tutorial 6: Barcoding expt. transformation of plasmid DNA Intro and Day 1 - plasmid extraction
1/31
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
32 Terms
what is agarose?
how does the % agarose effect pore size
solidifying agent
decrease % of agarose = increase gel pore size
therefore pick the % of agarose needed based on expected size/MW of protein
review agarose gel electrophoresis
what is it/what is it used for
how does it work?
how do different shaped molecules move through the gel?
Technique for resolving proteins or nucleic acids
Gel – semisolid open meshwork of interlinked linear strands
Wells are loaded with a sample
Electric field is applied across the gel
Charged macromolecules are driven together in the direction of the anode
The distance that a band moves depends on its mass and the size of pores within the gel
Smaller molecules travel further than larger ones
Tracking dye ( DNA Loading Dye, a charged, low molecular weight compound) is usually loaded into each well at the start of the run to monitor the progress of molecule movement on the gel
Bands are visualized by staining specific for the molecule under investigation (e.g. DNA)
Discrete bands are observed when
Enough molecules are present to bind the DNA dye to make the band visible
When individual macromolecules have distinctly different sizes
Agarose (polysaccharide from seaweed) is used for separation of nucleic acid molecules
After the gel solidifies, it is submerged in a buffer for conductivity
A direct current power source is connected to the gel chamber and an electrical current is applied
Molecules with a net negative charge (e.g. DNA from oxygen atoms on phosphate groups) move towards the anode
The buffer controls pH and is a conductor of electricity
Within limits, the greater the current the faster the charged molecules migrate
for linear molecules, the smaller the linear fragment, the faster it migrates through the gel
Shape also influences the rate of migration: compact molecules (e.g. a sphere – e.g. plasmids) will migrate faster through a gel than a linear molecule of the same size
Negatively charged particles always migrate towards the positive pole whereas positively charged particles always migrate towards the negative pole.
the charge to mass ratio is the same for different sized DNA molecules: the nucleotides
in DNA are linked together by negatively charged phosphodiester groups and therefore there are two charged phosphate groups for every base pair.-30,000 bp)
what % gel is used in the lab? how much agarose is needed?
1% agarose
1g
what are the 2 most common running buffers used on nucleic acid electrophoresis?
Tris acetate EDTA (TAE) and tris borate EDTA (TBE) are the two most common running
buffers used in nucleic acid electrophoresis. As buffers, they have a fairly constant pH and are able to conduct electricity because of their concentration of hydrogen ions
why might there be a fuzzy band at the bottom of the gel lane with samples?
this band is called primer-dimer and is not an amplified target DNA; it results from an association between primers, giving rise to a primer-dimer.
A primer-dimer consists of 2 primer molecules that hybridized (annelead) to each other because of complementary bases in their sequences.
why is DNA negatively charged?
DNA is negatively charged because of the presence of phosphate groups in nucleotides. The phosphate backbone of DNA is negatively charged, which is due to the presence of bonds created between the phosphorus and oxygen atoms. In DNA structure, a phosphate group comprises one negatively charged oxygen atom, which is responsible for the entire strand of DNA to be negatively charged.
what is pKAN
antibiotic selection marker
kanamycin resistance gene
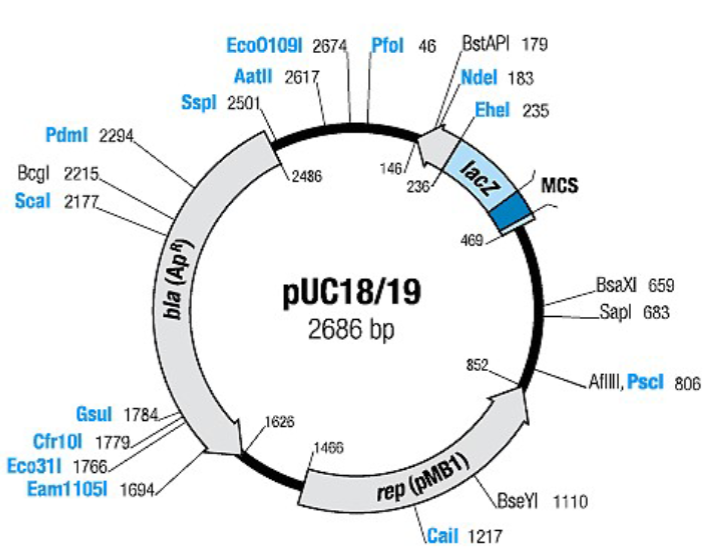
what is pUC18
antibiotic selection marker
ampicillin resistance gene (the β-lactamase (bla) gene
what is the purpose of using agarose gel electrophoresis in this experiment?
to confirm that plasmid DNA was extracted
breifly describe the steps/what is done in the exeriment
isolate plasmids (pKan, pUC18) DNA. Each of the plasmid vectors carries an antibiotic selection marker: pKan – kanamycin resistance gene; pUC18 – ampicillin resistance gene (the β-lactamase (bla) gene)
confirm that plasmid DNA was extracted using agarose gel electrophoresis
Digest plasmid DNA using restriction enzymes
Ligate restriction fragments into the multiple cloning site (MCS) of pUC18 with the aim of cloning the kanamycin resistance gene from pKan into the MCS
transform the new (recombinant) plasmid into the E. coli strain DH5α and plate transformed cells onto selective media
Select clones resistant to both ampicillin and kanamycin
what is the pUC18 (aka indirect) screening method?
employs the ‘blue/white’ screening technique to select colonies with a successful insert (=kanamycin resistance gene).
what is special about the pUC18 plasmid?
contains the lactamase (bla) gene which confers resistance to
ampicillinThe bla gene encodes the production of beta-lactamase, which hydrolyzes the β- lactam ring and thus inactivates the antibiotic
what do Beta-lactam antibiotics do? what are some examples?
bacteriocidal and inhibit synthesis of peptidoglycan:
The final step in peptidoglycan synthesis is mediated by transpeptidases (some penicillin binding proteins are transpeptidases)
β-lactam antibiotics are analogues of D-alanyl-D-alanine, an amino acid residue needed for peptidoglycan synthesis
β-lactam antibiotics bind to transpeptidases and this inhibits transpeptidation
Peptidoglycan precursors are built up and this triggers the release of autolysins (hydrolases that digest peptidoglycan) → cell death
examples: penicillin and ampicillin
describe the difference between gram-negative and gram-positive bacteria. what does this mean for beta-lactam antibiotics
Gram-positive bacteria like S. aureus or S. pyogenes have their cell membrane
surrounded by a thick peptidoglycan layer, without an outer membrane surrounding the peptidoglycan layer.Gram-negative bacteria like E. coli have a thin peptidoglycan layer surrounded by a thin outer membrane.
In contrast to gram-positive bacteria (absence of an outer membrane), beta-lactam antibiotics have to penetrate through porins of the outer membrane of gram-negative bacteria
what are some strategies of bacterial resistance to beta-lactam antibiotics?
modification of porins (permeability barrier)
modification of targets (low affinity of penicillin binding proteins) for the drug
production of inactivating enzymes (beta-lactamases)
Inhibition of release of autolytic enzymes.
describe the pUC18 plasmid. what leads to a high copy number?
The pUC plasmids contain an ampicillin resistance gene and origin of DNA replication, ligated to a portion of the lacZ gene of E. coli, as well as a multiple cloning site (MCS)
contains bla, lacZ, and rep (pMB1) genes.
The pMB1 replicon rep is responsible for the replication of plasmid (source – plasmid pBR322).
The high copy number of pUC plasmids is a result of the lack of the rop gene
and a single point mutation in the replicon rep of pMB1, enhancing its ability to initiate plasmid replication
what is the Rop protein?
the Rop protein is a negative regulator of plasmid replication within the ColE1 family: it’s absence will ‘loosen the control’ on plasmid replication leading to increased copy number.
describe X-gal
When the Rop protein is introduced into a suitable E. coli host strain carrying a lacZ product that is able to complement the portion on the plasmid, the cells are able to make beta-galactosidase and produce blue colonies on plates containing the substrate, X-gal.
X-Gal (5-bromo-4-chloro-3-indolyl-beta-D-galacto-pyranoside) is an inert chromogenic substrate for beta-galactosidase which hydrolyzes X-Gal into colorless galactose and 4- chloro-3-brom-indigo, forming an intense blue precipitate
what does cloning DNA fragments (kanamycin resistance gene) into the Multiple Cloning Site do? how are the colonies then analyzed?
Cloning DNA fragments (kanamycin resistance gene) into the Multiple Cloning Site inactivates the lacZ gene, therefore cells with these plasmids can not make beta- galactosidase and therefore produce white colonies on the agar plates.
The appearance of white colonies on X-gal plates after transformation should indicate the presence of the kanamycin gene inserted into the pUC18 vector.
These colonies will then be streaked onto kanamycin/ampicillin plates to verify the presence of the kan gene in the vector.
which restriction enzymes are used in this lab?
HinDIII and BamHI to digest both plasmids
what do nucleases do? what kinds are there?
digest polynucleotides by cleaving the 5’ to 3’ phosphodiester bonds that link
nucleotides:Endonucleases: nucleases that act within a strand
Exonucleases: act at the end of a nucleic acid
what are restriction endonucleases? does it have symmetry?
nucleases that block or restrict viral replication; act on DNA with specific recognition sequences and only when the recognition sequence is not
modifiedRecognition sites have a rotational symmetry (“ a palindrome”)
what is the restriction enzyme nomenclature system?
Restriction enzymes are identified with a 3-letter designation of the bacterium from which they are isolated, plus a single-letter strain designation (as needed) and a roman numeral showing the order in which it was identified. The 3-letter bacterium designation should begin with a capital letter and is italicized

plasmid definition
An autonomous, self-replicating, extrachromosomal DNA molecule
cloning definition
Incorporating a DNA molecule into a chromosomal site or a cloning vector
clone definition
A population of cells that carry a cloning vehicle with the same DNA insert
transformation definition
The uptake of extrachromosomal DNA in a bacterium or yeast cell in which the DNA often changes the phenotype of the recipient organism
transformant definition
A cell which has taken up extrachromosomal DNA and is expressing a change in phenotype
competent cells definition
Cells which have been treated by chemical or physical means in order to be sensitive to foreign DNA
function of Tris/EDTA in TE buffer?
EDTA chelates divalent ions such as Magnesium and calcium and this weakens the cell wall/outer membrane because it disintegrates the LPS and increases the permeability of the outer membrane: EDTA sequesters divalent cations that contribute to the stability of the outer membrane by providing electrostatic interactions with proteins and LPS.
Divalent cations (Mg2+, Ca2+) are essential for DNAse activity. EDTA chelates divalent cations in the solution preventing DNAses from damaging the plasmid and also helps by destabilizing the cell wall
function of solution II (SDS/NaOH)
The lysis buffer contains sodium hydroxide (NaOH) and the detergent Sodium Dodecyl Sulfate (SDS).
SDS is there to solubilize the cell membrane by breaking down protein-protein interactions.
NaOH helps to break down the cell wall, and it disrupts the hydrogen bonding between the DNA bases, converting the double-stranded DNA (dsDNA) in the cell, including genomic and plasmid DNA to single stranded DNA (ssDNA).
This process is called denaturation and is central part of the procedure, which is why it’s called alkaline lysis.
SDS also denatures most of the proteins in the cells, which helps with the separation of the proteins from the plasmid later in the process.
function of solution III (KOAc-potassium acetate)
Addition of potassium acetate (solution 3) returns decreases the alkalinity of the mixture.
Under these conditions the hydrogen bonding between the bases of the single stranded DNA can be re-established, so the ssDNA can re-nature to dsDNA