lec 10.1 - attenuation and operons
1/16
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
17 Terms
what is attenuation?
The mechanism that links the supply of the amino-acyl-tRNA to the ability of RNA polymerase to read through a termination site located at the beginning of a cluster of structural genes that code for the enzymes that synthesize the amino acid carried by the tRNA
and the supply of the amino-acyl tRNA is dependent on the supply of the amino acid
explain how attenuation can be used to control transcription once it is started
attenuation is a control system where the product of the operon can stop the production of the operon's mRNA
often seen in amino acid operons
e.g. the tryptophan operon (must most amino acid operons work the same way)
describe the tryptophan operon (what genes are there) and its regulation
has 5 genes (E, D, C, B, and A) encoding enzymes that synthesize tryptophan and an additional leader gene (L)
under negative regulation: had a repressor (coded by trpR gene) however the repressor cannot bind/repress the operon unless the tryptophan co-repressor is present (because if the cell has tryptophan there is no need to make it so we can repress)desc
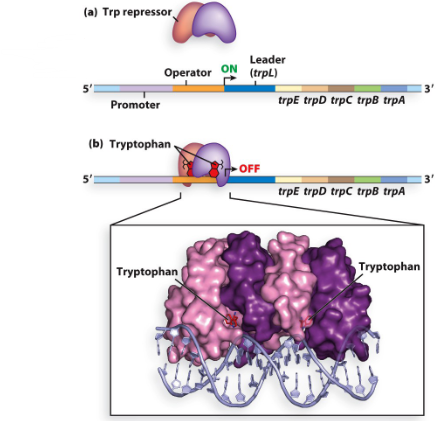
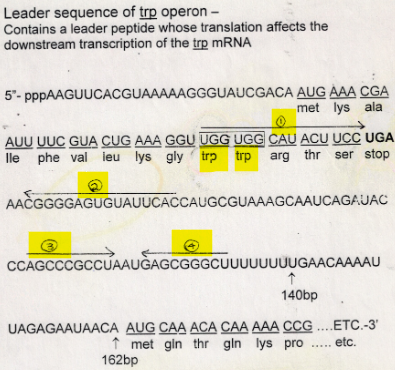
how many repeats does the trpL sequence have? how can they bind?
contains 4 repeat sequences: 1, 2, 3, and 4 that can bind to each other in 2 ways:
1 with 2 and 3 with 4
2 with 3
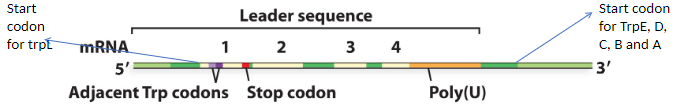
what happens with leader repeat sequences 2 and 3 bind?
then RNA piol can transcribe the mRNA of all the genes (polycistronic)
what happens with leader repeat sequences 1 and 2 bind while 3 and 4 bind?
then a terminator sequence (Rho-independent termination) is produced and RNA pol falls off DNA after only making 140 bases of mRNA and therefore the genes are not transcribed and the enzymes are not made
what determines which leader repeat sequences bind to each other?
the Trp codons adjacent to the start codon
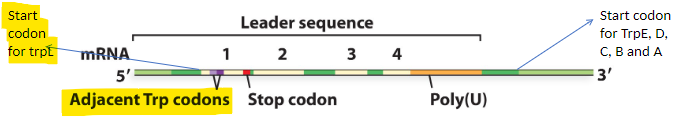
describe the trpL gene in the leader sequence
2 sets of inverted repeats (1 and 2; 3 and 4)
Both can make stem and loop structure
Stop codon for protein made from TrpL gene is between repeat 1 and 2
Stem and loop made from
repeat 3 and 4 looks like a independent terminator2 Trp codons that are right next to each other in this small protein
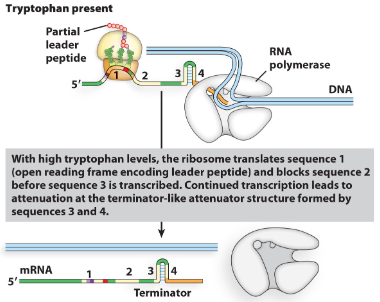
what happens to the trp operon when tryptophan is present?
Once RNA polymerase starts the mRNA of the leader sequence the ribosome attaches and starts translation.
If trp is, present trp will be put into the protein at the trp codons and the ribosome will go to the stop codon before stalling.
this causes sequence 3 to bind to sequence 4
creating a stem-loop terminator (Rho-independent) which causes the RNA polymerase to fall off after making only 140 bases of the mRNA.
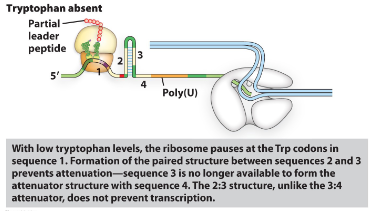
what happens to the trp operon when tryptophan is absent?
Once RNA polymerase starts the mRNA of the leader sequence the ribosome attaches and starts translation.
If trp is absent the ribosome will stall at the trp codons (in the first repeat sequence) waiting for the cell to find trp.
in the meantime, sequence 2 will bind to sequence 3 and therefore the terminator
sequence is not formed and the RNA
polymerase will be able to continue to
make the mRNA of all the genes (full operon is transcribed)
in the leader sequence, the location the ribosome stops at is dependent on what?
availability of trp-tRNA
what happens when there is lots of trp-tRNA?
When there is lots of trp-tRNA the Ribosome travels along mRNA until it reaches the stop codon UGA– which is between sequences 1 and 2.
this placement allows sequence 1 to bind
to 2 and 3 to bind to 4 which makes a termination structure and so RNA Polymerase (which is in front of ribosome) interacts with the terminator and stops transcription
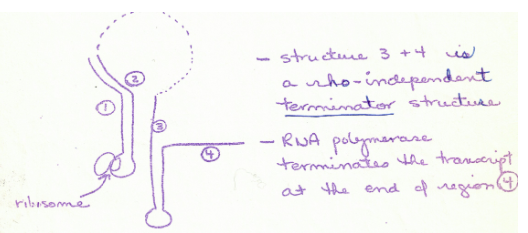
what happens when there is little trp-tRNA?
When there is little trp-tRNA the Ribosome travels along mRNA until it stalls at
trp codons in sequence 1 – therefore sequences 2 and 3 bins and RNA polymerase will continue to transcribe the rest of the genes because no terminator was formed

list other operons that are controlled by attenuation
His
Phe
Leu
Thr

what are regulons?
a group of several genes or operons that are turned on or off in response to the same signal by the same regulatory protein.
what is an example of a regulon? what are its 2 regulators?
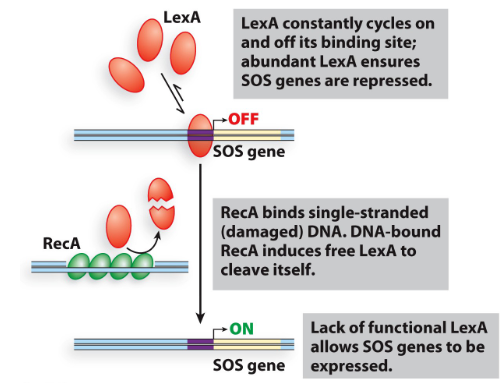
Ex: SOS system – to help with DNA damage
Uses 2 regulators: RecA protein and LexA protein
explain how LexA and RecA regulate the SOS gene
LexA is a repressor; it binds to the operator to turn off genes in SOS system
RecA binds to single stranded (damaged) DNA which causes free LexA to cleave itself so it can no longer repress this allows SOS genes to be expressed
essentially, DNA damage leads to LexA inactivation