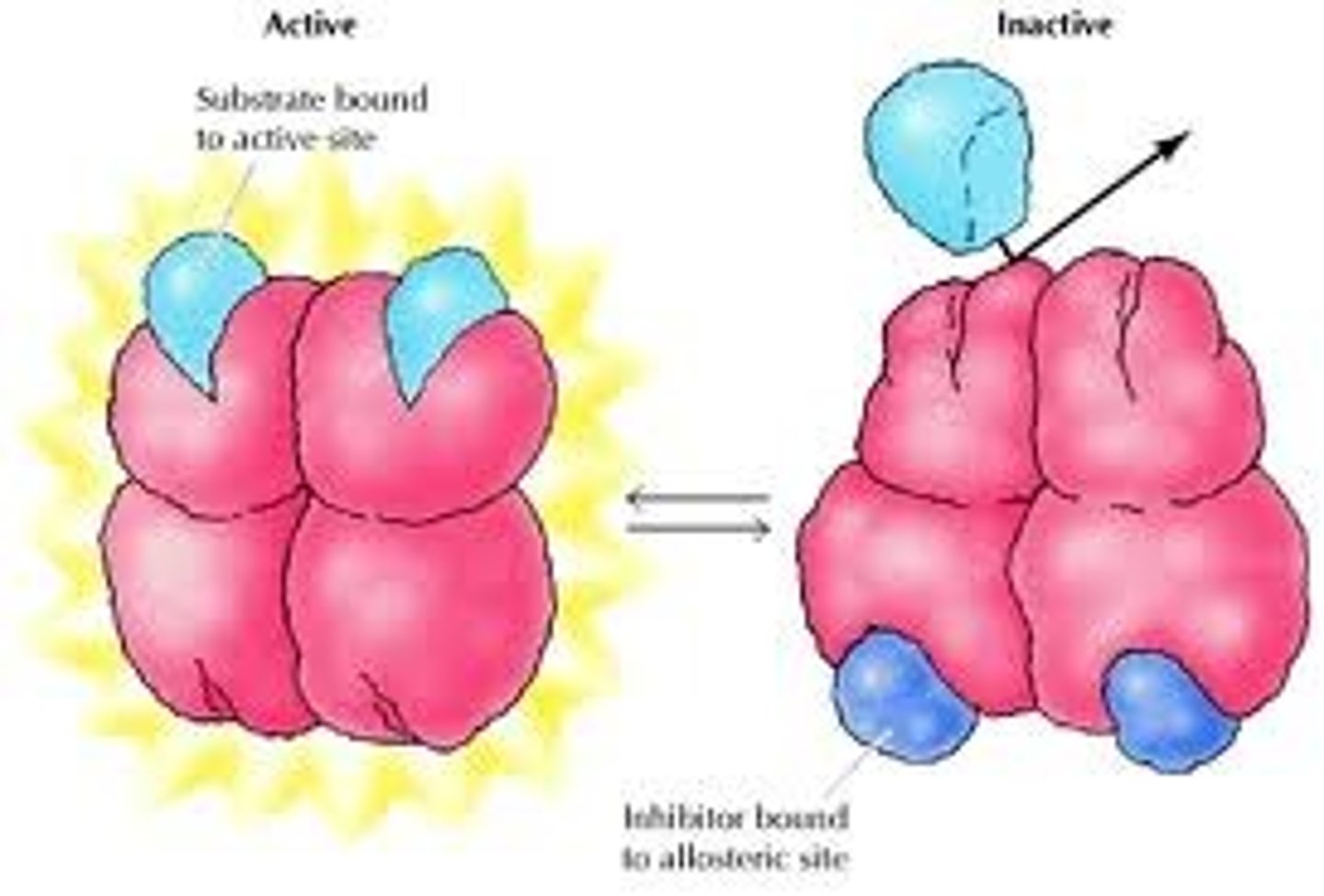
Allosteric Regulation
The form of enzyme regulation brought about when an effector molecule binds to an enzyme at it's allosteric site, thus bringing about changes in it's conformation and therefor effecting it's ability to function. This form of regulation is immediately effective and also immediately reversible.
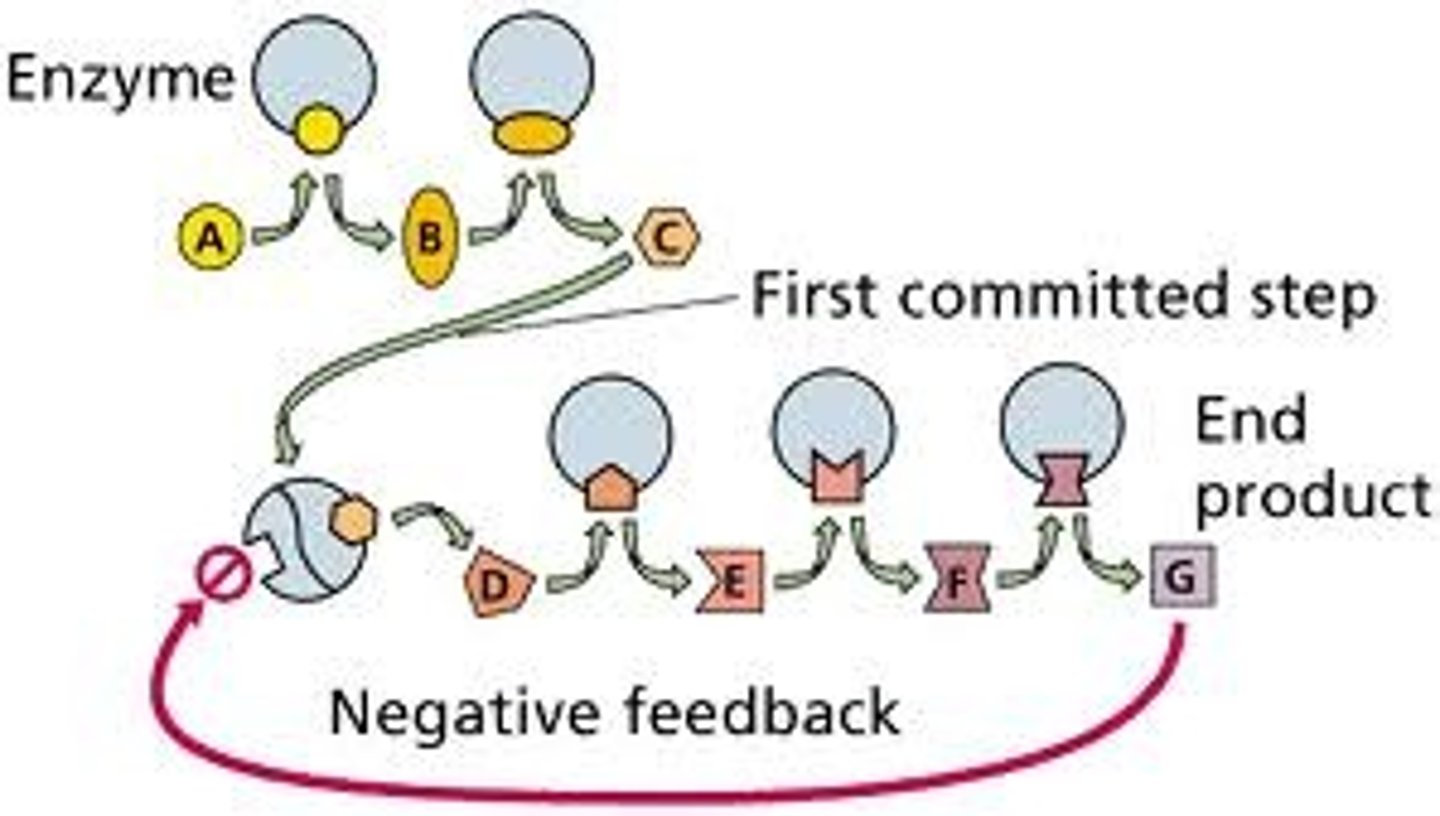
Feedback inhibition of regulatory enzymes
This is the mechanism by which the activity of an enzyme is allosterically effected by the later products in the catalytic pathway, thus preventing over production of the product. So the penultimate product of the enzyme also acts as the effector molecule at the enzyme allosteric site.
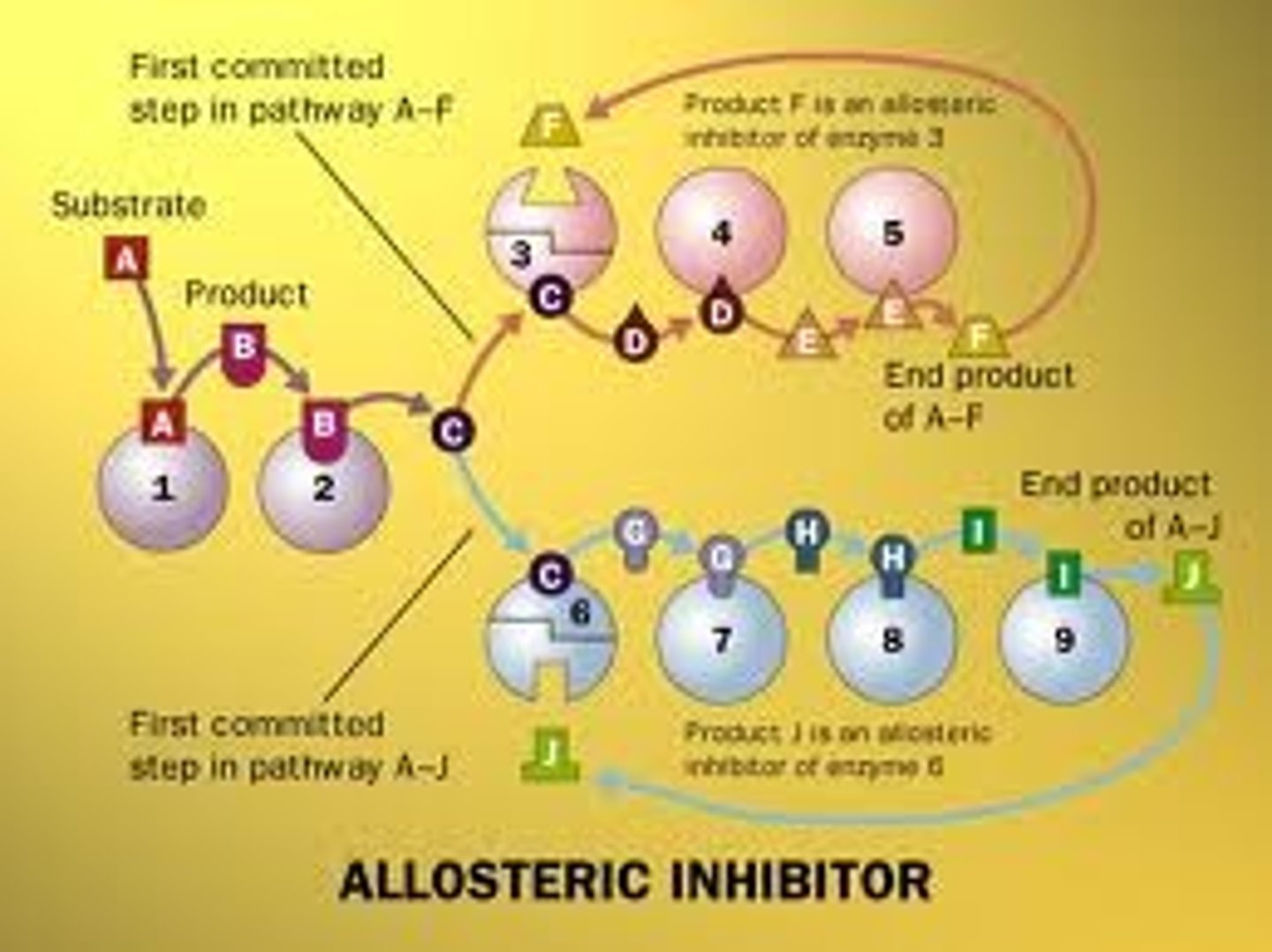
Allosteric Inhibitor
A product produced later in a catalytic pathway which inhibits the activity of enxymes earlier in the catalytic pathway.
Allosteric Activator
These effectors are often products of an earlier step in a catalytic pathway, they act to increase the activity of an enzyme at a time of high substrate availability.
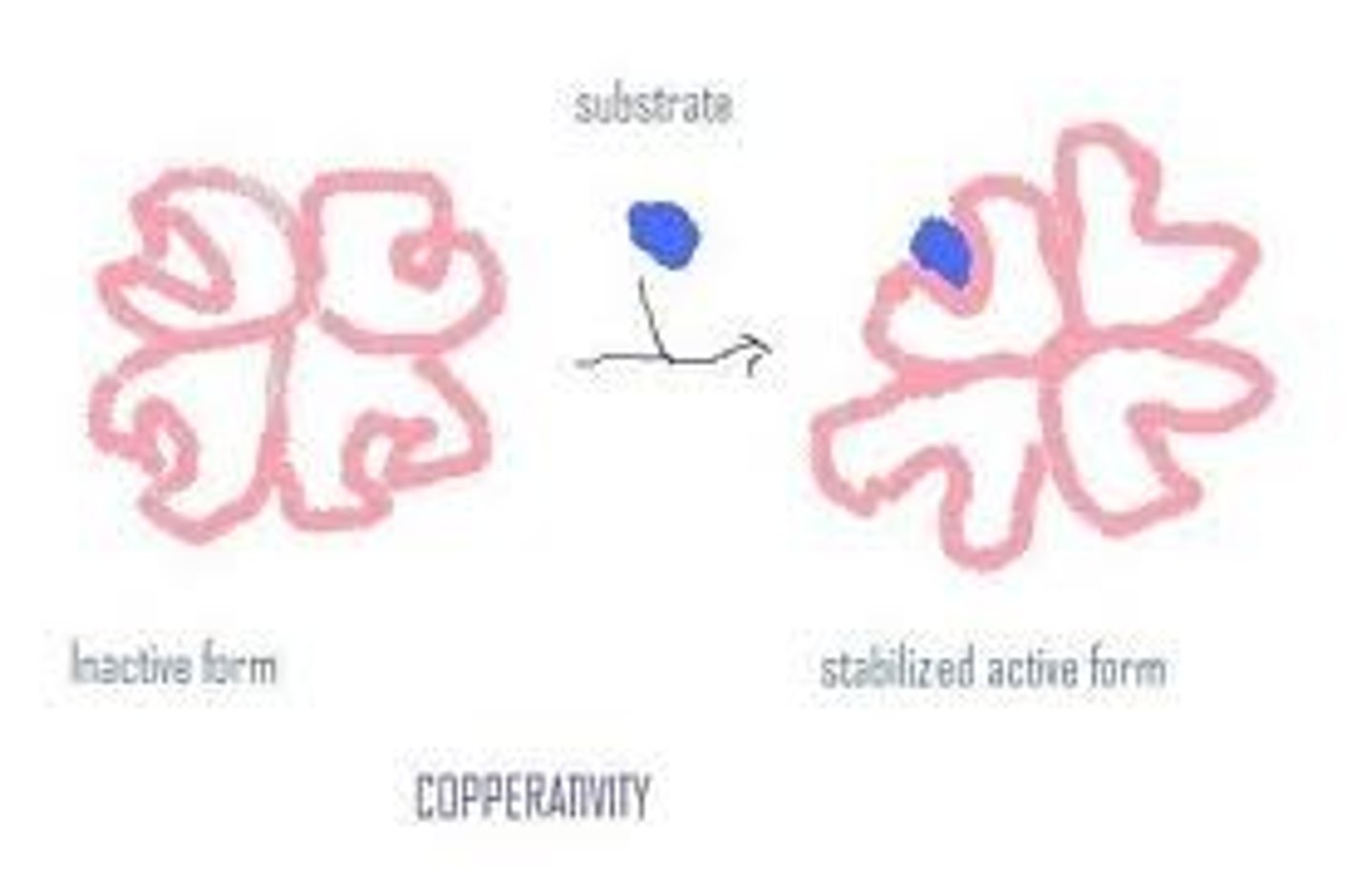
Cooperativity
This is the concept that allosteric enzymes have multiple active sites and that binding of a bustrate to one of these active sites changes the conformation of the other sites making ES binding more likely. This gives the enzyme greater sensitivity to change allowing better responsiveness to changes in substrate availability.
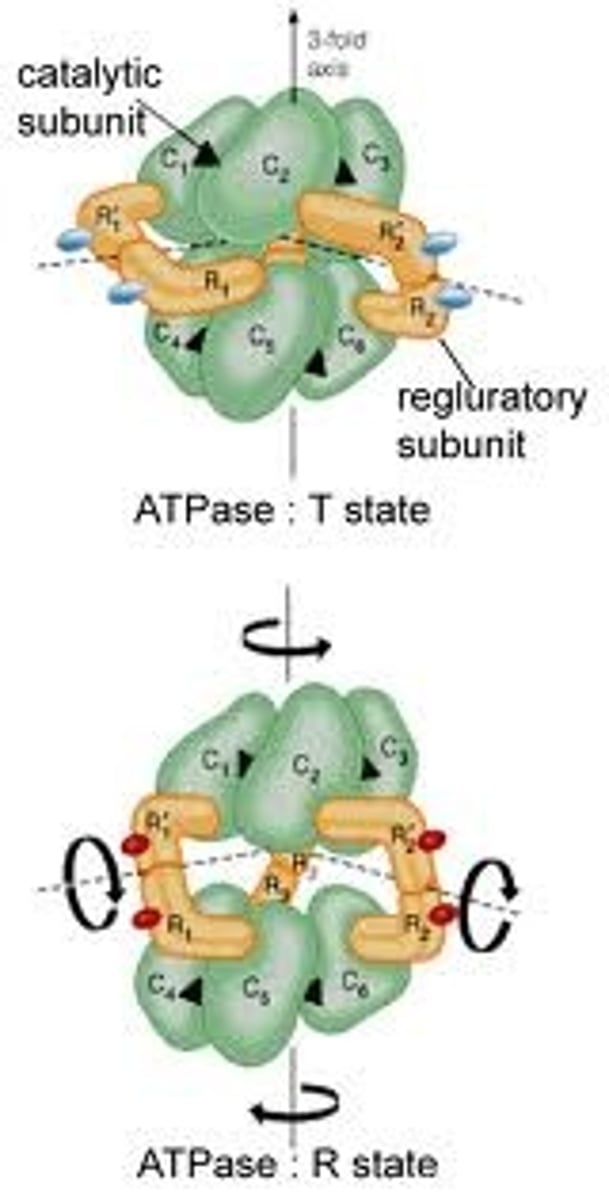
Allosteric enzymes
Five key characteristics (of this enzyme)
1.Larger, multi-subunit proteins consisting generally of two different subunits eg. catalytic and regulatory.
2. Substrates bind cooperatively to active sites on catalytic subunits.
3. A plot of v against s produces an S shaped sigmoid curve.
4. Effectors for these enzymes can be inhibiting or activating and their binding can also produce sigmoid curves.
5. Feedback inhibition can occur- the end product of the enzymes pathway can inhibit enzyme activity.
Reversible covalent modification
This process involves the reversible addition of a small chemical group (e.g phosphate, acetyl) to the side chain of a particular amino acid residue. The most common example of this modification is protein phosphorolation. This process plays a major role in cell functioning.
Plasmalemma
The name by which the cell membrane of a plant is sometimes known.
Spherical Micelle
The most stable structure for an aggregate of single tailed amphipathic liquid molecules e.g detergent in water.
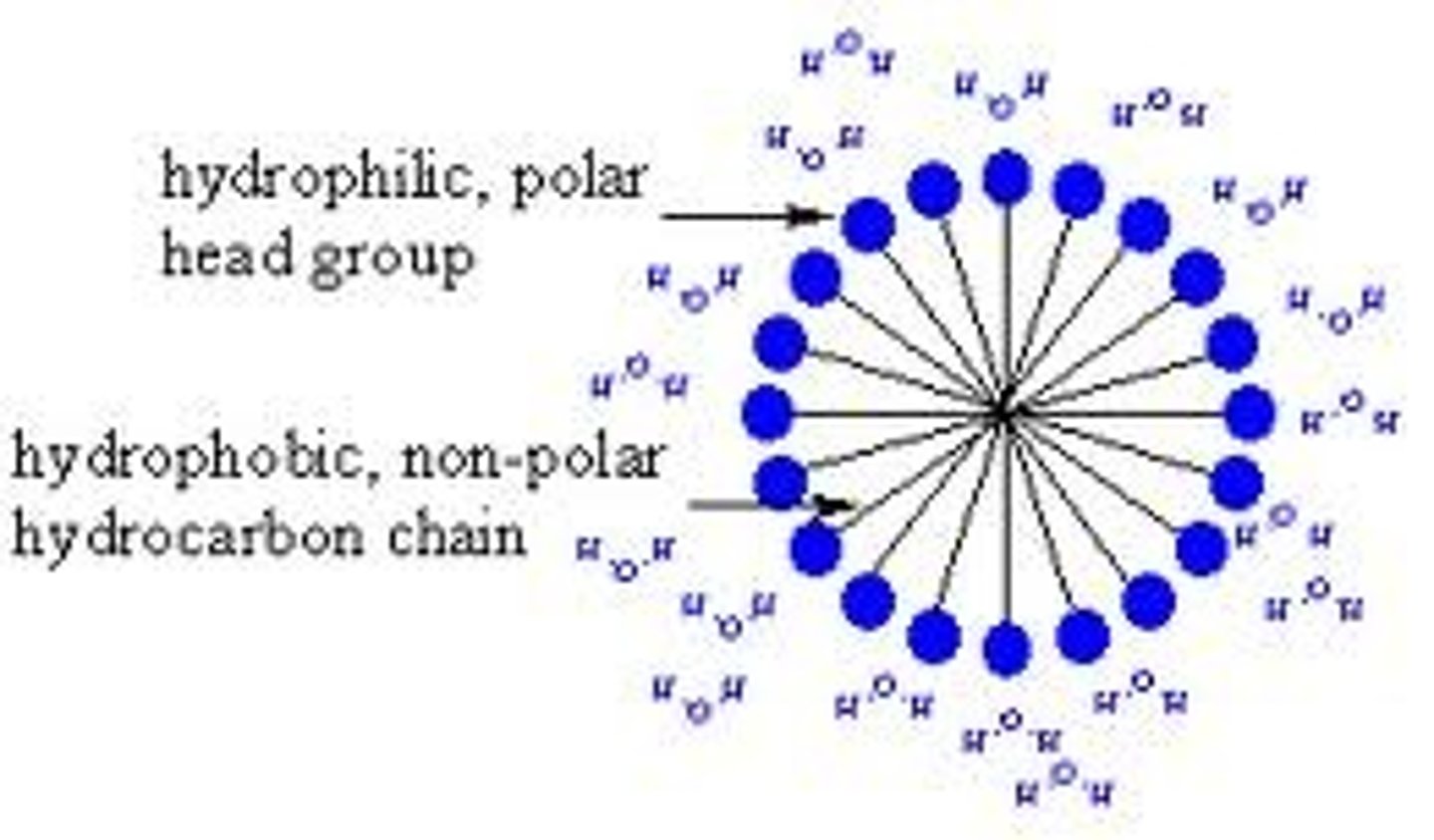
Amphipathic Lipids
These lipids are molecules that are mostly lipid-like (hydrophobic) in structure, but at one end have a region that is polar or ionic (hydrophilic). The hydrophilic region is usually referred to as the head group, and the lipid portion is known as the tail(s). Cell membranes typically consist of three separate classes of lipids of this type. These include phospholipids, glycolipids, and steroids.
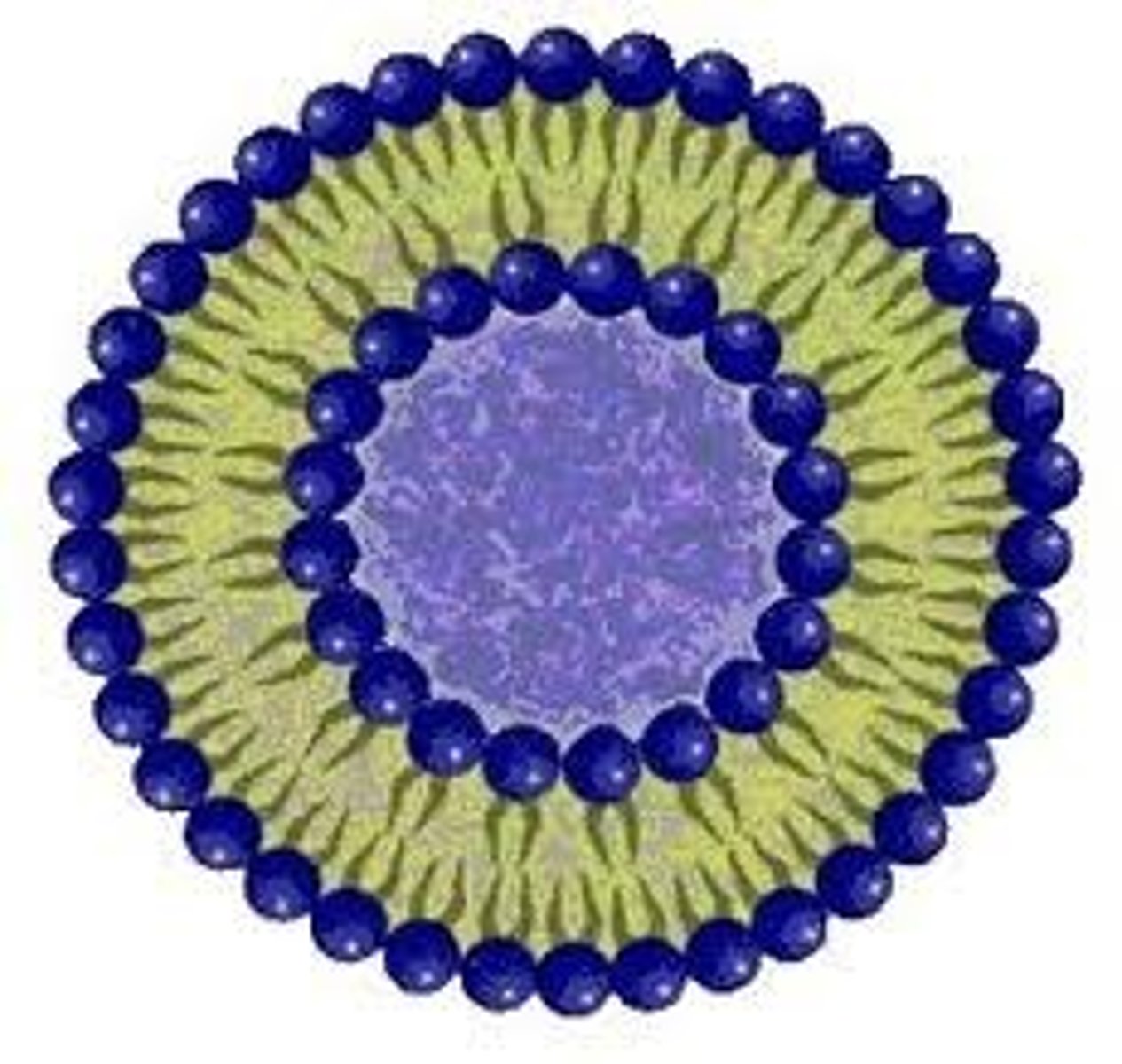
Liposomes
The structure formed by two tailed lipids when mixed with water, it consists of a spherical bilayer withthe hydrophobic tails pointing inwards and the hydrophillic heads pointing outwards in close contact with each other and the water.
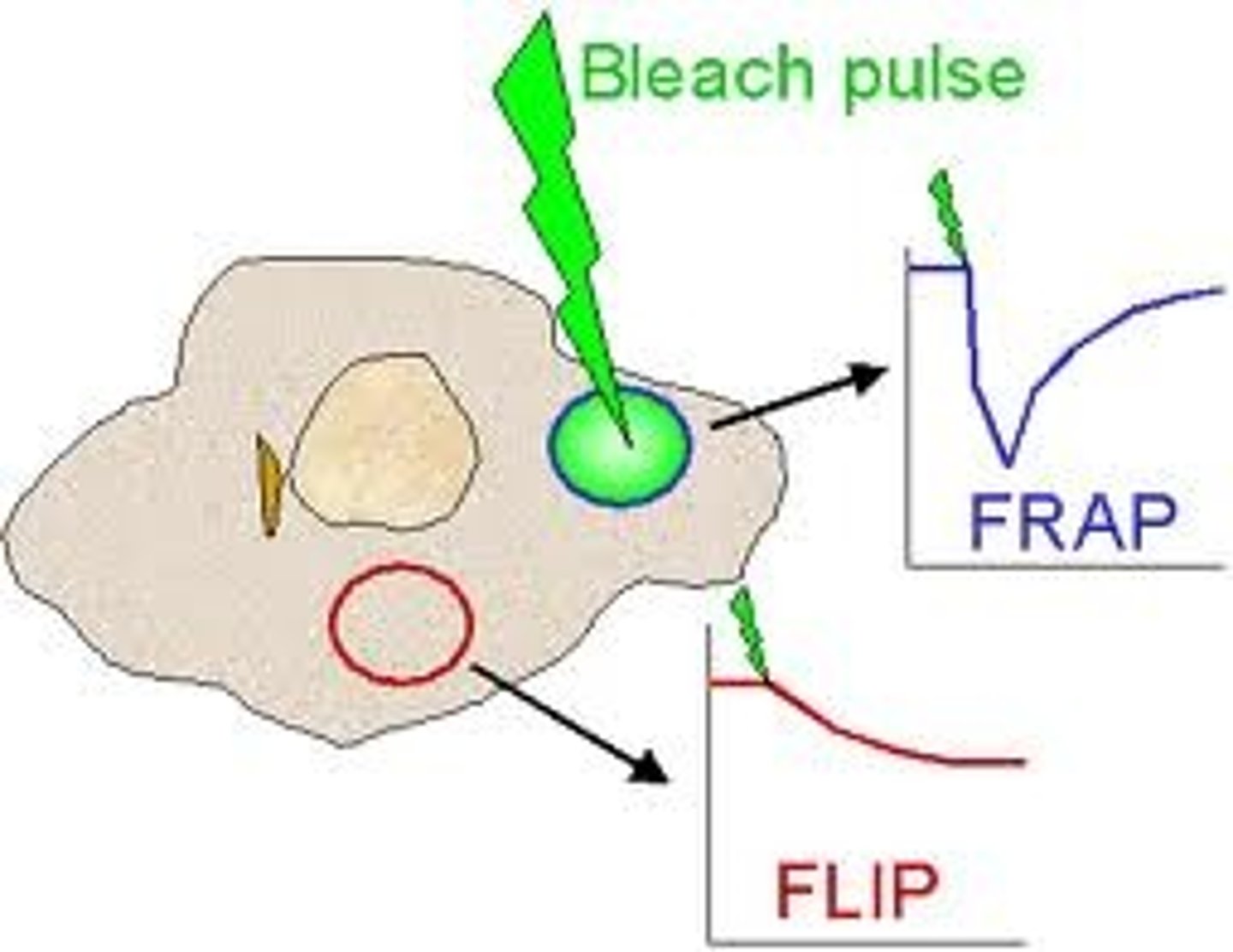
FRAP (Fluorescence recovery after photobleaching)
An optical technique capable of quantifying the two dimensional lateral diffusion of a molecularly thin film containing fluorescently labeled probes, or to examine single cells. This technique is very useful in biological studies of cell membrane diffusion and protein binding. In addition, surface deposition of a fluorescing phospholipid bilayer (or monolayer) allows the characterization of hydrophilic (or hydrophobic) surfaces in terms of surface structure and free energy. Similar, though less well known, techniques have been developed to investigate the 3-dimensional diffusion and binding of molecules inside the cell; they are also referred to as FRAP.
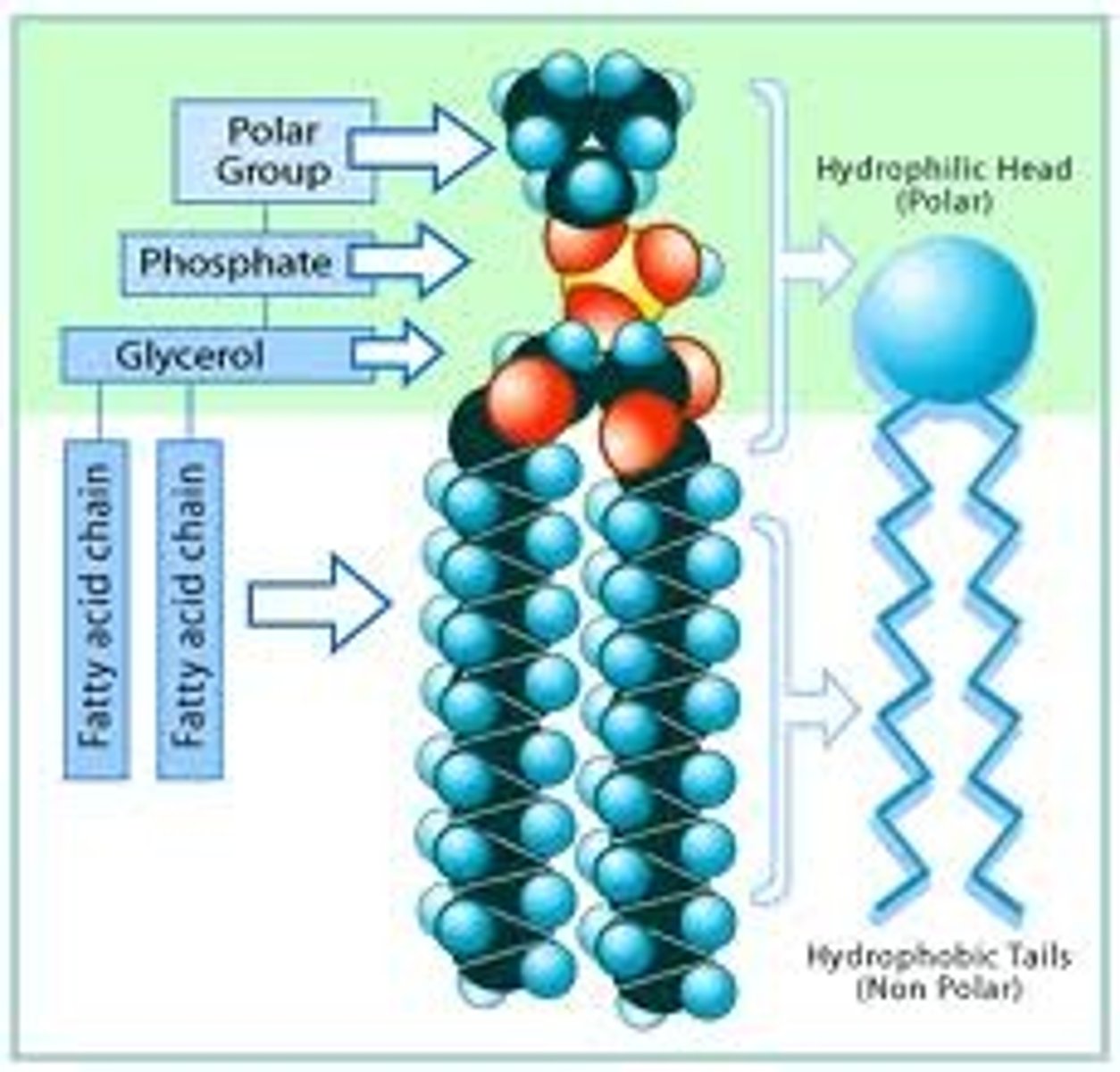
Phospholipids
Phospholipids are a class of lipids and are a major component of all cell membranes as they can form lipid bilayers. Most phospholipids contain a diglyceride, a phosphate group, and a simple organic molecule such as choline; one exception to this rule is sphingomyelin, which is derived from sphingosine instead of glycerol. The first phospholipid identified as such in biological tissues was lecithin, or phosphatidylcholine, in the egg yolk, by Theodore Nicolas Gobley, a French chemist and pharmacist, in 1847. The structure of the a phospholipid molecule consists of hydrophobic tails and hydrophilic heads, it also consists of cholesterol molecules which are found in-between the spaces of the phospholipid.
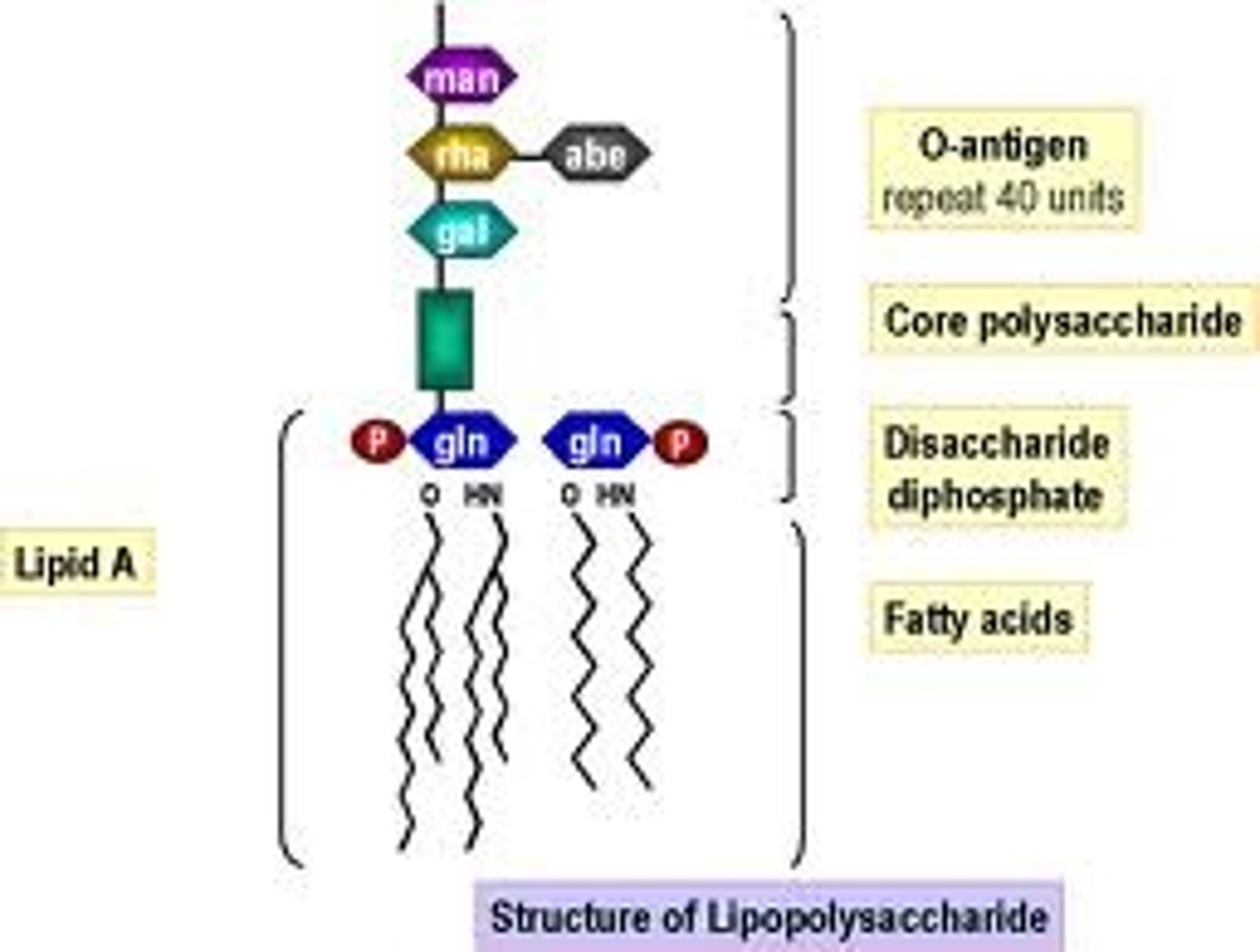
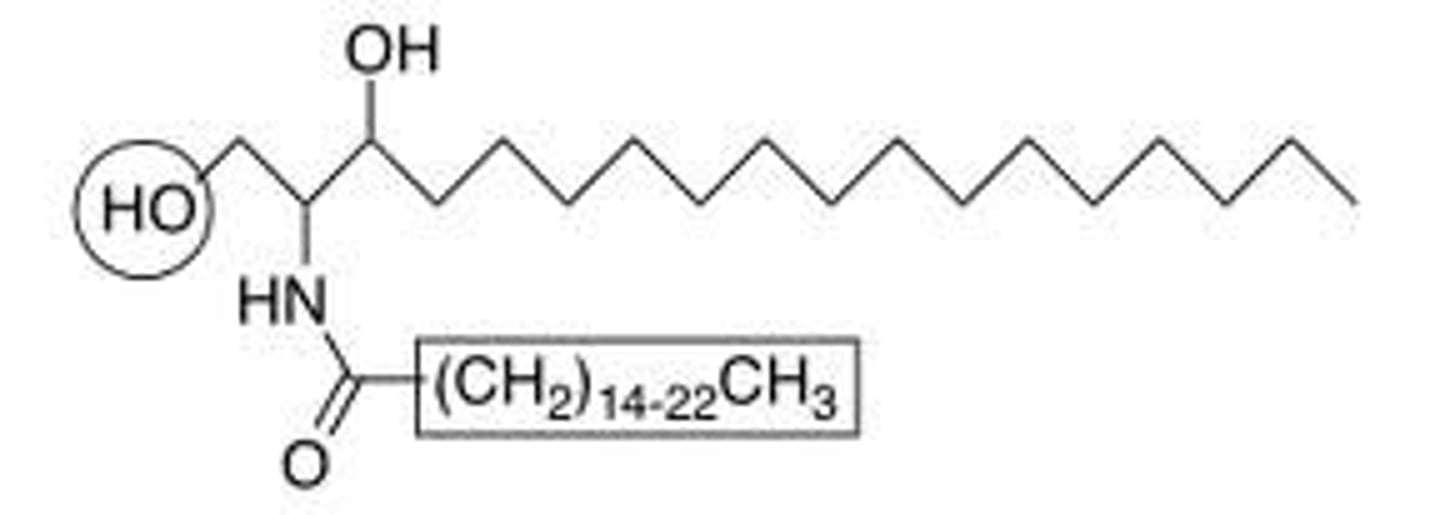
Lipopolysaccharides
Abbreviated as(LPS), also known as lipoglycans, are large molecules consisting of a lipid and a polysaccharide joined by a covalent bond; they are found in the outer membrane of Gram-negative bacteria, act as endotoxins and elicit strong immune responses in animals.
Sphingolipids
A class of lipids containing a backbone of sphingoid bases, a set of aliphatic amino alcohols that includes sphingosine. They were discovered in brain extracts in the 1870s and were named for the mythological Sphinx because of their enigmatic nature. These compounds play important roles in signal transmission and cell recognition. Sphingolipidoses, or disorders of sphingolipid metabolism, have particular impact on neural tissue. A sphingolipid with an R group consisting of a hydrogen atom only is a ceramide. Other common R groups include phosphocholine, yielding a sphingomyelin, and various sugar monomers or dimers, yielding cerebrosides and globosides, respectively. Cerebrosides and globosides are collectively known as glycosphingolipids.
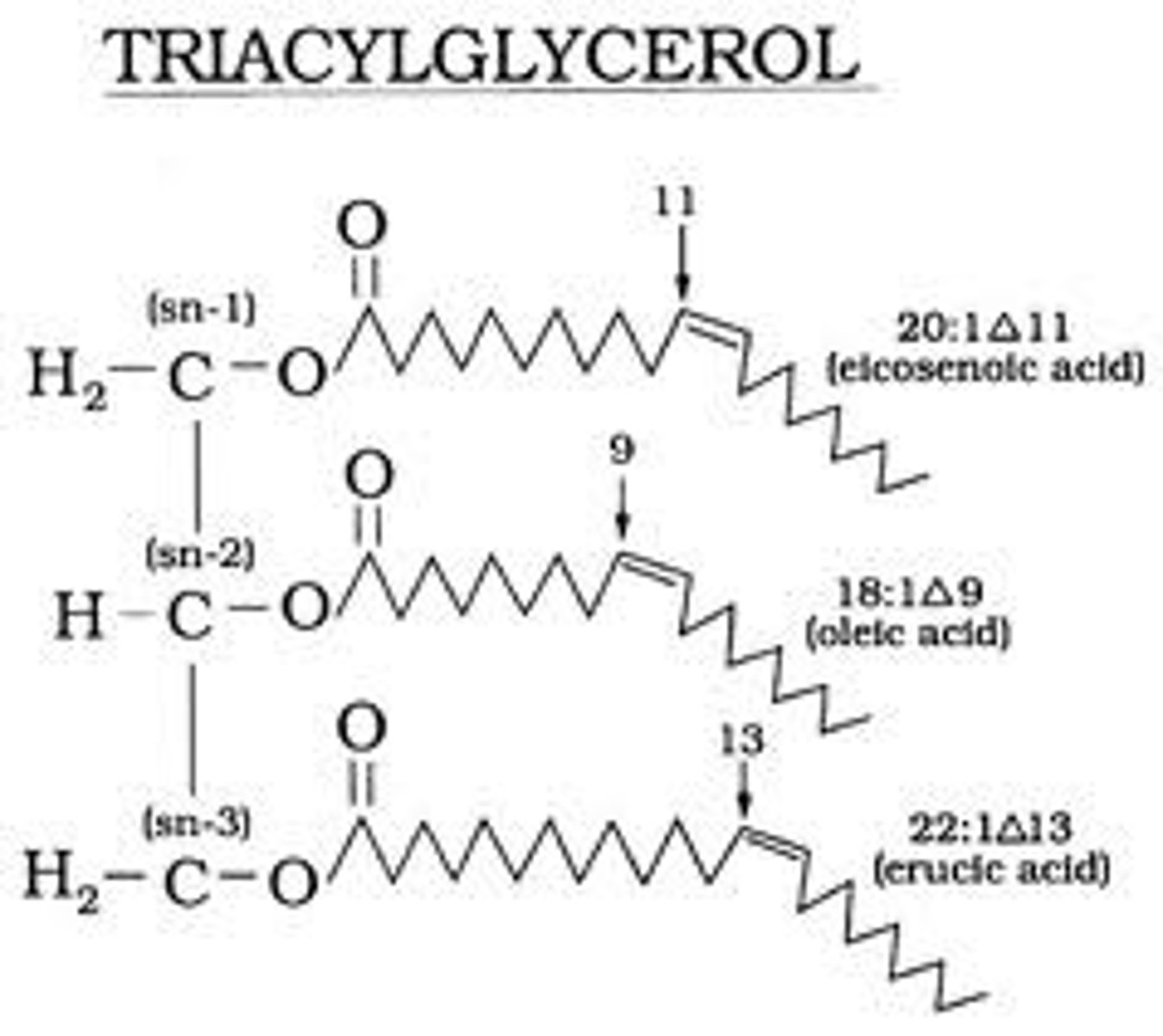
Triacylglycerols
Also known as triglyceride (TG, triacylglycerol, TAG, or triacylglyceride)an ester derived from glycerol and three fatty acids.It is the main constituent of vegetable oil and animal fats.