Histone Code and Chromatin Remodeling
1/75
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
76 Terms
What is chromatin?
Chromatin is 1/3 DNA and 2/3 proteins by mass. Think of DNA bound to histones with non-histone proteins (chaperones) attached.
What is the energetically favorable conformation of a histone?
Dimer
What structure is highly conserved across species?
Histone fold
How is an octameric nucleosome constructed?
H3-H4 dimer tetramizes without DNA.
DNA wraps around the H3-H4 tetramer.
Two H2A-H2B dimers are formed
The two H2A-H2B dimers are inserted into the already bound H3-H4-DNA complex to form an octameric nucleosome.
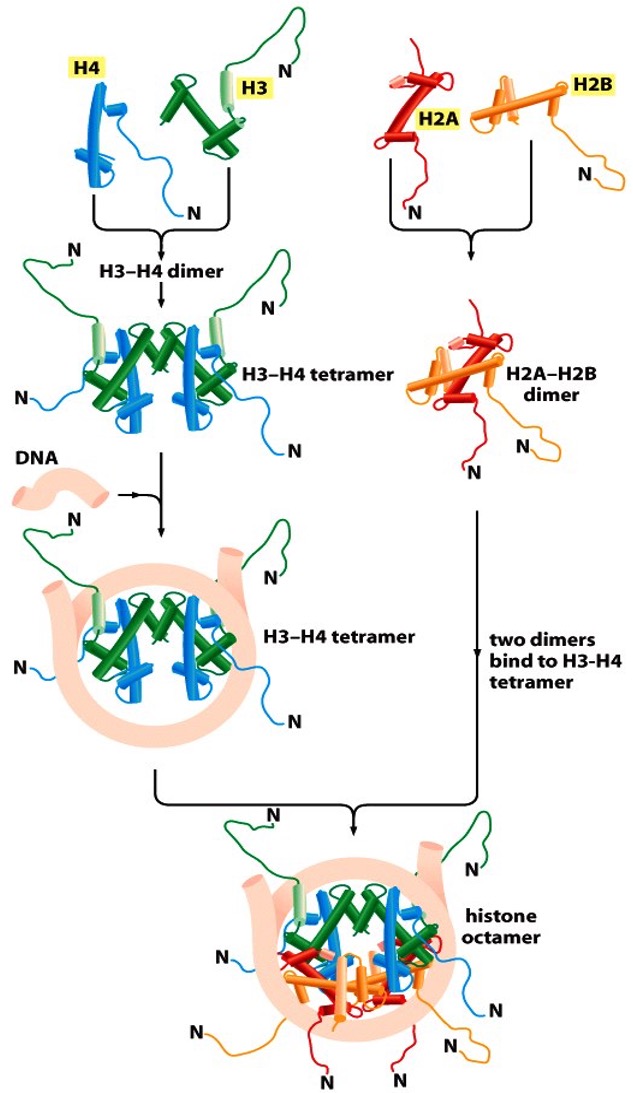
Properties of a nucleosome
8 flexible N-tails
Tails are accessible to modifying enzymes
Assembly requires histone chaperones (NAP1 and CAF1)
How many times does DNA wrap around a histone?
dsDNA wraps two times around one nucleosome (2 gyres)
Which direction does dsDNA wrap around a nucleosome?
Left-handed
Half nucleosome
7 helical turns/DNA axis (1 gyre)
10 bps/turn = 70 bps total for half a nucleosome
Full nucleosome
14 helix turns/DNA axis (2 gyres)
140 bps total to form a complete nucleosome
How does DNA bend around a histone octamer?
Compression of minor grooves inside, facing nucleosome (A=T) and expansion of minor grooves outside, not facing nucleosome (G=C).
The A=T basepairs of the minor groove are preferred to face the nucleosome because there are only 2 H bonds in an A=T bp. This makes it easy to disrupt the bonds when loosening the nucleosome, compared to the 3 H bonds in G=C).
What interacts with the nucleosome?
Narrow minor groove, inside, facing the nucleosome, AT-rich.
What is the broad minor groove?
The minor groove facing the outside of the nucleosome where GC regions are located. It is broadened because when DNA wraps around the nucleosome, the inner minor grooves are narrowed due to curving around the octamer.
What is the role of the flexible histone tails?
Keeps nucleosomes dynamic, gives operational meaning to the cell
List the types of covalent nucleosome tail modifications
Acetylation (loosens)
Deacetylation (tightens)
Methylation (tightens)
Demethylation (loosens)
Describe histone tail-tail interactions
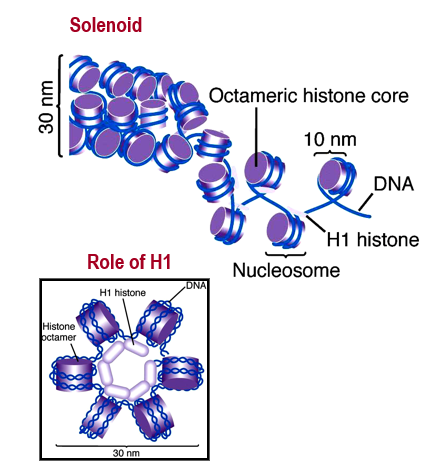
Helps in forming 30 nm fiber that folds in on itself to become longer.
Describe the solenoid structure
Histones are linked together via H1, bringing 6 nucleosomes together to make a symmetrical structure. These structures are 30 nm wide and form the solenoid.
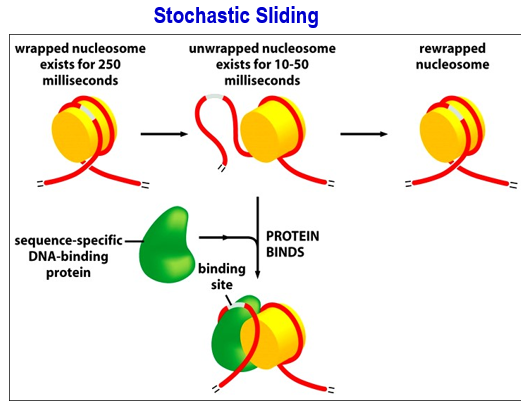
Why are nucleosomes dynamic?
To allow TFs to bind
Example of dynamic nucleosome: Stochastic sliding
Stochastic sliding = Nucleosome breathing.
Unwrapped nucleosome exists for 10-50 ms, exposing the binding sites. Sequence specific DNA-binding proteins/TFs bind. There is a higher chance of this happening if it happens in the condensate.
Can happen simultaneously with the ATP-dependent remodeling
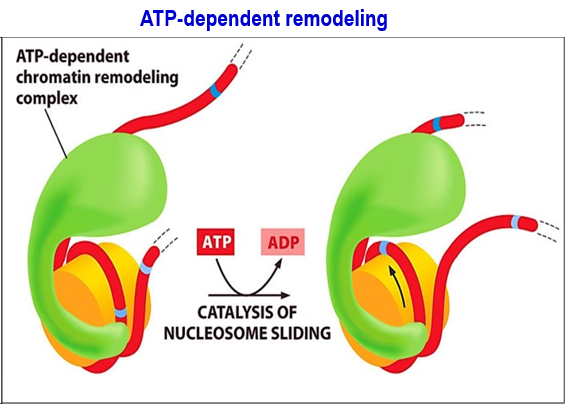
Example of dynamic nucleosome: ATP-dependent remodeling
ATP-dependent remodeling: ATP-dependent chromatin remodeling complex directly targets nucleosome next to the gene where a TF needs to bind → catalysis of nucleosome sliding using ATP energy → leads to shifting of DNA on nucleosome, exposing the binding sites.
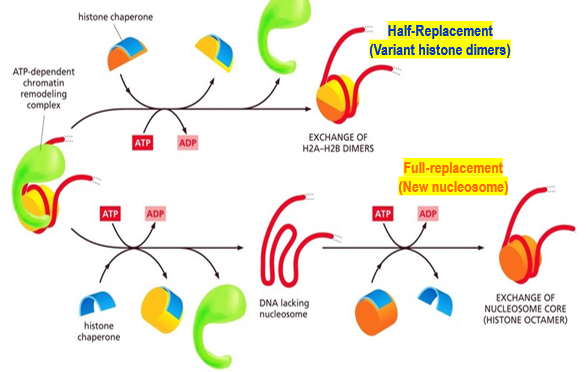
What is the role of histone chaperones
To assist with chromatin remodeling. Use either a half-replacement or full-replacement method
Half-replacement method
Histone chaperone removes the H2A-H2B dimers. (Remember, in histone remodeling, 100% of the H2A & H2B must be removed and replaced). Histone chaperones then replace with variant histone dimers H2AZ and H2AB
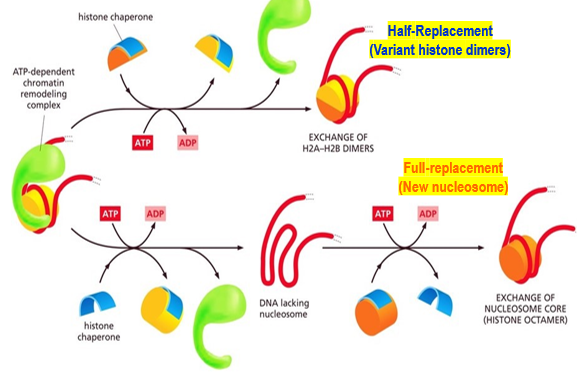
Full-replacement method
Histone chaperones remove the histone octamer completely (H2A, H2B, H3, H4) leaving naked DNA. Replaces with modified histone octamer created a new nucleosome.
What are the three types of chromatin-remodelers?
Structural remodelers (ATP dependent enzymes)
Histone chaperones (remove/insert histones)
Chemical remodelers (HAT, HDAC, etc)
HATs
Histone Acetyl Transferases (activation)
HDACs
Histone Deacetylases (repression)
HMTs
Histone Methyl Transferases (repression)
HDMs
Histone demethylases (activation)
DNMTs
DNA methyl transferases (repression)
DNDMs
DNA Demethylases (activation)
Histone Code Writers
HATs (Histone Acetyl Transferases)
HKs (Histone Kinases)
HKMTs (Histone Lysine Methyl Transferases)
PRMTs (Protein Arginine Methyl Transferases: PRMT -1 to 9)
Histone Code Erasers
HDACs (Histone Deacetylases)
HKDMs (Histone Lysine Demethylases) HKDM -1,2,3,4,5,6
JUMONJIs (HDMS/Histone demethylases): Arg demethylases
H-PPTases (Histone Phosphatases)
PADIs (Peptidyl Arginine Deiminases): PADI-1,2,3,4,6, → citrullination (arginine converted to citrullin)
Histone Code Readers
Chromodomains
Bromodomains
Common Histone Code
Multiple marks on the same reside are mutually exclusive.
Most modifications are on accessible N-tails and C-tails (H2A/H2B).
Few modifications occur in structured globular domains.
Operational meanings of the histone code
Mark for newly replicated DNA.
Mark for damaged DNA that need repair.
Mark to activate transcription.
Mark to repress transcription.
Mark to permanently silence transcription.
Mark to initiate replication (licensing).
Mark that signifies heritable imprinting.
Mark for compaction of the chromatic (heterochromatin).
Mark to open the chromatin (Euchromatin).
The dynamic histone code
Writers, erasers, readers, movers, shapers
H3 Histone Code
This is how cells read given info on tails:
H3K9Me1: Histone H3, K9 (Lysine, position 9, methylated (with one methyl group). Gene silenced.
H3K4Me1K9Ac: Histone H3, K4 (Lysine, position 4, methylated. K9Ac (Lysine, position 9, acetylated). Gene expressed
H3S10PhK14Ac: Histone 3, S10Ph (Serine, position 10, phosphorylated). K14Ac (Lysine, position 114, acetylated. Gene expressed.
H3K27Me3: Histone 3, K27Me3 (Lysine, position 27, methylated with three methyl). Gene silenced
Histone Tail Modifications: Lysine Modifications
Acetylation
Methylation
Sumoylation
Uniquitylation
Lysine acetylation and methylation are competing reactions.
Histone Tail Modifications: Serine Modifications
Phosphorylation → phosphoserine
Histone Tail Modifications: Arginine Modifications
Methylation
H4K4me3
Highly accessible, open chromatin.
Gene expression is ON.
1% abundance
H3K9ac
Highly accessible, open chromatin.
Gene expression is ON.
1% abundance
H3k9me3
Heterochromatin (either constitutive or facultative)
Gene expression is OFF.
25% abundance
H3K27me3
Facultative heterochromatin
Gene expression is OFF.
13% abundance
Why is there higher abundance for heterochromatin than open chromatin?
Because protein coding regions are small.
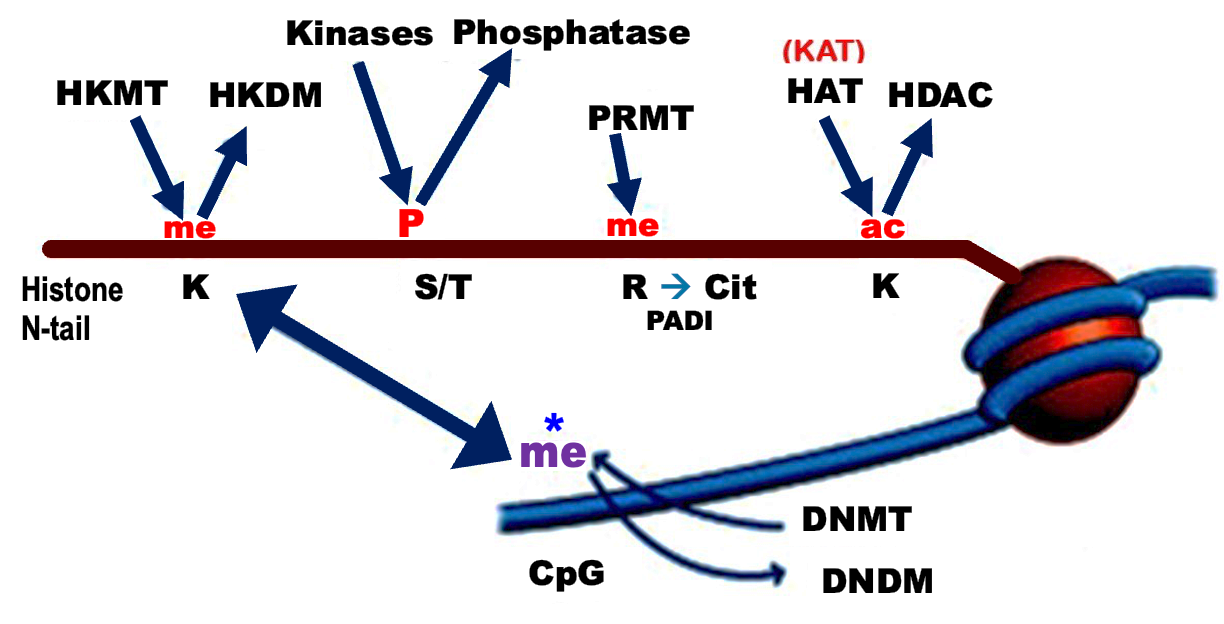
Important photo to remember.
Main histone code writers and erasers.
Six Histone Code-readers
Chromodomain: Binds K-me
Bromodomain: Binds K-ac
14-3-3 proteins: Binds S-p
PHD: K-me
MBT: K-me
Tudor: K-me
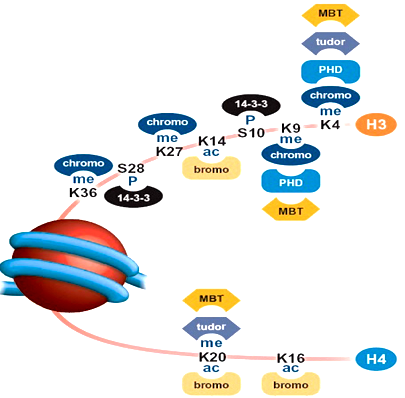
What is the code reader-remodeler complex?
Reads multiple distinct marks on different histones - an example of high fidelity regulation.
Uses a scaffold protein with protein readers imbedded in it, which bind to specific histone modifications on nucleosome surface.
After binding, remodeling complexes (set of proteins: writers, eraser, ATPases, assembly factors) will bind to the scaffold. This will go ahead of the reader and remodel upcoming histones by bringing in TFs and HATs.
Leads to gene expression/silencing
PHD histone reader
It takes a PHD to read a histone code!
PHD Domain recognizes Lysine Trimethylation (K-me3) marks on histone tails
What is the chromodomain?
Chromatin Organization Modifier (Chromo) domain: 30-70 amino acid protein that helps condense heterochromatin.
What structures contain the chromodomain?
HP1 and Polycomb Group (PRC 1/2) proteins have chromodomains that bind methylated Lysines on H3-tails.
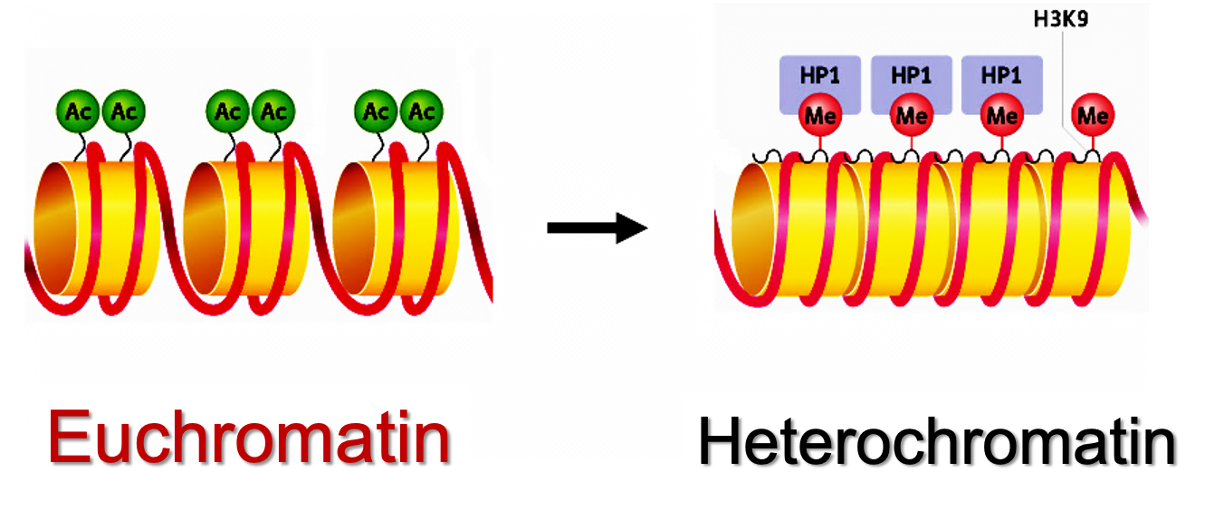
What is the role of HP1 on H3?
The role of HP1 (Heterochromatin-1) on H3 Lysine 9 methylation (H3K9me3) is the conversion from euchromatin → heterochromatin.
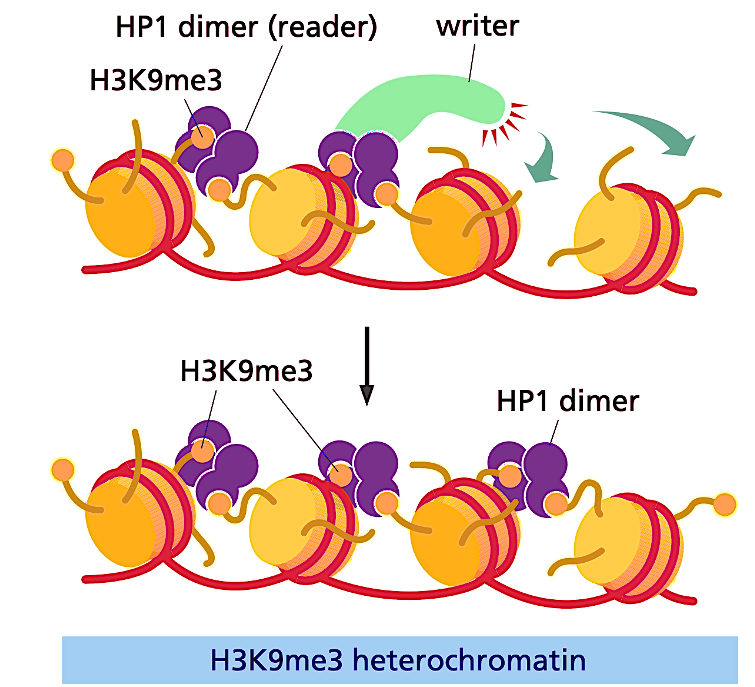
How does HP1 read/write methylation marks on H3K9me3?
HP1 dimer (reader) recruits the writer, moving the modifications down and adding more methyls as the complex moves.
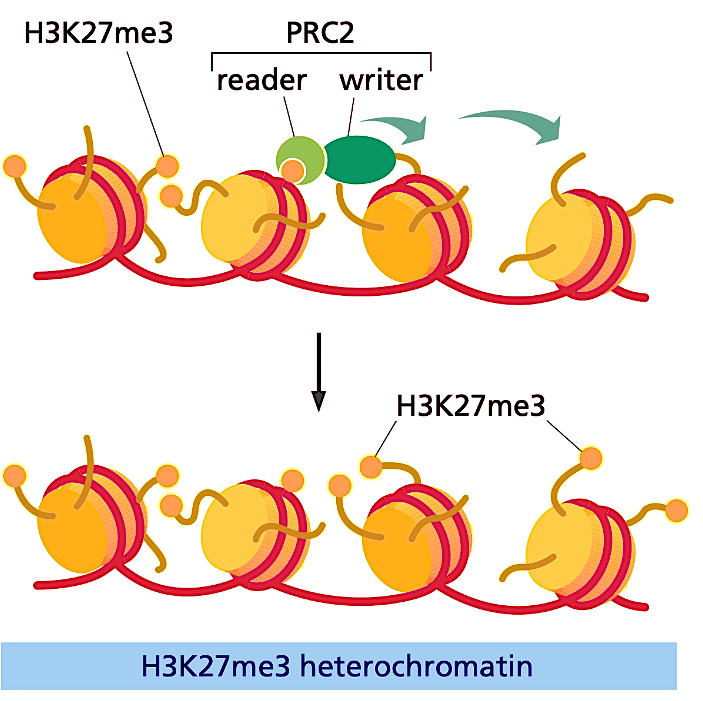
How’s does PRC2 read/write methylation marks on H3K27me3?
PCR2 recruits the writer and together they add methylation marks.
Initiation of methylation marks on heterochromatin
Sequence specific transcription factor binding
Writer (HMT) recruitment and histone modification
Code-reader protein HP1 recruitment
Code-reader-writer complex spreads locally
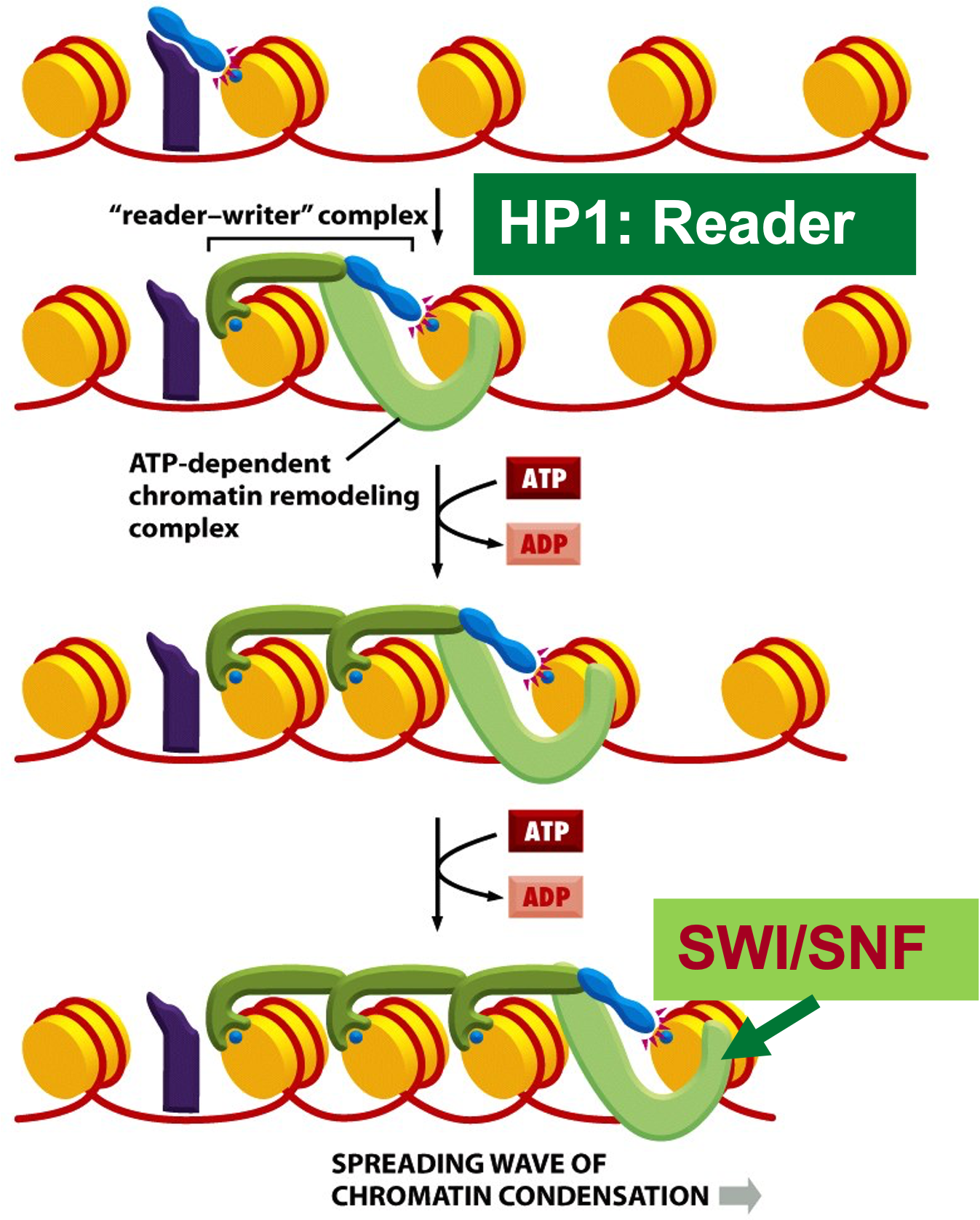
Chromatin remodeler SWI/SNF remodels histones ahead of the Writer-reader-complex
Remember: the HP1 will continuously add the writer along the histones, adding methylation. But transcription factors add the FIRST writer, which recruits the FIRST reader!
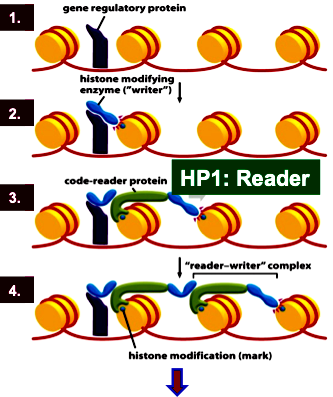
Summarize the heterochromatin gene-silencing mechanism
HMT (writer) puts on methyl marks
HP1 (reader) recognizes H3K9me3
SWI/SNF remodels histones and requires ATP.
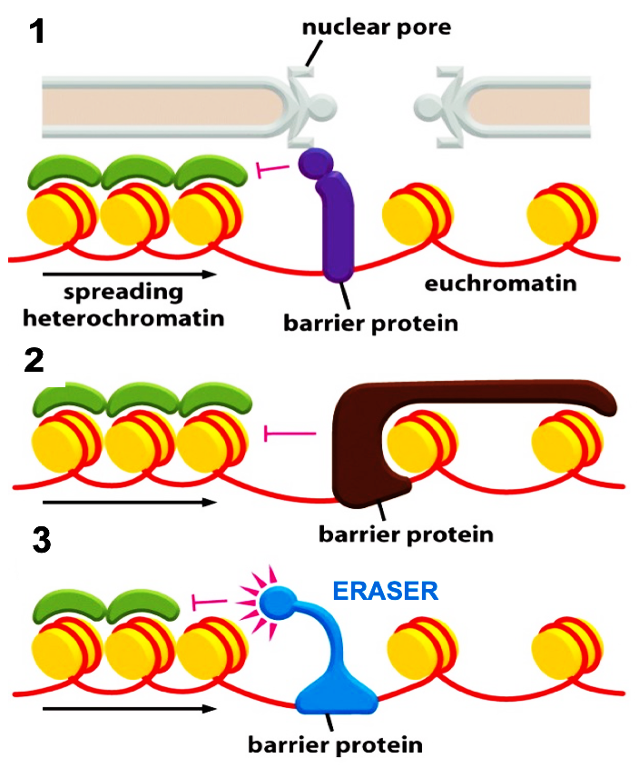
What are the mechanisms of a barrier protein?
STOPS heterochromatin spreading!
Tethering of barrier protein onto nuclear pore and stops heterochromatin spreading
Tight wrapping of a barrier protein to the nucleosome on the euchromatin side
Eraser recruitment: barrier protein (HDMs) continuously erase H3K9me3 on heterochromatin side and stops its spread.
These can exist at the same time!!
What is the bivalent histone code of H3?
Activation and repression of H3 will go back and forth due to the Trithorex-Complex (TRX) and Polycomb Group PRC-1/2.
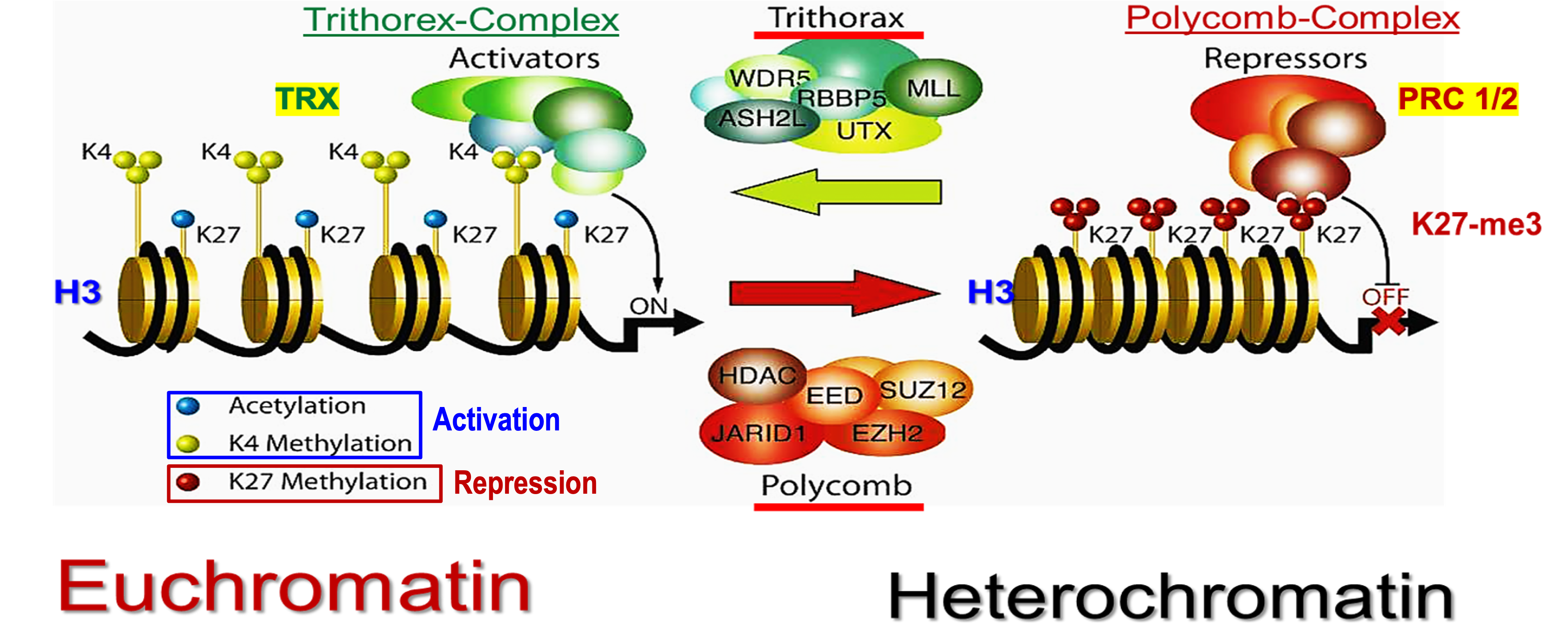
Trithorax (TRX)
Activating group on H3.
Activates formation of euchromatin via acetylation and K4 methylation.
Polycomb Group PRC-1/2
Repressive group on H3
Forms heterochromatin via K27 methylation.
Describe the mechanism of SAGA-HAT and SWI/SNF in yeast
Activator recruits SAGA-HAT (writer) and acetylates histones near gene promoters.
[TA + Ac-Histones] recruit the SWI/SNF remodeling complex via SWI/SNF bromodomain that recognizes and binds acetylated histones.
SWI/SNF + ATP displace acetylated histones near the promoter and form a nucleosome free region.
TBP + TFIID + PIC assemble at the nucleosome free “naked DNA” and initiate gene transcription
![<ol><li><p>Activator recruits SAGA-HAT (writer) and acetylates histones near gene promoters. </p></li><li><p>[TA + Ac-Histones] recruit the SWI/SNF remodeling complex via SWI/SNF bromodomain that recognizes and binds acetylated histones. </p></li><li><p>SWI/SNF + ATP displace acetylated histones near the promoter and form a nucleosome free region. </p></li><li><p>TBP + TFIID + PIC assemble at the nucleosome free “naked DNA” and initiate gene transcription </p></li></ol><p></p>](https://knowt-user-attachments.s3.amazonaws.com/85e68dea-d2bb-4c77-a823-2f8124d9c205.png)
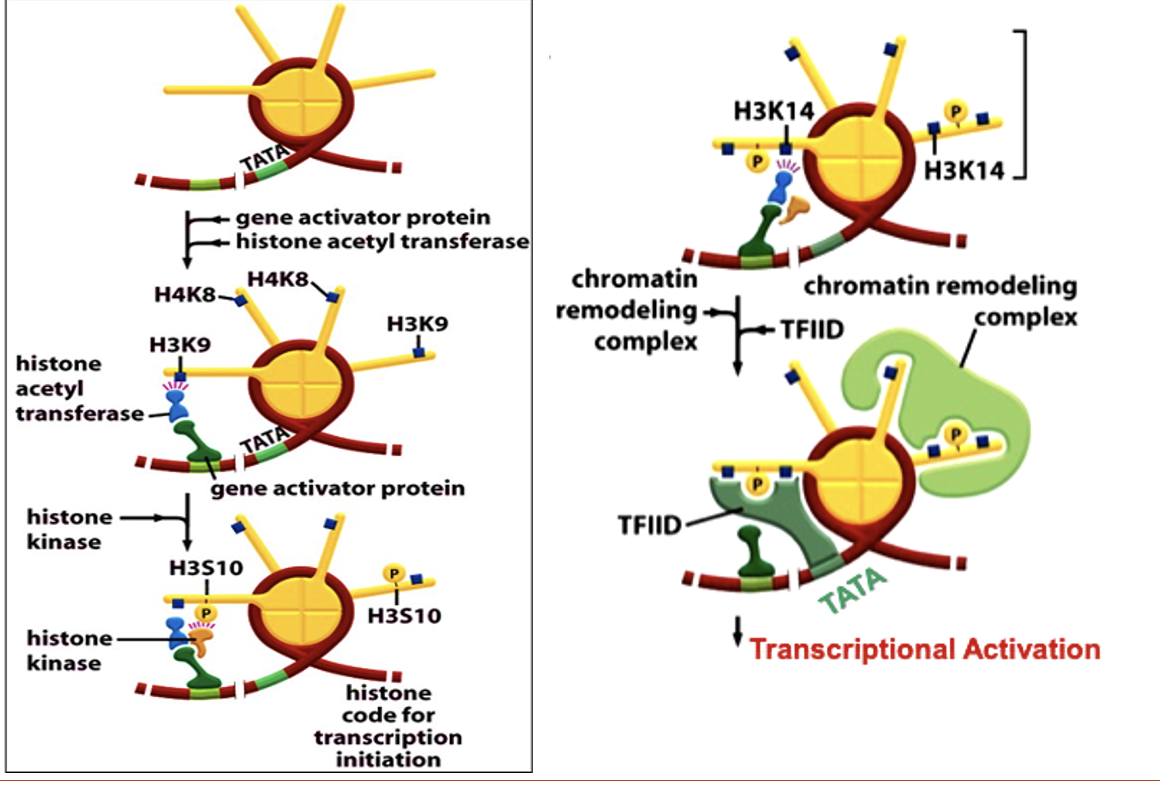
Describe interferon gene expression in humans
Activator binds to DNA-consensus site
Activator recruits HAT
HAT acetylates (H3K9) and (H4K8) symmetrically
Activator attracts histone kinase
Histone kinase phosphorylates serine at position H3S10.
Phospho-H3S10 orients HAT to cause H3K14 acetylation
SWI/SNF bind acetyl-histone via bromodomain, remodel nucleosome and recruit TBP-TFIID complex
TBP-TFIID nucleates PIC at the TATA box
RNA Pol is recruited and interferon gene is expressed.
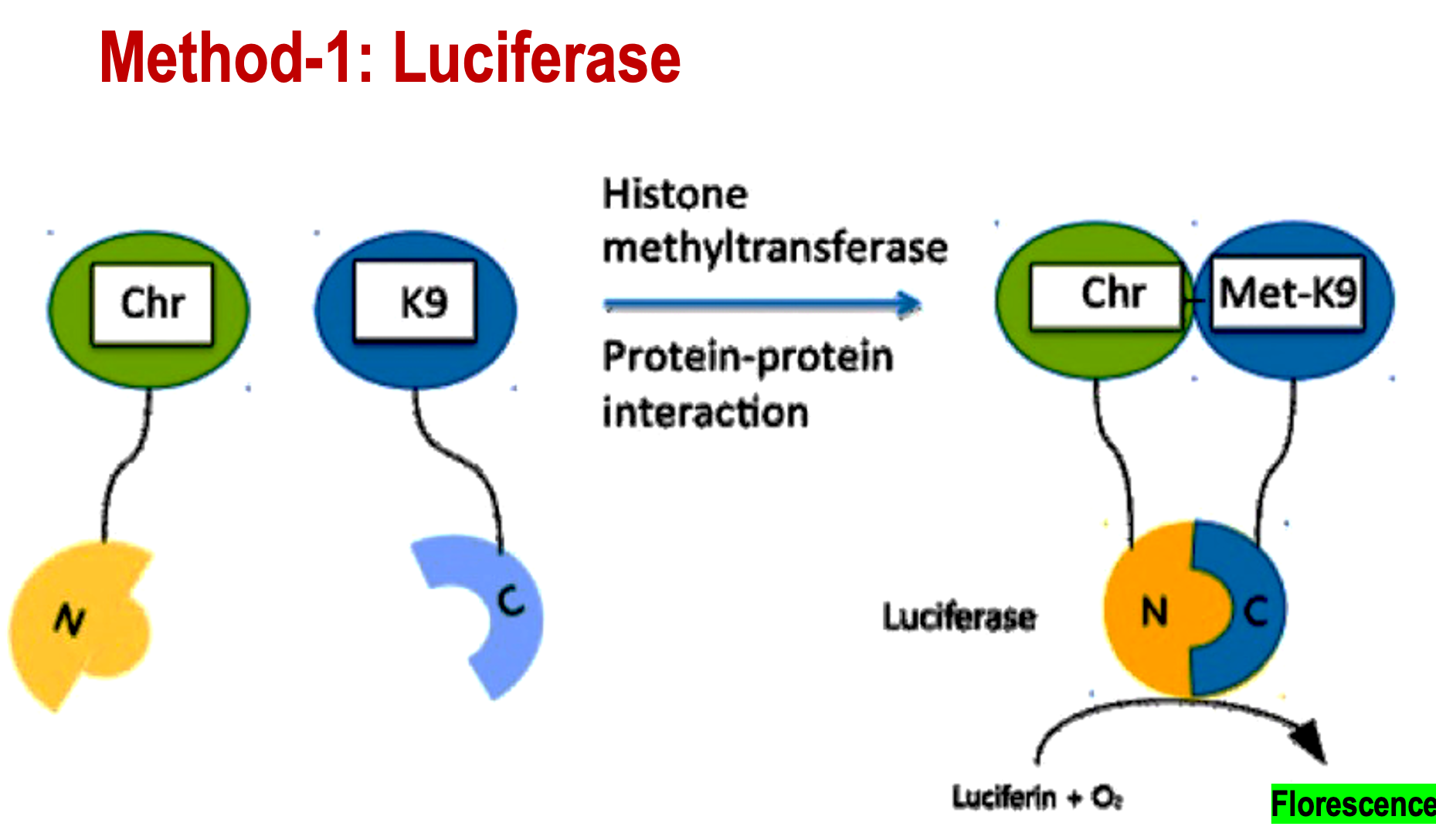
What is a way to detect the direct interaction of histone PTMs with readers?
FRET (fluorescence resonance energy transfer)
Detects protein-protein interactions.
Describe the FRET method of Luciferase
Chromodomain binds to the N terminal of Luciferase. Lysine (K9) binds to the C-terminal of Lucifer's.
After the histone is methylated by a methyltransferase, the Chr-N will bind to the Met-K9. This will create a fluorescent signal from luciferase, allowing you to detect the reader complex.
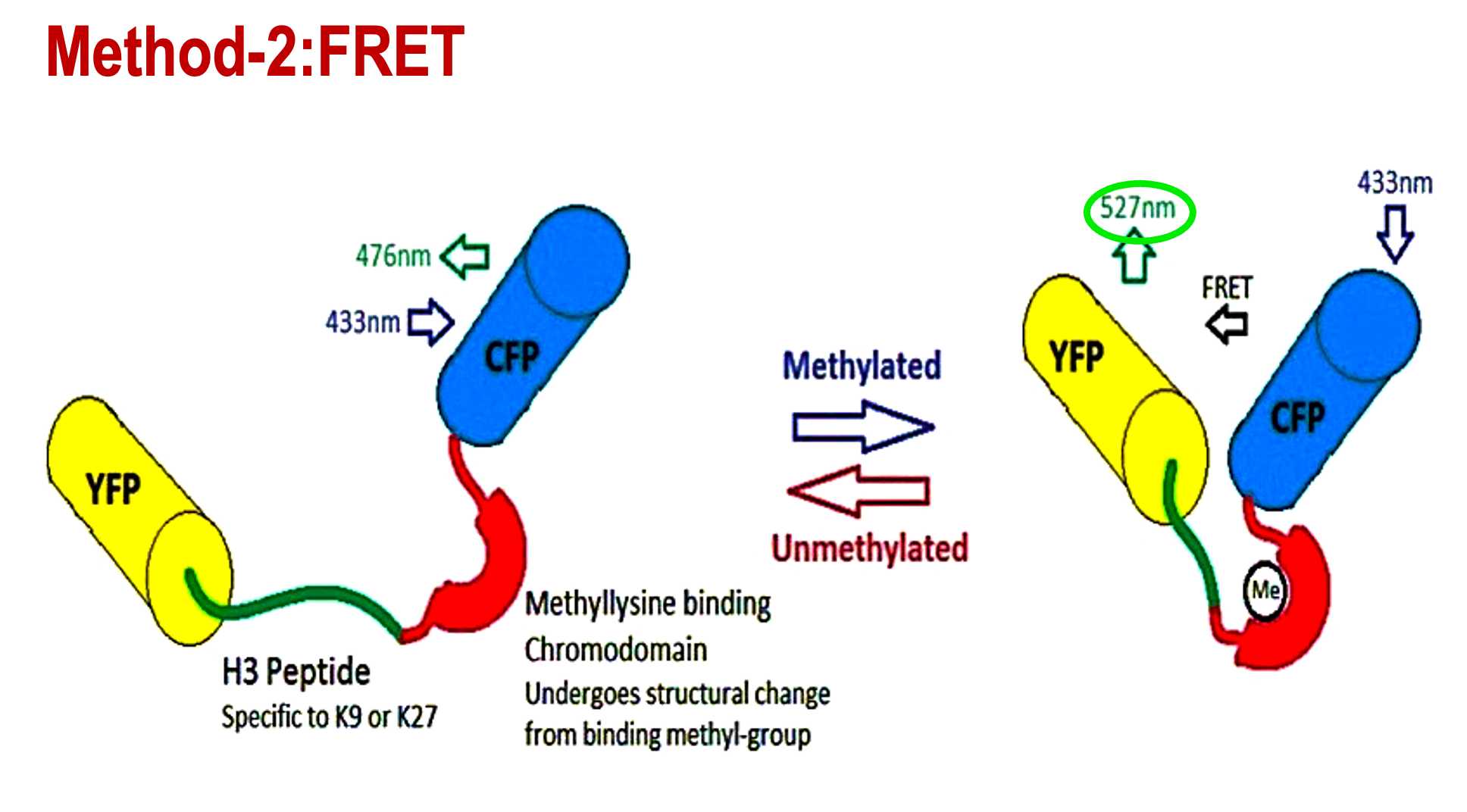
Describe the FRET method using fluorescent protein
YFP and CFP tethered to an H3 peptide. If the H3 peptide becomes methylated, YFP and CFP will be brought together to close proximity, creating a detectable FRET signal.
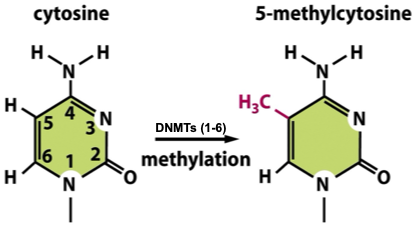
Functional role of DNA methylation at C5-methylcytosine
C5-methylcytosine (5mC)
Helix stability: increased (tighter, compact)
Functions:
immense system
gene silencing
X chromosome inactivation
imprinting/epigenetic memory
stem cell pluripotency
differentiation
DNA recombination
Silences invading parasitic DNA sequences in the genome as a defense mechanism
70-80% of the CpG sites are methylated in the genome
~50% of proximal gene promoters have methylated CpG islands → silence gene transcription
MOST COMMON
Functional role of DNA methylation at N4-methylcytosine
N4 - methylcytosine (4mC)
Helix stability: decreased
Functions:
Immunse system
DNA replication
DNA repair
Gene expression
Functional role of DNA methylation at N6 - methyladenosine
N6 - methyladenosine (6mA)
Functions:
Immune system
DNA damage response
transcription
nucleosome positioning
cell cycle regulation
Describe the inheritance of DNA methylation
This is a way cells maintain cell identity after cell division.
GC base pairs adjacent to the methyl on the old strand becomes methylated as well after DNA replication.
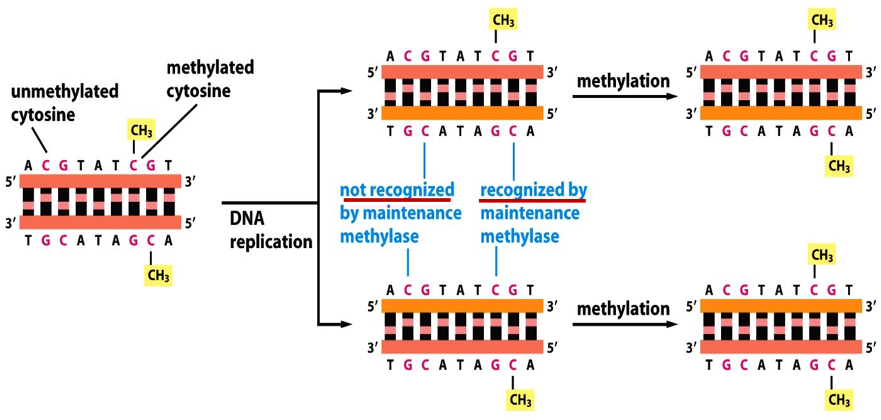
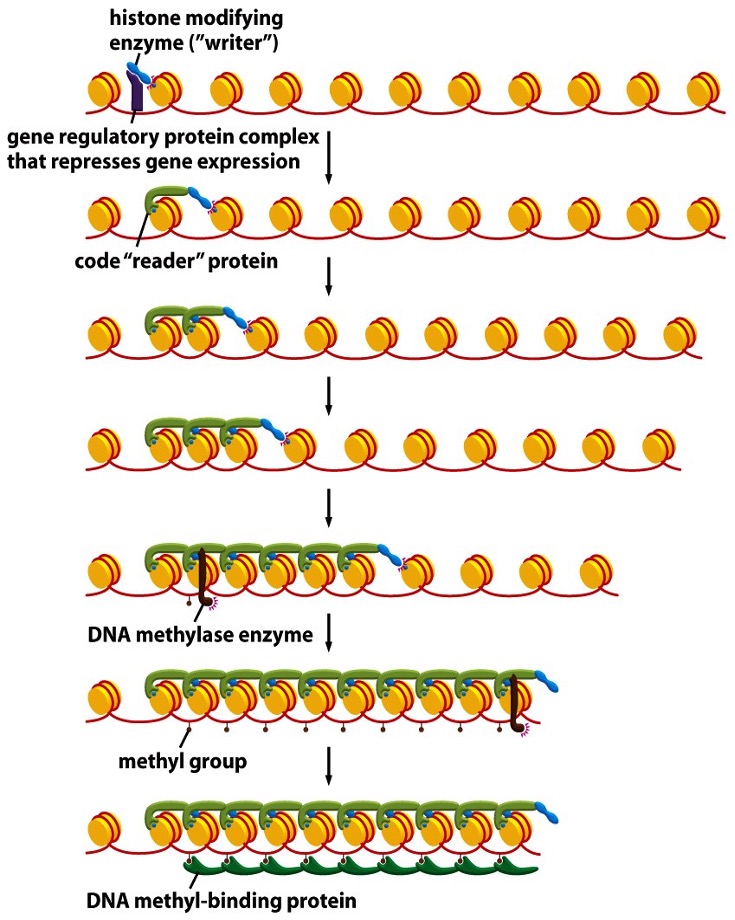
Describe genomic imprinting by DNA methylation
A repressor binds DNA and recruits a histone writer, which puts a histone mark
Histone mark is read by a histone-reader
Reader-writer complex spreads the mark
DNA methylate (DNMT) is attracted by the reader
DNMT adds 5-methyl groups on cytosones
Methyl groups bind DNA-methyl binding proteins in sequence and silences a locus
Leads to formation of a compact, dark, silenced locus.
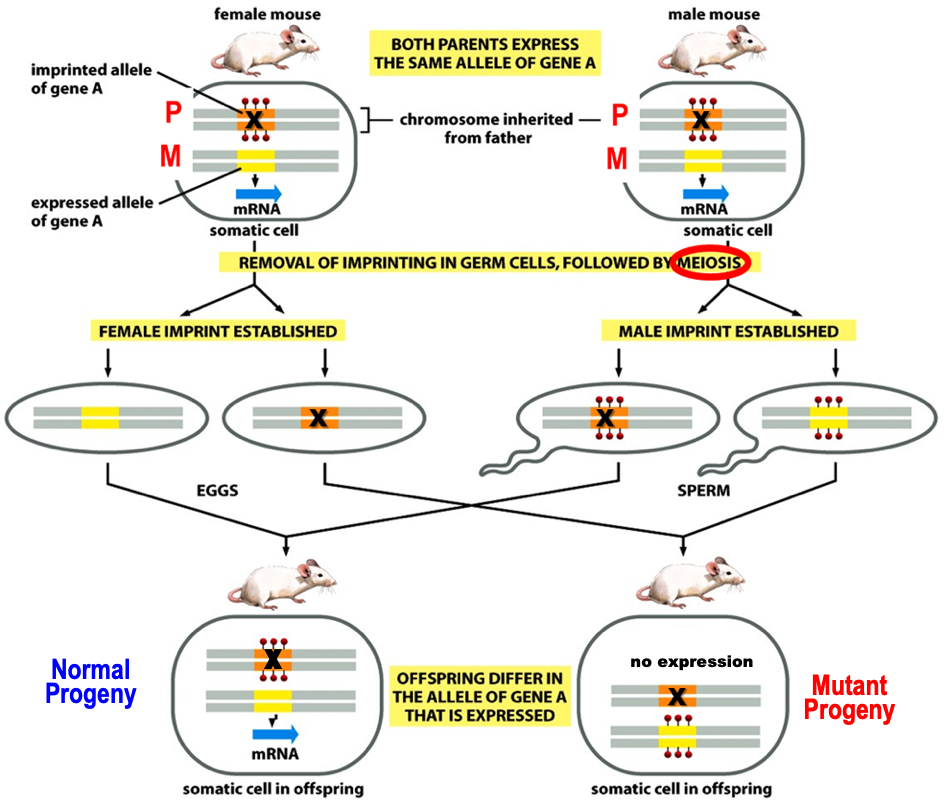
Genomic Imprinting on diploid genomes
Diploid genome will behave like a haploid.
Imprinting by methylation silences specific gene copy in a gender specific manner in germ cells. So, unprinted copy from the other parent is expressed.
The genome behaves like a haploid for imprinted genes as only one copy is expressed.
Mutation (X) in the expressed copy causes genetic diseases in a non-mendelian ratio.
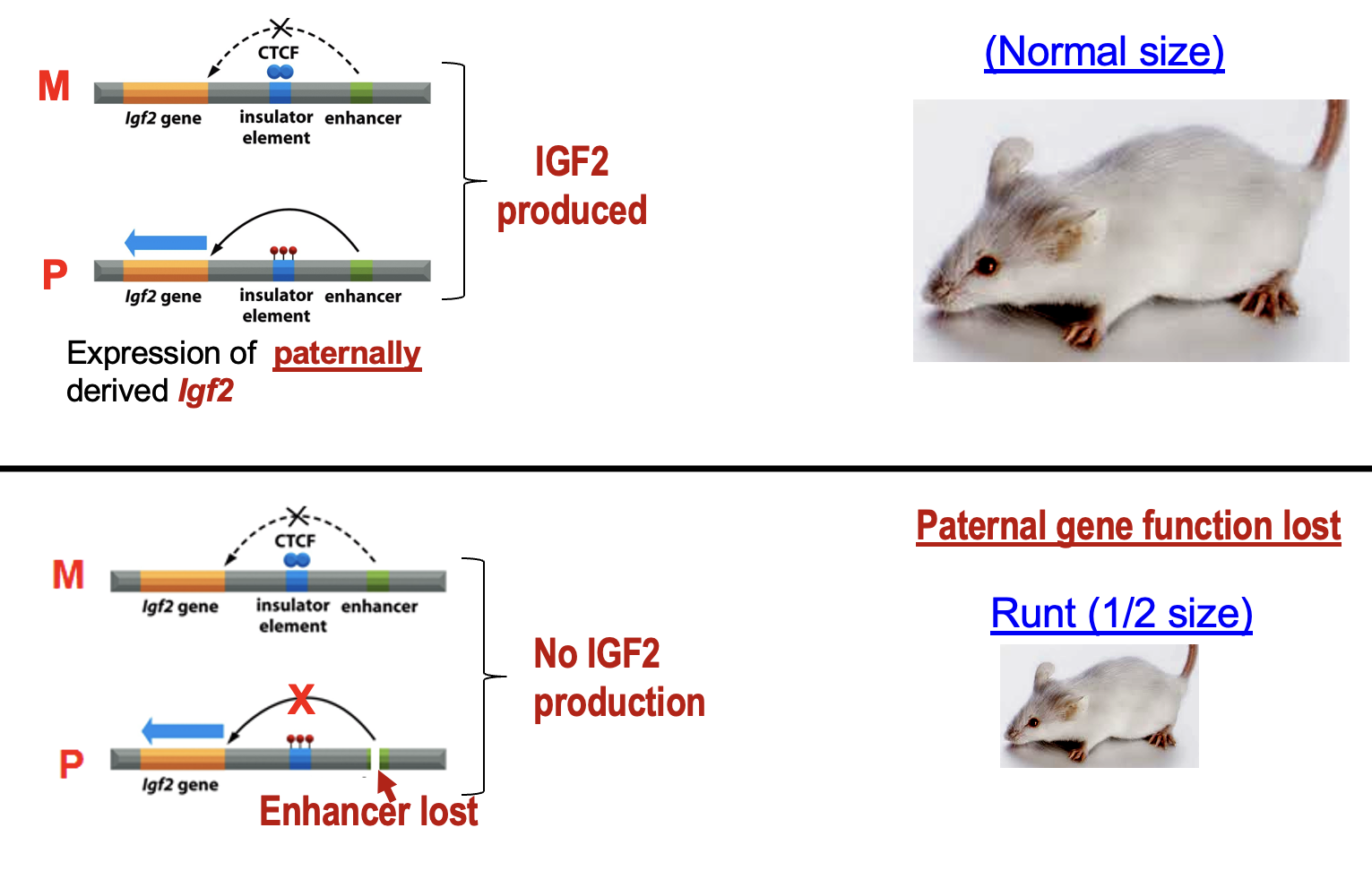
Genomic imprinting at Igf2 gene
Normal size rat becomes small rat when Igf2 is not produced and paternal gene function is lost.
In normal rat, expression of Igf2 (insulin growth factor 2) is produced.
In the mutation, an enhancer is lost, therefore the gene for Ifg2 cannot be activated, leading to a small rat.
Female random X-inactivation
All females will shut down one of their X-chromosomes randomly during embryogenesis to maintain gene dosage.
X-linked mutations is more severe in males than females because of this.
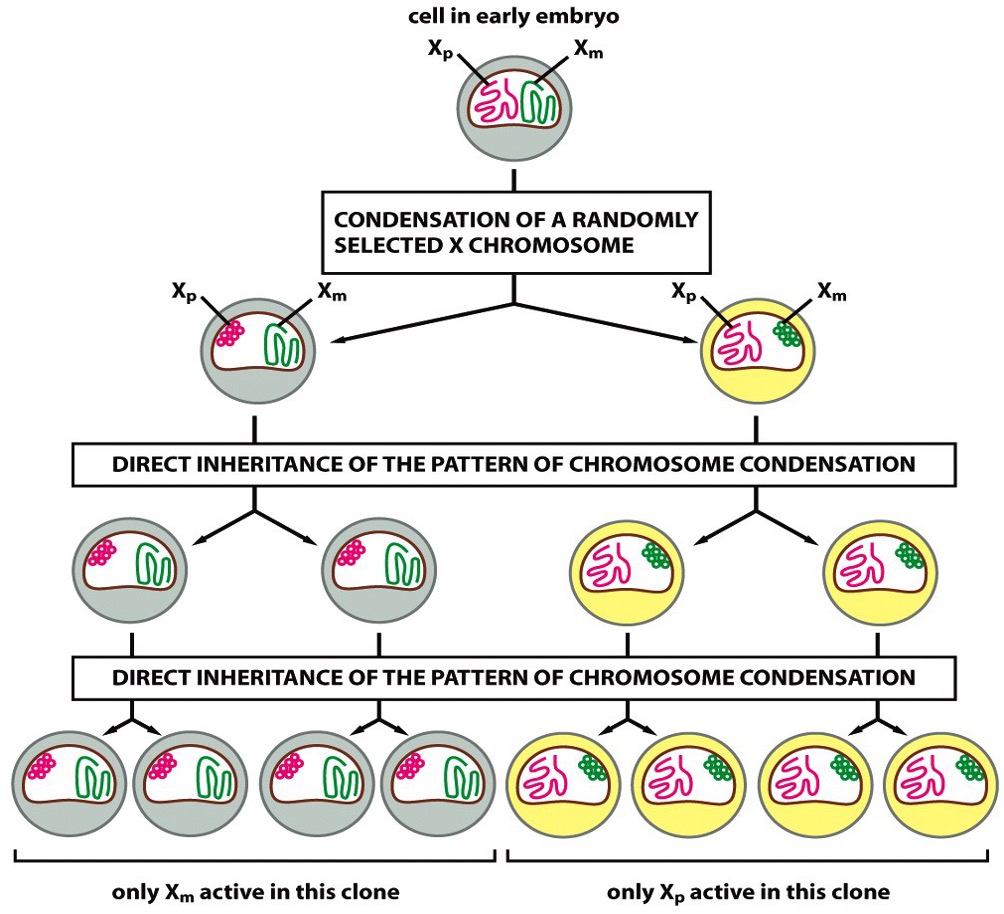
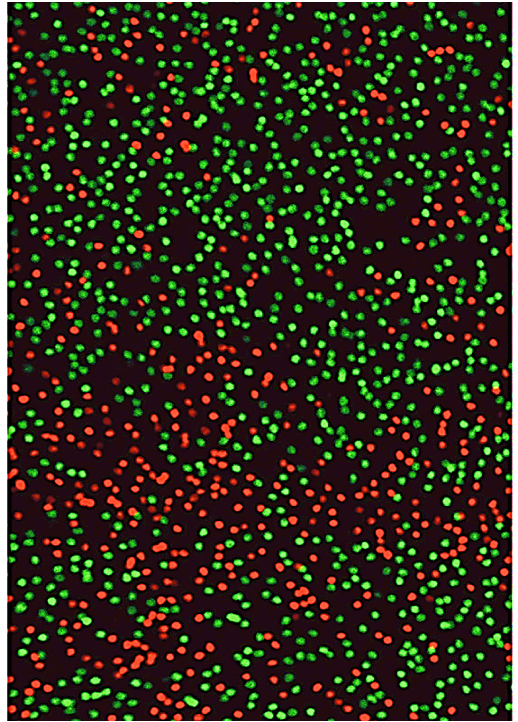
Monoallelic Gene Expression on X-chromosome
Photoreceptors in the retina show patterns of X-chromosome inactivation.
Can use lineage tracing with GFP-reporter on Xm (green) and RPF reporter on Xp to see the female mosaic (two different X chromosomes in females).
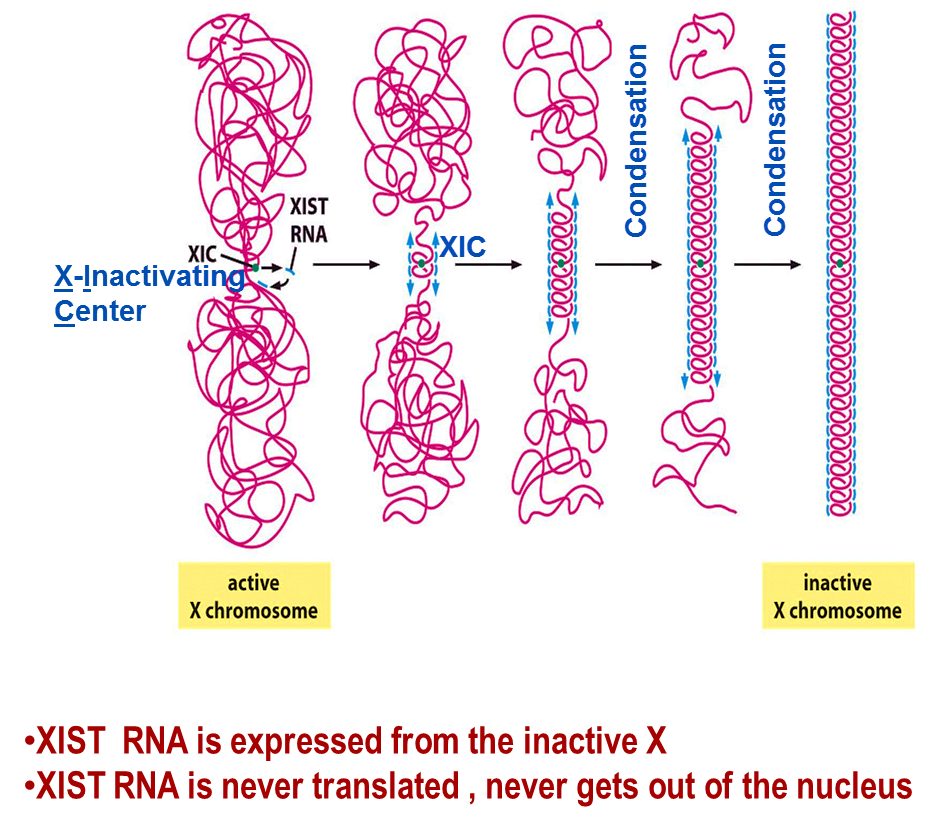
What is the role of XIST RNA
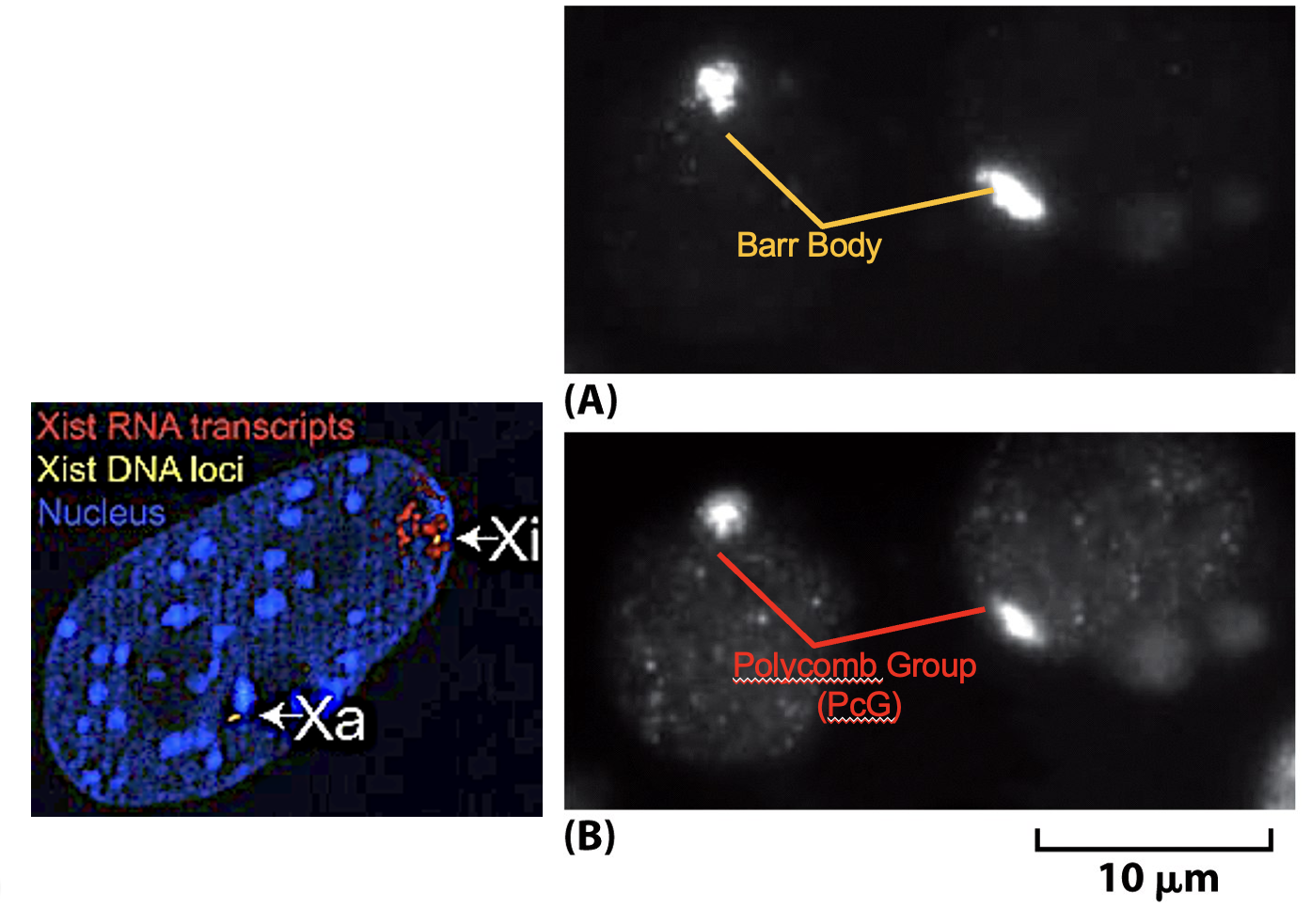
XIST RNA plays an important role in X-chromosome inactivation.
First, the active X-chromosome becomes inactive and condensed with variant hypo-acetylation, H3&H4 ubiquitylation, H3 methylation, DNA methylation and association of HP1. Then, XIST RNA will coat the X-chromosome.
How can you know the sex of a fetus?
The XIST gene encodes a large XIST RNA. The X-chromosome containing the XIST gene will be inactivated.
Barr Bodies are inactive X-chromosomes painted with XIST RNA; RNA FISH (fluorescent in situ hybridization test)
Polycomb (PcG) Antibodies bind to Baar Bodies.
If there is an active XIST signal, then the baby is a female 🙂