Hereditary Hemochromatosis (HH) and RFLP
1/10
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No study sessions yet.
11 Terms
How is PCR-RFLP used to diagnose HH
PCR-RFLP to detect HFE variants (C282Y mutation)
Used to genotype genetic disorders caused by point mutations, insertions, or deletions
After PCR, amplified products are cut with restriction enzymes at specific locations
Analyzed results can determine if point mutation is present, and the classification of the individual as heterozygous (carrier) or homozygous (mutant or wild type)
There are 2 alleles for each gene (potentially different, one from the mother, one from the father)
Band patterns on a gel will indicate the genotype based on how particular enzymes cut the PCR product
Molecular diagnostics of hereditary hemochromatosis
Any molecular method that can detect a single nucleotide variant can be used to diagnose HH
Methods include...
PCR, PCR-RFLP, Sequencing (Sanger, pyrosequencing, NGS), & Hybridization assays
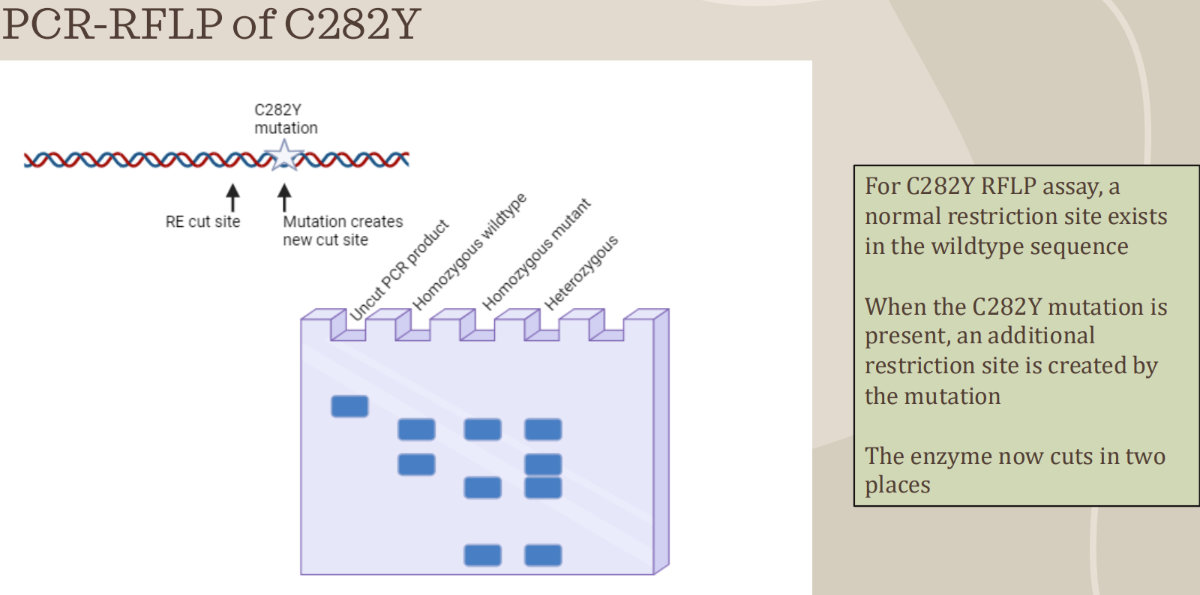
How does a C282Y mutation look like on RFLP
Uncut = 390 bp
WT = 2 bands
250bp & 140bp
MT = 3 bands
250bp, 111bp, & 29 bp
Hetero. = 4 bands
250, 140, 111, & 29bp
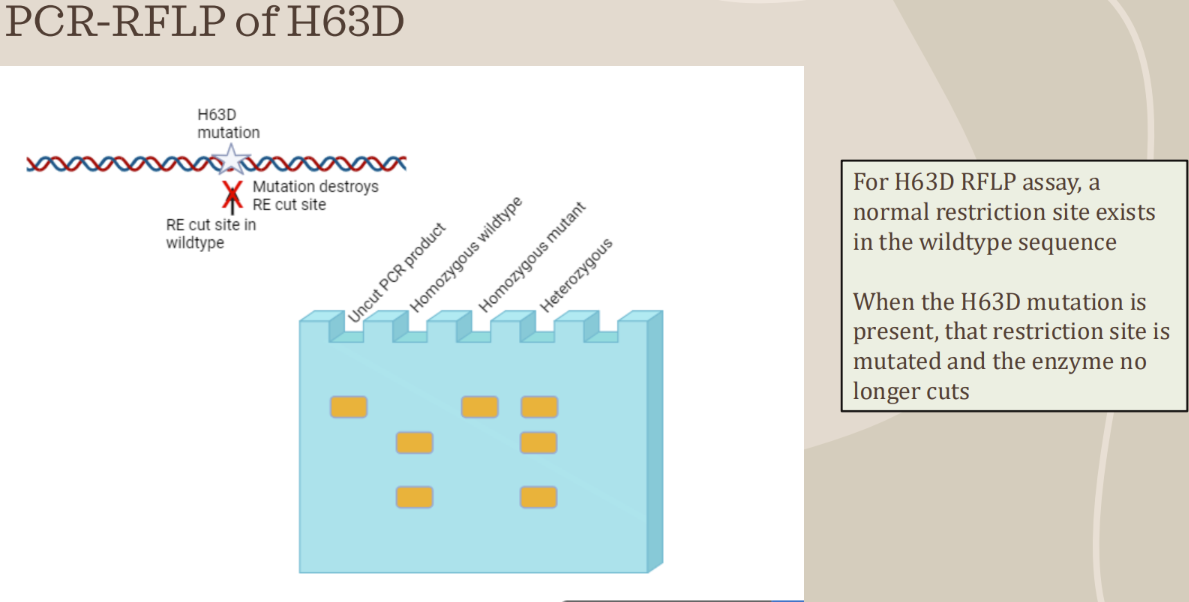
How does a H63D mutation look like
Uncut = 208 bp
WT = 2 bands
138bp & 70 bp
MT = 1 band
208 bp
Heterozygous = 3 bands
208, 138, & 70 bp
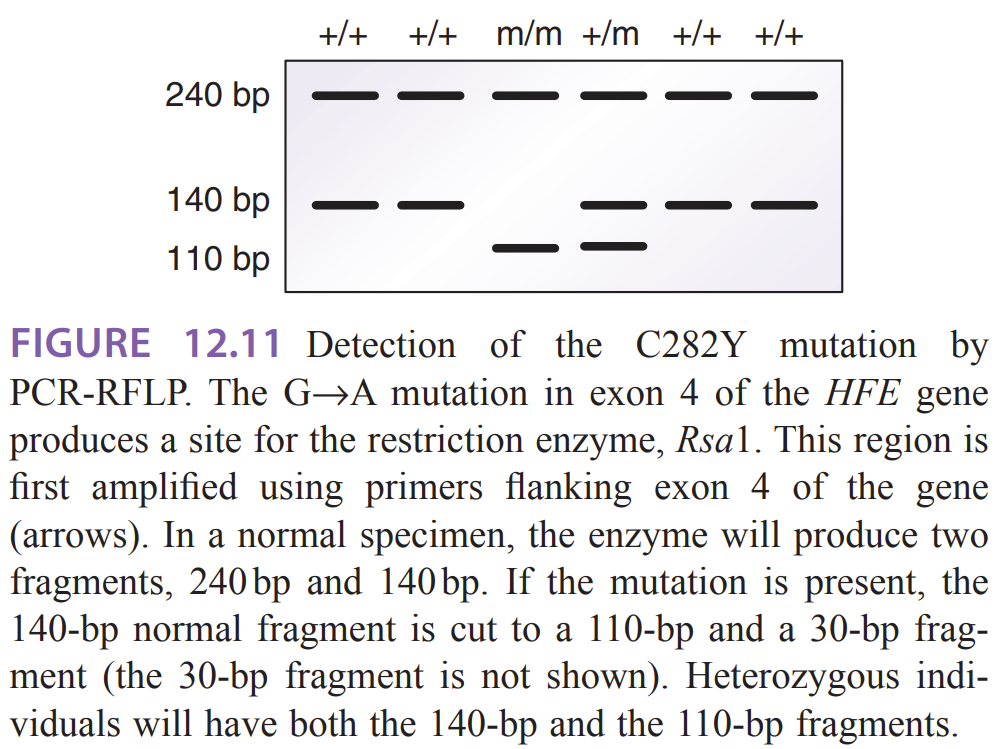
How does HH look like on a gel
Normal (wild-type, +/+) — two fragments 240 bp and 140 bp after RsaI digestion.
Heterozygous (+/m) — shows a combination of the normal 140 bp band and the mutant 110 bp band (plus the 240 bp fragment). The 30-bp fragment from the cut 140→110+30 is usually not visible on a standard agarose gel
Homozygous mutant (m/m, C282Y/C282Y) — the 140-bp normal fragment is cut to 110 bp (and 30 bp not shown), so bands are 240 bp and 110 bp (no 140 bp)
Common restriction-digestion problems
A. Partial digestion (incomplete cutting) → can mimic heterozygote
B. Star activity (nonspecific cutting)
C. No digestion observed
D. Smearing or extra bands on gel
E. Small fragment not visible (e.g., the 30-bp fragment)
Assessing predisposition from genotype:
Homozygous C282Y (m/m): highest genetic predisposition to hereditary hemochromatosis — this genotype is the one classically associated with iron-overload disease. The reading notes C282Y is the most frequent mutation and is detectable by PCR-RFLP.
Heterozygous (C282Y carrier, +/m): typically a carrier state; most heterozygotes do not develop the full disease phenotype, though biochemical penetrance/expressivity is variable. The reading states population carrier frequency and that clinical symptoms and iron studies are indicators for testing.
Compound heterozygotes (e.g., C282Y/H63D): may have intermediate or increased risk compared with simple heterozygotes; the reading lists H63D and S65C as other common variants and implies multiple variants can be present. Clinical correlation recommended.
Relate patient results to likelihood of being affected with hemochromatosis
Population and disease frequency (from the reading):
The C282Y mutation is common in people of Caucasian descent (mutation present in ~10% of the Caucasian population); disease frequency is about 2–3 per 1,000.
Clinical correlation (how to translate genotype into likelihood):
Homozygous C282Y (m/m): the genotype most strongly associated with clinical hereditary hemochromatosis. A patient with m/m has a substantially increased risk and should have iron studies (serum ferritin, transferrin-iron saturation) and clinical evaluation for organ involvement (liver, pancreas, heart, skin). The reading specifically states clinical symptoms and increased serum ferritin/transferrin-iron saturation are indications for mutation testing and diagnosis.
Heterozygous C282Y (+/m): usually a carrier; most carriers do not develop overt hemochromatosis but may warrant biochemical follow-up if clinically indicated. Penetrance and expressivity are variable (so a carrier alone is not diagnostic).
Normal genotype (+/+) — low genetic predisposition due to C282Y (other causes of iron overload should be considered if clinical suspicion remains).
Put simply (clinical action guide based on reading):
If PCR-RFLP shows m/m → consider the patient at high genetic risk for HH; order/interpret iron studies (serum ferritin, transferrin saturation) and arrange clinical follow-up.
If +/m → patient is a carrier; usually low immediate risk, but correlate clinically; monitor iron studies if symptoms or family history.
If +/+ → genotype for C282Y does not indicate HFE-related risk; if suspicion remains, evaluate other HFE variants (H63D, S65C) or non-HFE causes. The reading lists other HFE variants that can also be tested.
Explain how iron might play a role in COVID-19 pathogenesis and relate that to the hereditary hemochromatosis genotype
In essence
COVID-19 promotes iron trapping and oxidative damage.
Hereditary hemochromatosis already predisposes to iron overload.
Together, they could amplify one another, worsening inflammation, organ injury, and clotting risk—supporting the article’s view that iron may be a “vicious culprit” in COVID-19 pathogenesis.
How iron might play a role in COVID-19 pathogenesis
The article explains that COVID-19 infection triggers a cytokine-driven inflammatory response, known as a cytokine storm, in which interleukin-6 (IL-6) stimulates increased production of ferritin and hepcidin
Hepcidin traps iron inside macrophages and enterocytes, preventing its release into the bloodstream.
This sequestration causes intracellular iron accumulation and elevated ferritin levels (hyperferritinemia).
The excess iron reacts with oxygen to form reactive oxygen species (ROS) and reactive nitrogen and sulfur species, leading to oxidative stress and cellular damage.
These processes can induce ferroptosis, a form of programmed cell death dependent on iron-driven lipid peroxidation, damaging multiple organs including the lungs, liver, kidney, heart, and nervous system
Iron overload also contributes to mitochondrial dysfunction, gut and lung microbiota imbalance, and coagulopathy—all features observed in severe COVID-19 patients.
The article further links oxidized iron to abnormal blood clotting, suggesting that iron-driven oxidative and coagulative stress aggravates disease severity and mortality risk
In summary, the article proposes that altered iron homeostasis and hyperferritinemia form a vicious cycle: inflammation raises ferritin and hepcidin, which increase intracellular iron, leading to more oxidative injury and inflammation.
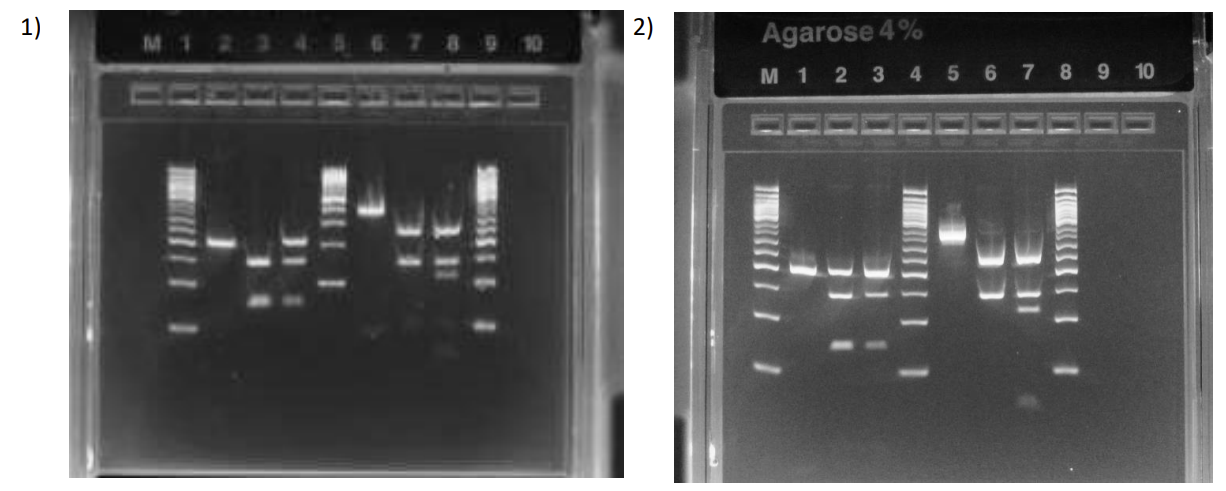
What are these results
Left side it H63D, right side is C282Y
Patient is WT in both mutations
Patient is hetero for H63D and WT for C282Y
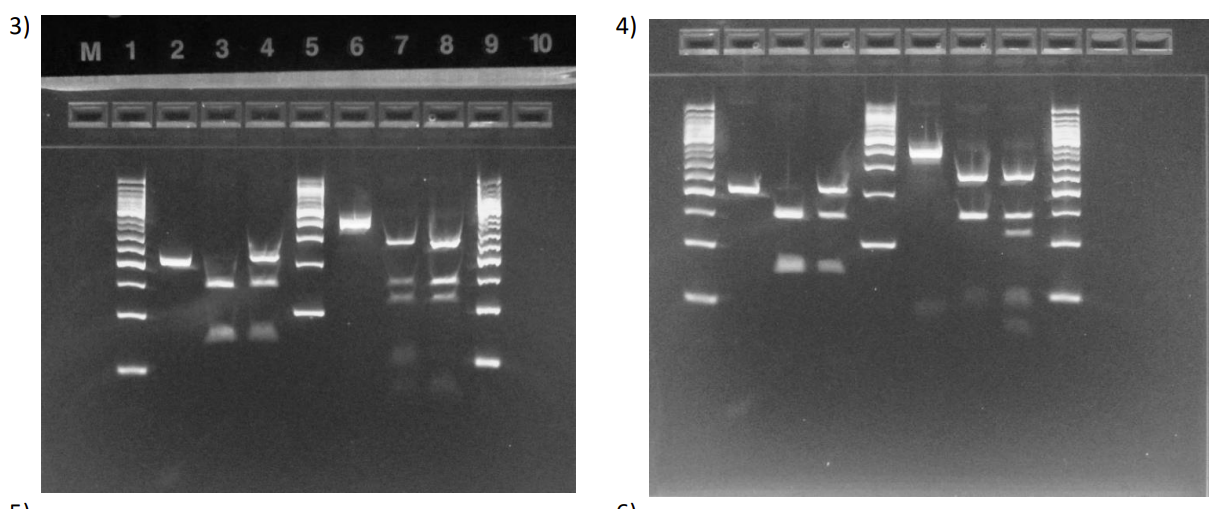
What is each lane
Left side it H63D, right side is C282Y
Patient is WT H63D and Hetero C282Y
Patient is WT for both mutations
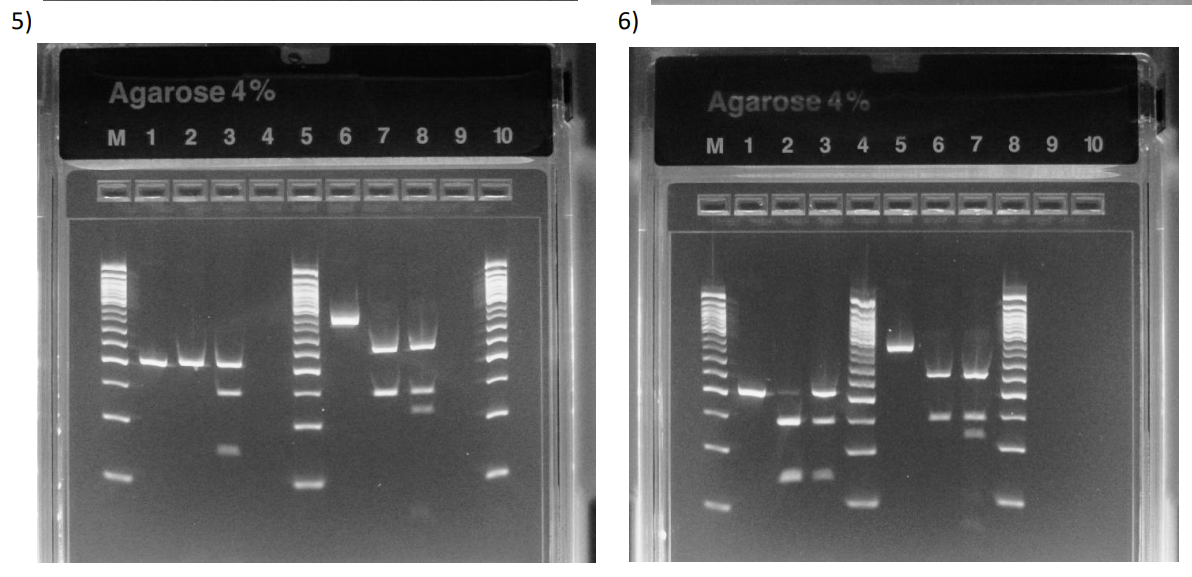
what is each lane? they sick?
Left side it H63D, right side is C282Y
Patient is Homozygous MT for H63D and WT for C282Y
There’s a possibility that the enzyme didn’t cut because the RE didn’t work or you placed the uncut in 2 lanes (needs follow up to confirm H63D MT)
Patient is WT for H63D (with partially uncut fragments) and WT for C282Y
The top band in H63D pt. lane can be from uncut fragments because if it was hetero, then it wouldn’t be as faint (want to follow up with more sequencing assays)
Can be considered a false hetero.