1 & 2- Kearns - Cell Cycle Regulation and Gene Expression
1/63
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
64 Terms
Overall, disruptions in the cell cycle result in _______________.
cancer
What are cell cycle non-specific disruptions?
abnormal cell division rate
dysregulation of progression from one phase to the next
could be G1—> S or M—→G1 etc. etc.
All of the following are examples of non-specific cell cycle disruptions except _________.
a. DNA mutations during S phase.
b. the progression from S phase to G2.
c. abnormal cell division rate.
d. the progression from G1 to S phase.
a
What are cell cycle specific disruptions?
DNA mutations
DNA replication mistakes
microtubule assembly disruptions
All of the following refer to disruptions in cell cycle specific regulation in cancer except ____________.
a. DNA mutations
b. disruption in microtubule assembly
c. abnormal cell division
d. mistakes in DNA replication
c
Describe the M phase of the cell cycle:
“MITOTIC PHASE”
mitosis and cytokinesis occurs
cell physically divides here
Describe the Interphase phase of the cell cycle:
broader term that refers to the G1, S, and G2 phase all together
G1- organelle development, growth
S- DNA REPLICATION (2n—>4n)
G2- safety check genome integrity prior to next division
Between each phase of the cell cycle there are _____________. This is also a drug target in cancer.
checkpoints
What’s the difference between cyclins and cyclin dependent kinases (CDK)? What do these both combine to do?
CYCLIN- PROTEINS that bind to and activate cyclin dependent kinases
CYCLIN DEPENDENT KINASES (CDKs)- ENZYMES (special protein) that PHOSPHORYLATE
both of these combine to drive the cell cycle through each phase
True/False: Different cyclins are expressed equally in concentration throughout each stage of the cell cycle.
false- each cyclin fluctuates based on what phase of the cell cycle
Are cyclins anticancer targets? Why or why not?
Cyclins ARE anticancer targets
Why? remember CDK+Cyclin= phosphorylate in order to continue the cell cycle, so if we stop this we will DEPHOSPHORYLATE, and we STOP the cell cycle and STOP cell growth!
How do healthy cells maintain their genomes?
tumor suppressor genes stop the formation of tumors
When we say “maintain the genome” what are we referring to?
how a cell maintains 46 chromosomes
Explain how the p53-p21-RB signaling pathway NORMALLY works in response to DNA damage.
DNA damage activates p53
p53 activates p21
p21 inhibits CDK/Cyclin complex= block cell cycle progression
no phosphorylation of RB/E2F further prevents cell cycle progression
What are the 2 fates of DNA?
Replication (DNA—> DNA)
Protein Synthesis (DNA—> RNA —→ Proteins)
Which is the building block of DNA?
a. nucleoside
b. nucleotide
b.
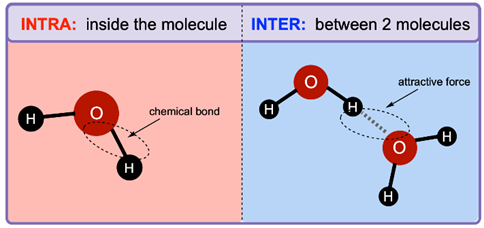
In general, what’s the difference between intramolecular and intermolecular bonds?
intra= inside the molecule
inter= between 2 molecules
In DNA, where are intramolecular bonds found and give an example:
bonds in the BACKBONE
example—> PHOSPHODIESTER BONDS BETWEEN NUCLEOTIDES
In DNA, where are intermolecular bonds found and give an example:
bonds between 2 separate strands
example- Hydrogen bonds between A and T
In cancer, would we want to target intra or intermolecular bonds?
intramolecular
How do we unwind DNA when it’s supercoiled?
TOPOISOMERASE I or II
What is the chemical reaction between Top I and DNA? What bond is formed and between what?
Summary of steps- “reversible ester bond between a tyrosine on top and the phosphate group on the DNA”
Detailed Steps-
1.Top binds to the supercoiled DNA
2.The tyrosine on top has an -OH group that acts like a nucleophile and attacks the phosphodiester bond of the DNA backbone
3.This breaks the one strand of DNA and creates a nick that relieves torsional strain
4.To repair the DNA strand the -OH group on the DNA performs a second nucleophilic attack on the phosphotyrosine bond which makes top dissociate
What are the 3 important enzymes involved in DNA replication? What are each of their roles and state whether or not they are drug targets in cancer.
TOP- unwinds DNA
HELICASE- separates (“unzips”) the 2 DNA strands
DNA polymerase- adds nucleotides to the dividing strands
ALL 3 ARE DRUG TARGETS IN CANCER
Nucleotide biosynthesis no matter if it’s a purine or pyrimidine always begins with what reaction? Using what enzyme?
___________ ——> _________?
Ribose-5-P ———> PRPP using PRPS
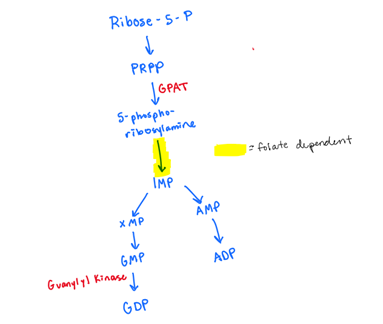
Explain how guanine is made de novo (not the salvage pathway):
include which step is folate dependent
which step is where oxidation occurs?
PRPP —> 5-phosphoribosylamine using GPAT
5-phosphoribosylamine to IMP (FOLATE DEPENDENT)
IMP—> XMP —> GMP (WE OXIDIZE IMP to GMP)
GMP—> GDP using GUANYL KINASE
GDP—> dGDP using ribonucleotide reductase
reduction of OH to H
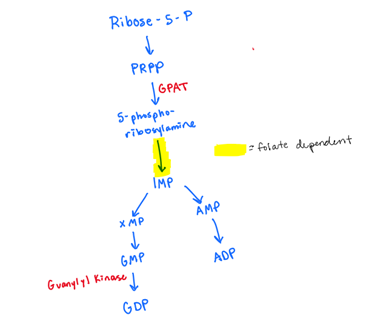
Explain how adenine is made de novo (not the salvage pathway):
include which step is folate dependent
which step is where aminated occurs?
PRPP —> 5-phosphoribosylamine using GPAT
5-phosphoribosylamine to IMP (FOLATE DEPENDENT)
IMP—>AMP (WE AMINATE IMP to AMP)
AMP—> ADP
ADP—> dADP using ribonucleotide reductase
reduction of OH to H
Explain how cytosine is made de novo:
PRPP+ Orotate—> UMP
UMP—> UTP
UTP—> CTP (amination reaction)
CTP—> dCTP using ribonucleotide reductase
Explain how Thymine is made de novo:
PRPP+ Orotate—> UMP
UMP—> dUMP using ribonucleotide reductase
dUMP —> dTMP using thymidylate synthase (folate dependent)
dTMP—> dTTP
What are the primary targets in cancer if we want to inhibit MITOSIS?
microtubules synthesis
What do microtubules bind to? What does this signify?
bind to the kinetochore of chromosomes
once this occurs—> cell division WILL OCCUR there is NO GOING BACK
What are some functions of microtubules?
chromosome segregation in mitosis
mechanical support (cell movement)
transport
intracellular transport
A microtubule is a heterodimer with what kind of symmetry? Explain.
has inherent asymmetry meaning that is has a -alpha terminus and a +b terminus
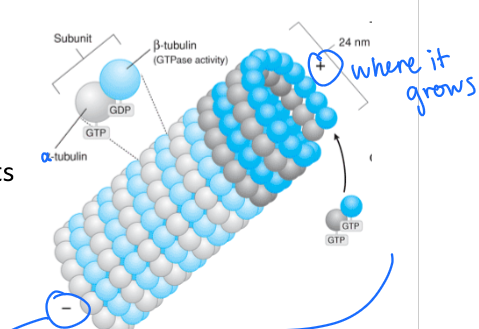
What would high concentrations of GTP do to microtubules?
GROW!!!! microtubule lengthens
rate of breakdown (GTP hydrolysis) = rate of growth (polymerization)
What would low concentrations of GTP do to microtubules?
SHRINK
rate of breakdown (GTP hydrolysis) > rate of growth (polymerization)
If we were designing an anti-cancer drug, would we want the drug to depolymerize or polymerize microtubules?
DEPOLYMERIZE
What is gene expression?
“the level to which our proteins are transcribed” (basically how our genes and DNA are used to make proteins)
What’s the difference between RNA polymerase and DNA polymerase?
RNA polymerase—> deals with protein synthesis
DNA polymerase—> deals with DNA replication
What is a transcription factor?
Regulatory proteins that bind to the DNA in the nucleus and effect transcription
The binding of _____ is required for transcription to start.
RNA polymerase
What does RNA polymerase bind to start transcription? Where is this region located?
promoter region, located right before the gene
Upstream of the promoter is a distal regulatory region that contains what 3 regions where transcription factors bind? What is their effect on gene expression?
a. insulator- protects from inappropriate signals
b. silencer- inhibits gene expression
c. enhancer- promotes gene expression
What does the term “Basal level of transcription” mean? What determines this?
it’s the normal or “base” level of transcription aka the amount of transcription we would have without any of the enhancers/silencers/or anything promoting/inhibiting transcription.
determined by the promoter sequence
Can RNA polymerase bind to the promoter region on its own?
no, needs TFs
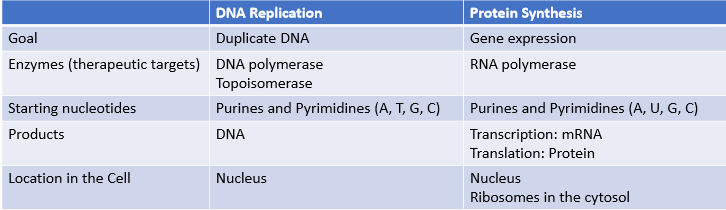
What’s the difference between DNA replication and protein synthesis in terms of…
goal
enzymes that are therapeutic targets
starting nucleotides
products
location in the cell where it occurs
DNA is stored in highly structured complexes called…
chromatin
In order for a gene to be transcribed what must happen to chromatin?
make accessible chromatin regions—→ modify/unwind nucleosomes (DNA+histone) in order to make the DNA accessible to RNA polymerase and other TFs
What are epigenetic changes?
By altering the DNA/Chromatin Structure, this affects how easy it is for enzymes needed for transcription to interact with and transcribe genes aka effects GENE EXPRESSION
How are epigenetic changes different from DNA mutations?
DNA mutations change the DNA sequence, but epigenetic changes alter the chromatin (DNA) STRUCTURE
Epigenetic changes may be caused by what?
development
environmental changes
drugs/pharma
aging
Diet
Can drugs that treat cancer be an epigenetic factor?
yes
Where in the cell does transcription take place?
nucleus
Where in the cell does translation take place?
cytoplasm/ribosomes
What are the steps in protein synthesis?
transcription
remove histones and uncoil DNA
separate DNA strands
assemble nucleotides into a single strand of mRNA using RNA polymerase
post-transcription modification
mRNA before leaving nucleus needs INTRONS cut out and the remaining exons are spliced together
translation
codons of mRNA binds to anticodons on tRNA
each tRNA carries a specific AA
rRNA then strings the AA together
Now you have a protein :)
3 mRNA nucleotides are called a_______.
codon
True or False: Introns are the coding sequence of mRNA.
false
What are housekeeping genes?
genes required AT ALL TIMES and are expressed CONTINUOUSLY
Expression of housekeeping genes can be modulated by…
regulatory proteins aka TRANSCRIPTION FACTORS by enhancing or repressing the interaction between RNA polymerase and the promoter
What is a specificity factor?
alter the specificity of RNA polymerase for a given promoter or set of promoters
What is a repressor?
impedes access of RNA polymerase to the promoter
What is an activator?
enhances the RNA polymerase—promoter interaction
Which of the following do epigenetic regulation of gene expression control:
a. TATA sequence
b. promoter region
c. regulatory transcription factors
d. enhancer sequences
c, d (WHY? remember that epigenetic influences how accessible a gene is by messing with the structure, so it would control how TFs bind to these regions)
Which of the following do genetic regulation of gene expression control:
a. TATA sequence
b. promoter region
c. regulatory transcription factors
d. enhancer sequences
a, b (WHY? genetic regulation refers to the DNA sequence itself and the TATA sequence and promoter region are actual parts of the gene)
mRNA synthesize is regulated by phosphorylation and dephosphorylation. What are 2 methods of phosphorylation and dephosphorylation? What are the enzymes involved?
protein phosphorylation
protein kinase= phosphorylate
protein phosphatases= dephosphorylate
GTP-binding phosphorylation
GTP binding= phosphorylate
GTP hydrolysis= dephosphorylate
What are the 3 steps of translation?
initiation- mRNA binds to P site, tRNA binds to P site carrying an AA
elongation- the tRNA moving from the P site moves it’s AA to the tRNA on the A site and then that tRNA moves from the P site to the E site and is released
termination- stop codon on mRNA, new polypeptide