Bio 97 Midterm 1
1/103
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
104 Terms
which organisms contain double stranded DNA genomes?
most life, except some viruses
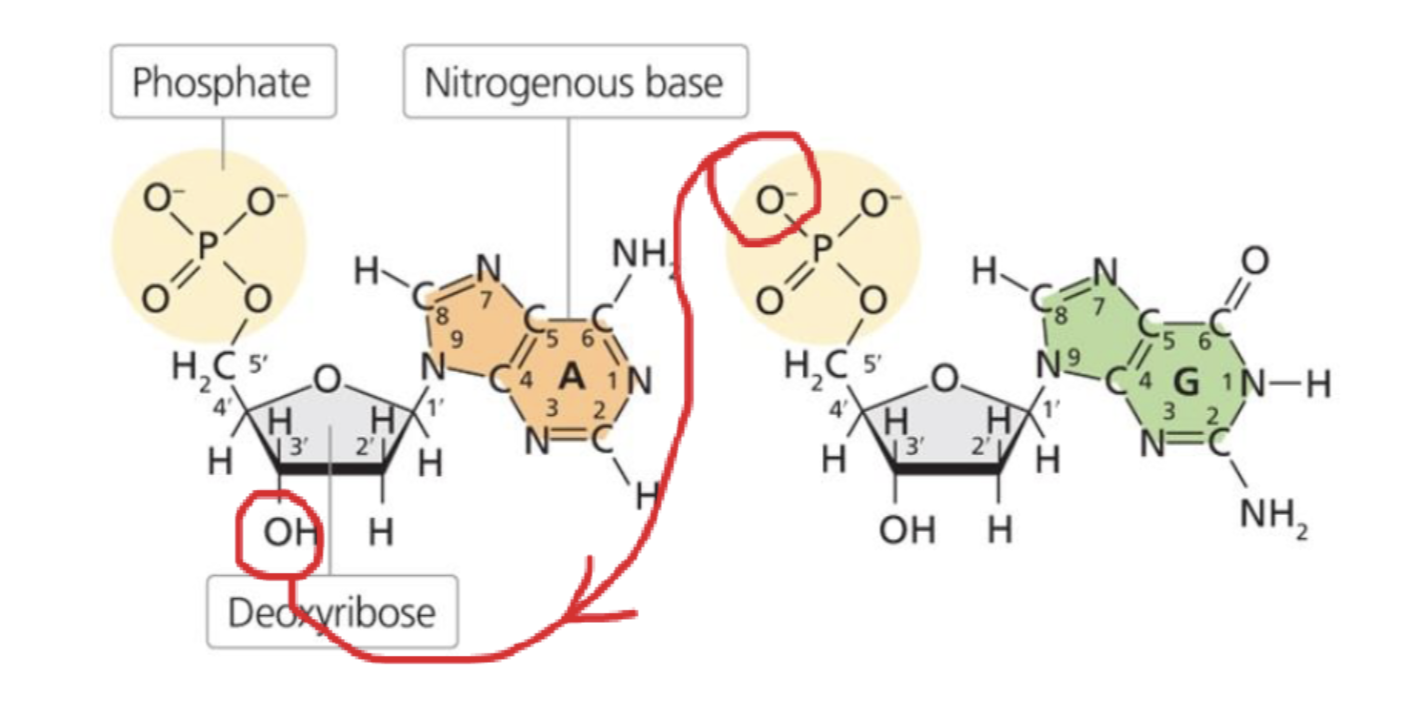
where do phosphodiester bonds link?
forms between the phosphate group on the 5’ carbon of one nucleotide’s sugar and the hydroxyl (-OH) on the 3’ carbon of the next nucleotide’s sugar —> creates the sugar phosphate backbone
which organisms contain single stranded DNA genomes?
parvoviruses and circoviruses
which organisms contain single stranded RNA genomes?
coronaviruses, poliovirus, coxsackie virus, rhinoviruses, Hep A virus
which organisms contain double stranded RNA genomes?
rotaviruses
which organisms experience change in genome (ssRNA —> dsDNA —> ssRNA genome)?
retroviruses
components of a nucleotide (what DNA and RNA consists of)
phosphate group, sugar, nitrogenous base
what are the two types of nitrogenous bases and how to differentiate them?
purine —> guanine and adenine (pure as gold) —> 2 rings
pyrimidine —> cytosine, uracil, thymine (CUT the pie) —> 1 ring (pie shaped)
which bases are found in DNA and RNA?
cytosine, guanine, adenine
which base is found only in DNA?
thymine
which base is found only in RNA?
uracil
how are nucleotides linked?
phosphodiester bonds
what reaction is involved in phosphodiester bond formation?
dehydration synthesis/condensation —> water molecule released
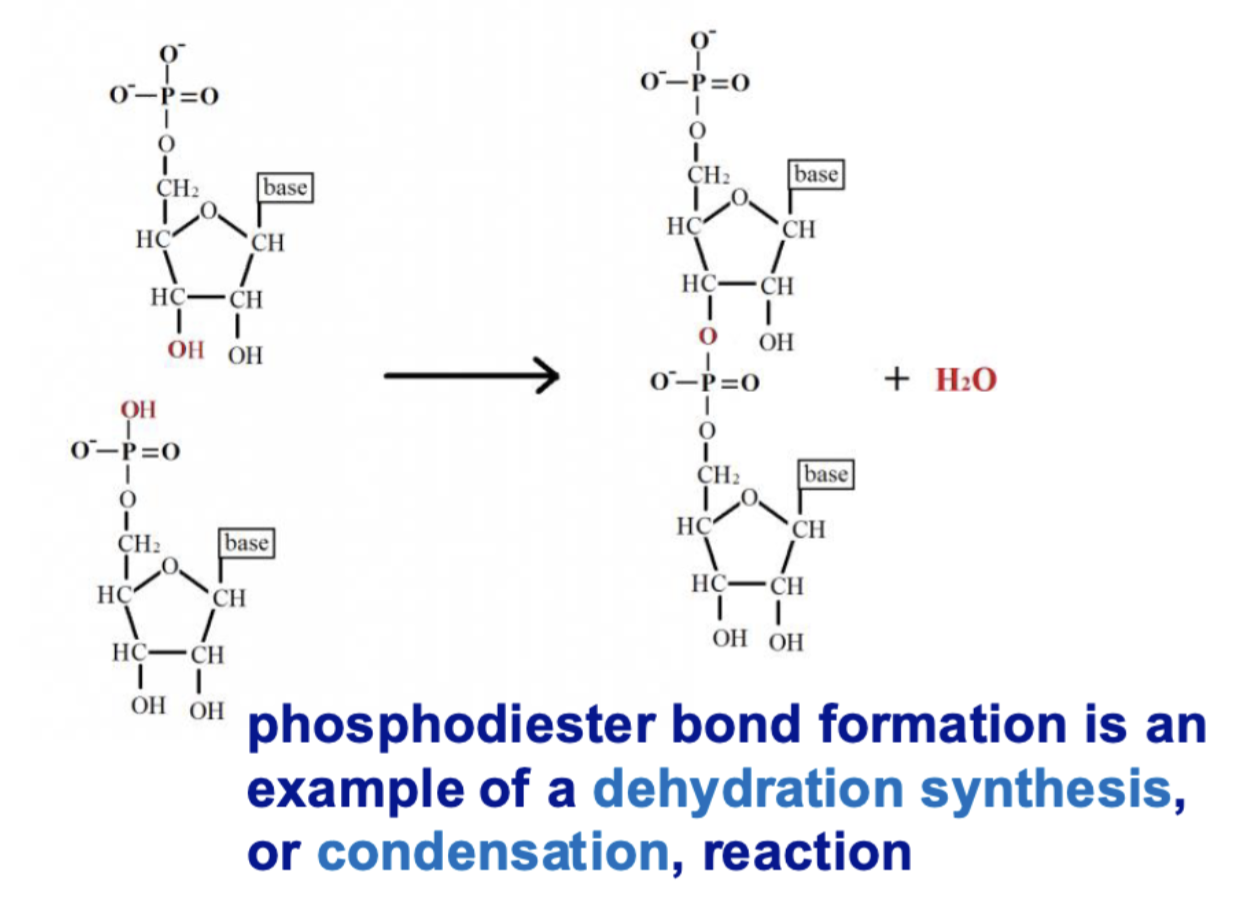
how do ester bonds relate to phosphodiester bonds?
a phosphodiester bond = phosphate group linking two sugars via ester bonds
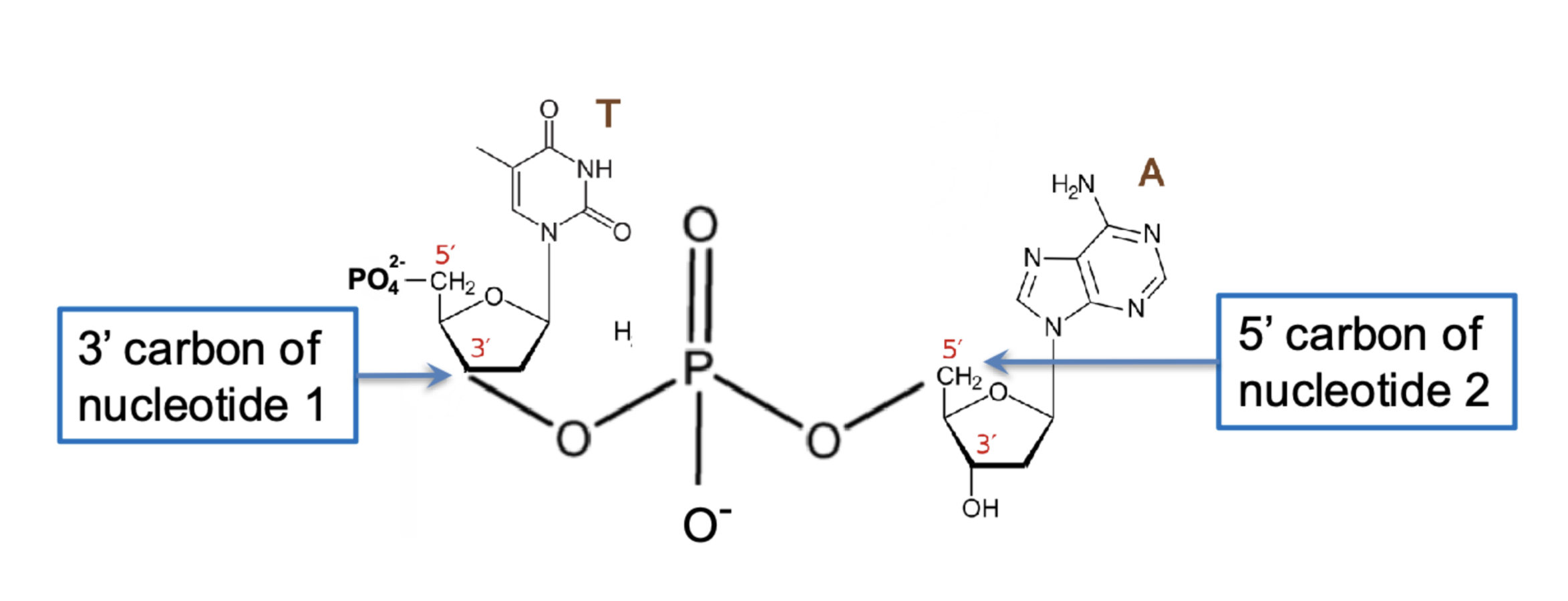
where is the next nucleotide being attached to?
the 3’ OH of the previous nucleotide
DNA is a double helix of ____ strands
antiparallel
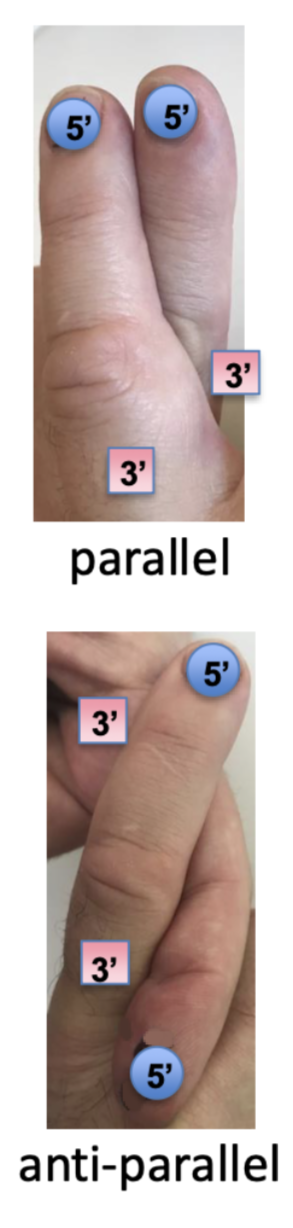
how are two strands of DNA held?
h-bonds formed between complementary base pairs
what base pairs with what?
A pairs with T (apples in the tree) —> 2 h-bonds
G pairs with C (cars in the garage) —> 3 h-bonds
A pairs with U (RNA)
why are h-bonds used between complementary base pairs?
cuz they are non covalent and relatively weak —> can separate at high enough temperatures or unzipped for DNA replication
why are partial charges necessary for h-bonding between base pairs in DNA?
h-bond donor has slightly positive hydrogen attached to electronegative atom
h-bond acceptor has slightly negative atom with a lone pair
attractive between slightly positive H and slightly negative (N/O) form h-bonds holding base pairs
5’ AGCTTCCC 3’
3’ TCGAAGGG 5’
is the new strand of replicated DNA the same as the old strand?
no cuz of complementary base pairing (different base pairs) but the new strand can be used template to make the old strand
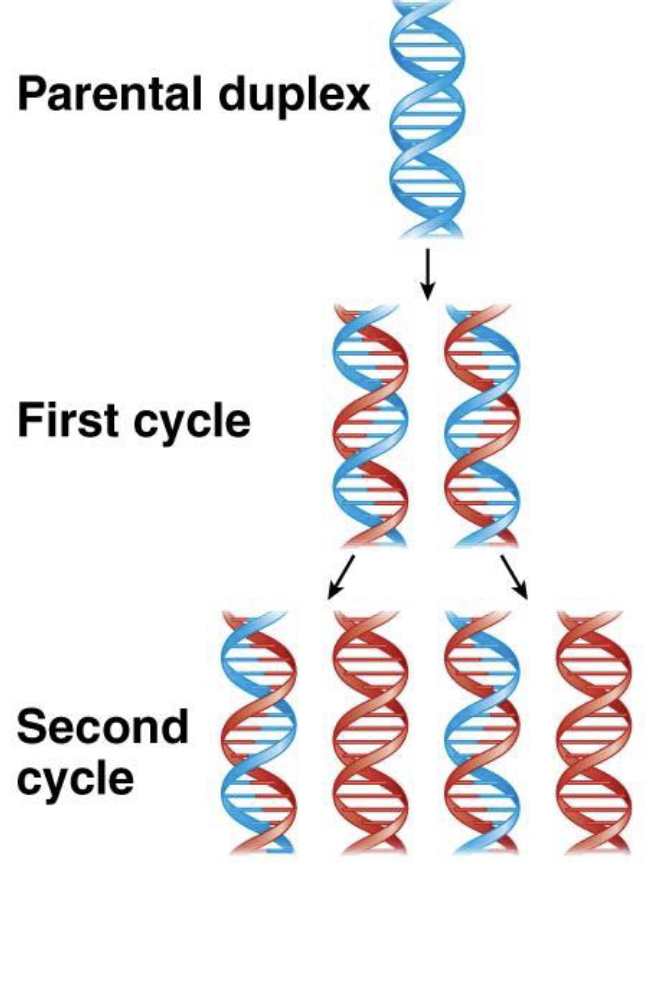
what happens if you start with a ds DNA molecule and make a complement of each strand?
you end with two double-helical daughter molecules that are identical copies of their mother —> each daughter strand contains one strand from mother and one newly synthesized strand
semi conservative replication
DNA replication
each DNA separates and is a template for a new complementary strand
after 1st cycle —> each DNA molecule has one old and one new strand
after 2nd cycle —> half of the molecules are hybrid (one old and one new) and the other half are completely new
confirmed by Meselson-Stahl experiment
how is a new DNA strand synthesized?
synthesized one nucleotide at a time
deoxynucleoside triphosphates (dNTPs) used to make reaction thermodynamically favorable
polymerization 5’ to 3’ (remember that nucleotides are added to 3’ OH)
done by DNA polymerase
what are dNTPs?
deoxynucleoside triphosphates
has a nitrogenous base, deoxyribose sugar, three phosphates
when DNA polymerase adds nucleotide to 3’ OH it uses the energy stored in dNTP ‘s phosphate bonds to break off two phosphates
remaining phosphate links the new nucleotide through phosphodiester bond
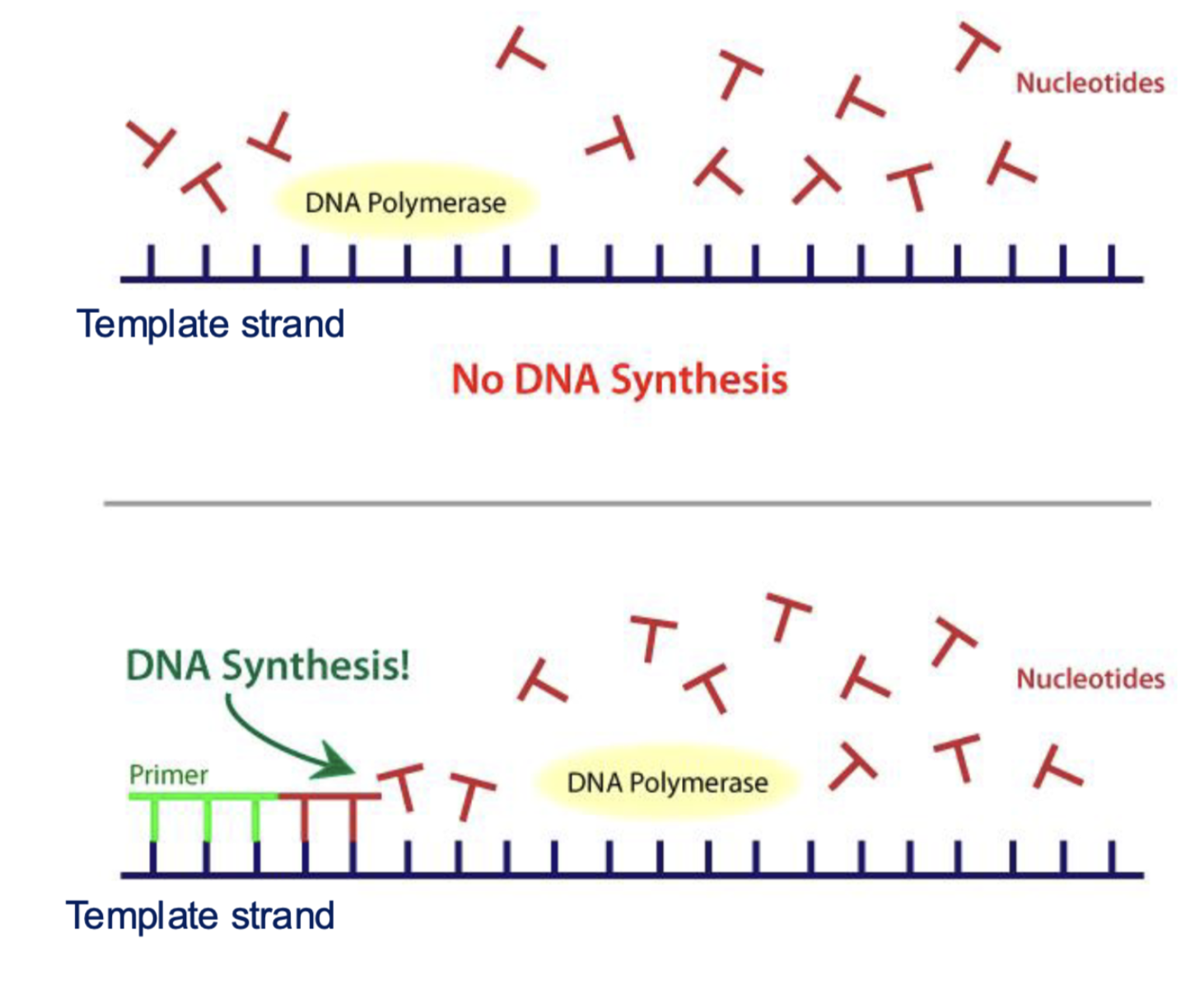
can DNA polymerase start off by itself?
no they need an RNA or DNA primer
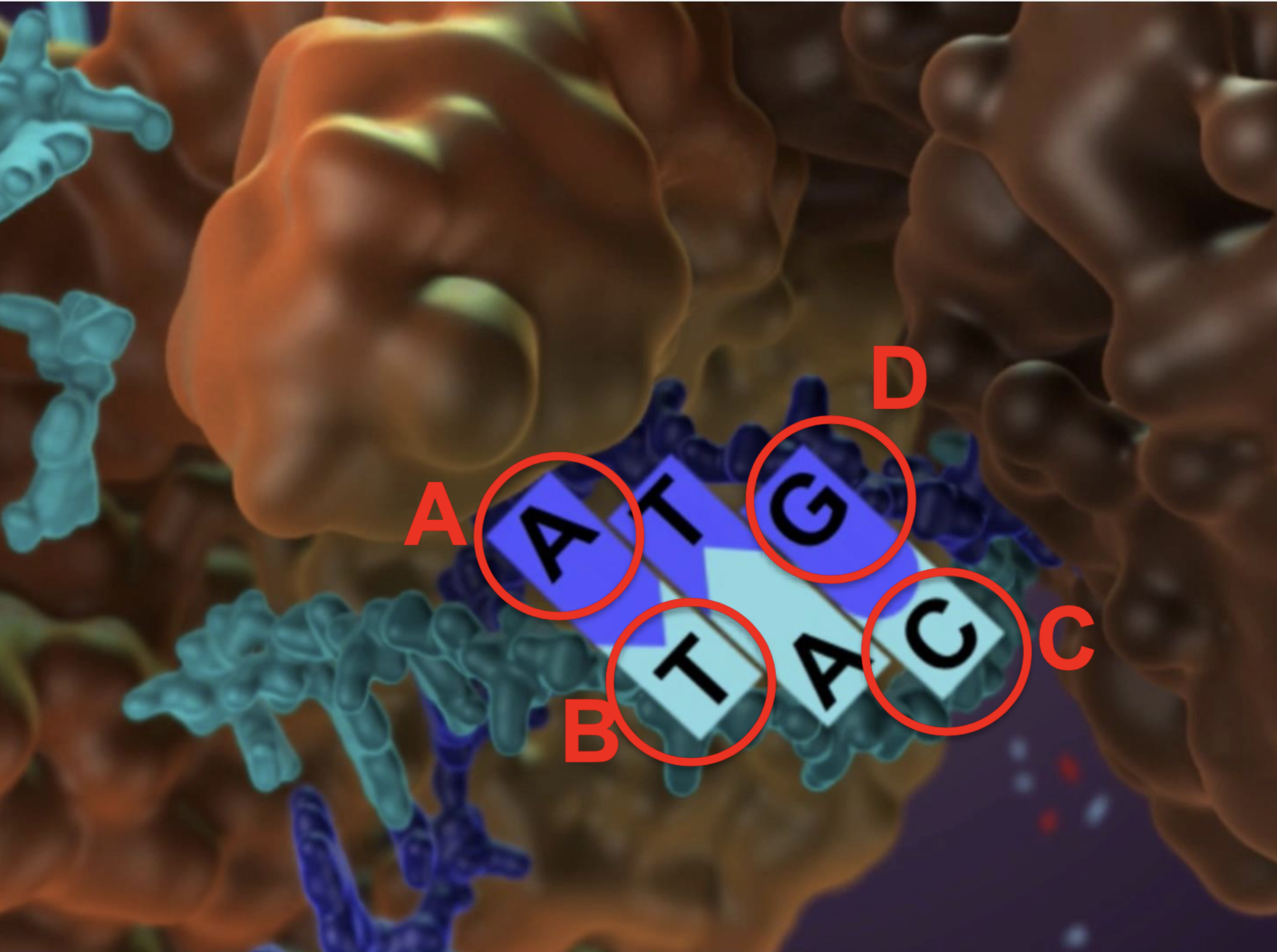
if the dark blue strand is the template and the light blue is the growing, which of the bases was recently added to the growing strand?
B (the thymine) cuz it is the 3’ OH
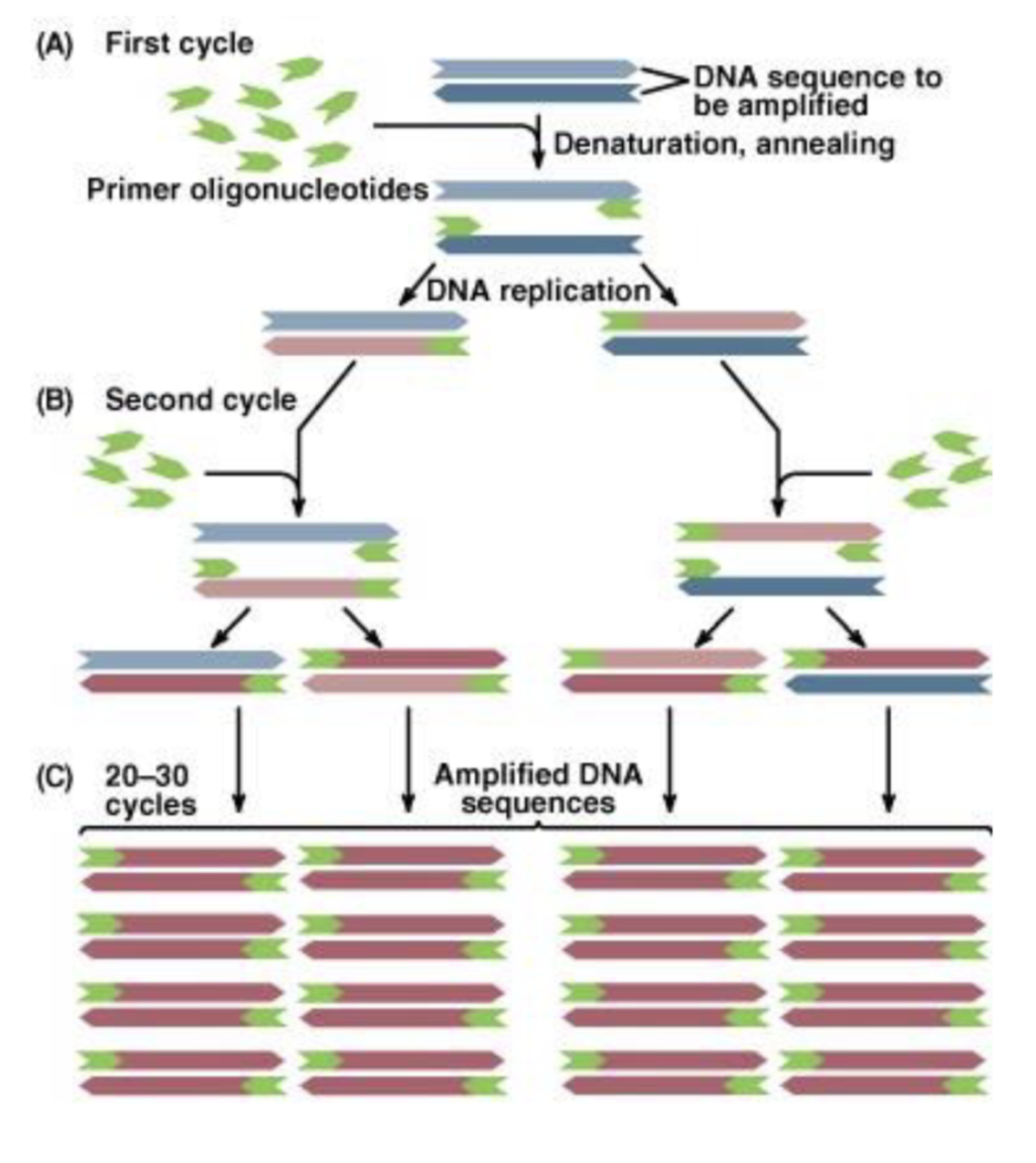
what is PCR?
polymerase chain reaction
DNA replication in a test tube
number of copies of the molecule that’s being replicated doubles each cycle
what is PCR used for?
pre-implantation genetic diagnosis
forensics
screening blood products for diseases
viral infections in monkeys by collected feces
sequencing the neanderthal genome
test for presence for virus causing COVID-19
what does PCR require?
sequence-specific primers (must know part of the DNA sequence)
primer must be made of DNA
special polymerase
is PCR semiconservative?
yes
how much DNA is made per cycle using PCR?
doubles amount of DNA each cycle —> cycles of polymerization must repeat to amplify DNA
what are the three steps of PCR?
denaturation, annealing, extension
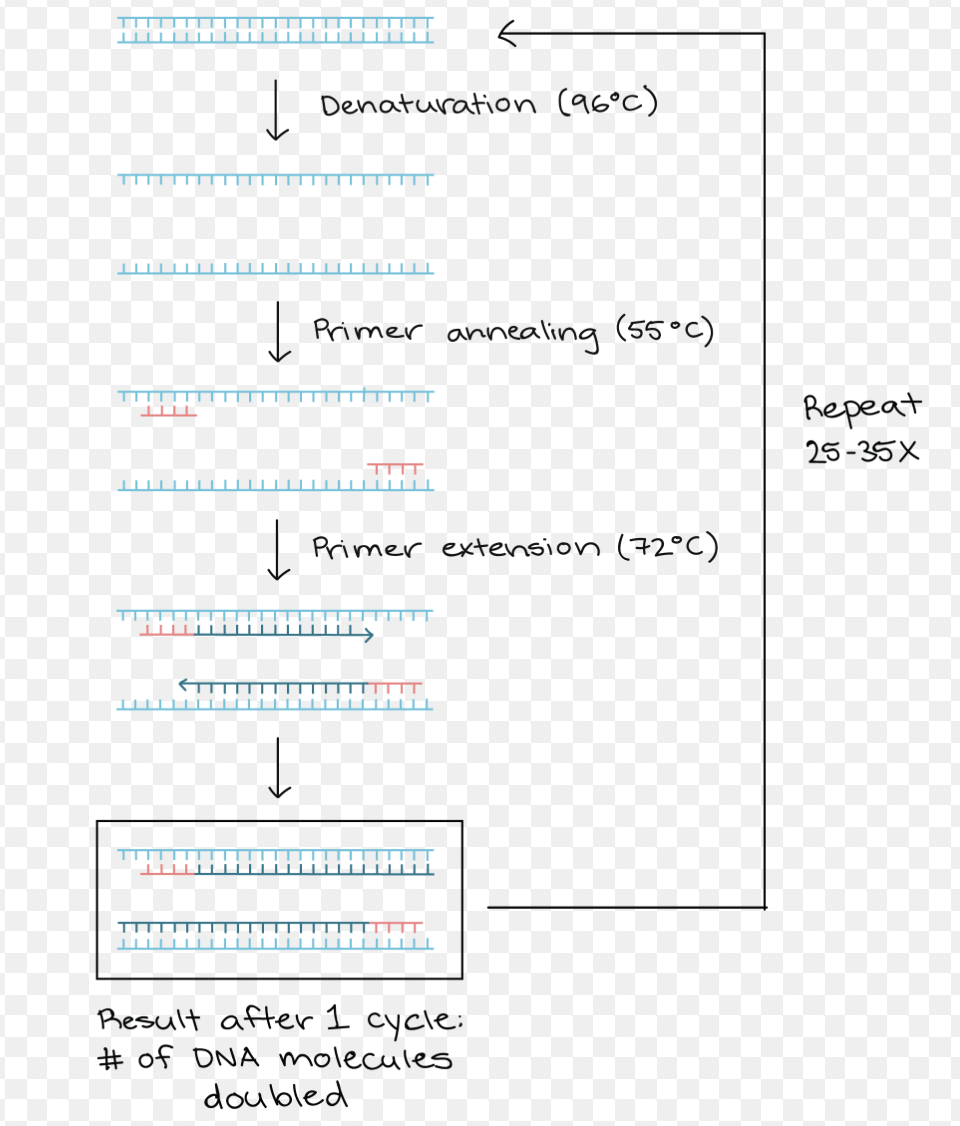
denaturation (step 1 PCR)
95C (heat added)
DNA strands separate/melt into single strands
annealing (step 2 PCR)
50C (depends on primer) (heat removed)
primers anneal/bind to complementary sequences on the template strands
extension (step 3 PCR)
72C (heat added)
polymerase does it things (replication starts)
why do we need PCR?
we need more DNA and we need to measure the size of a piece of target DNA
what is sanger sequencing/dideoxy DNA sequencing?
method used to determine exact nucleotide sequence of a DNA strand
how does dideoxy DNA sequencing work?
dideonucleotide triphosphate (ddNTP) lacks a 3’ OH (hydrogen is there instead)
without the OH, phophodiester bond cannot form
DNA replication stops (chain termination)
DNA is copied in presence of other normal dNTPs and some fluorescently labeled ddNTPs
ddNTP gets added sometimes instead of dNTP, stops the chain at specific base
DNA fragments of different lengths made
the glowing fragments lets scientists read the different DNA sequences
are there more ddNTPs than dNTPs?
no ddNTPs are present at lower concentrations for better control (we need more dNTPs cuz they build the strand)
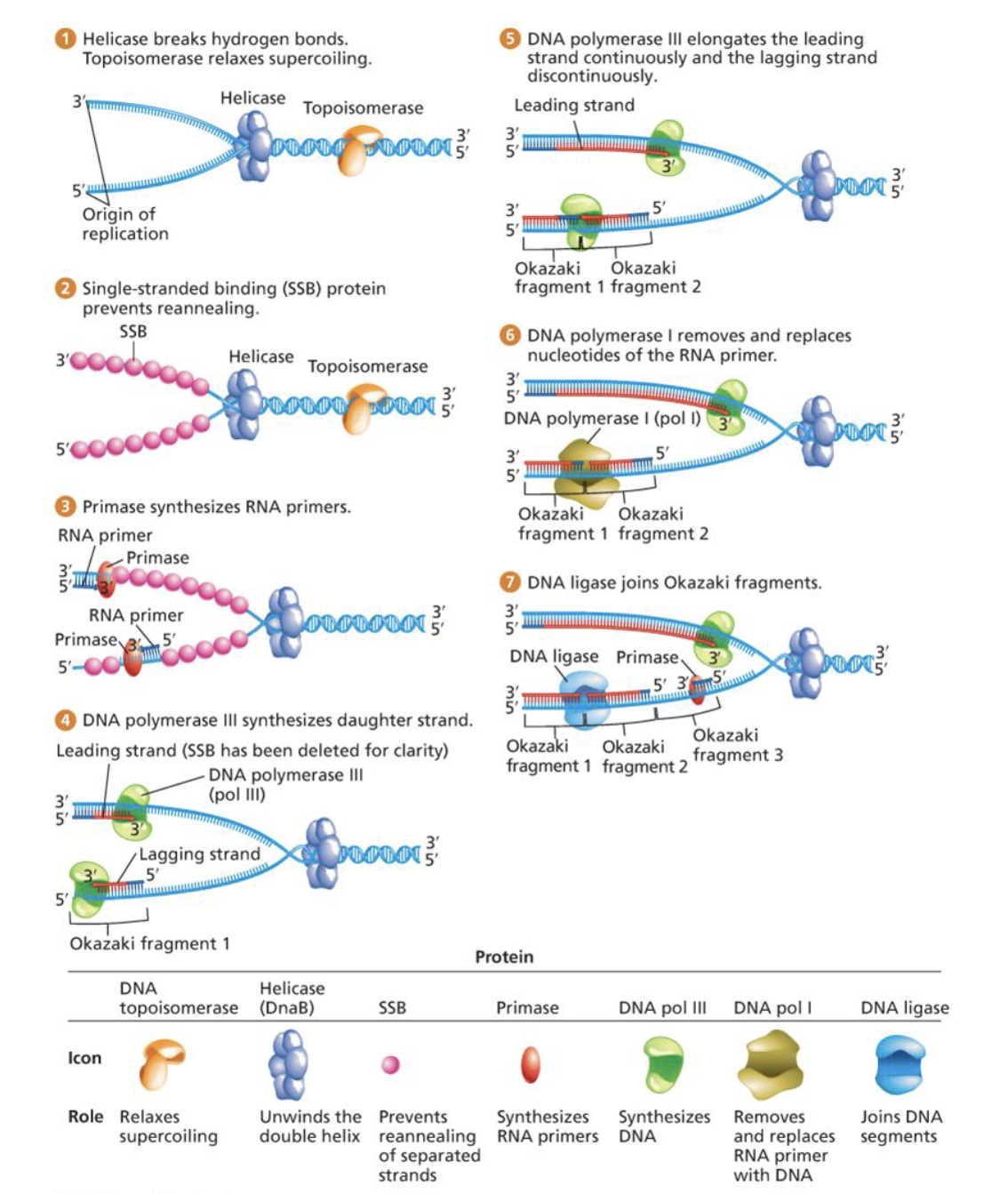
DNA replication steps
helicase breaks h-bonds and topoisomerase relaxes supercoiling
single-stranded protein prevents reannealing
primase synthesizes RNA primers
DNA polymerase III synthesizes daughter strand
DNA polymerase III elongates the leading strand continuously and the lagging strand discontinuously
DNA polymerase I removes and replaces nucleotides of the RNA primer
DNA ligase joins Okazaki fragments
what are replication origins?
sites where DNA synthesis initiates in cells
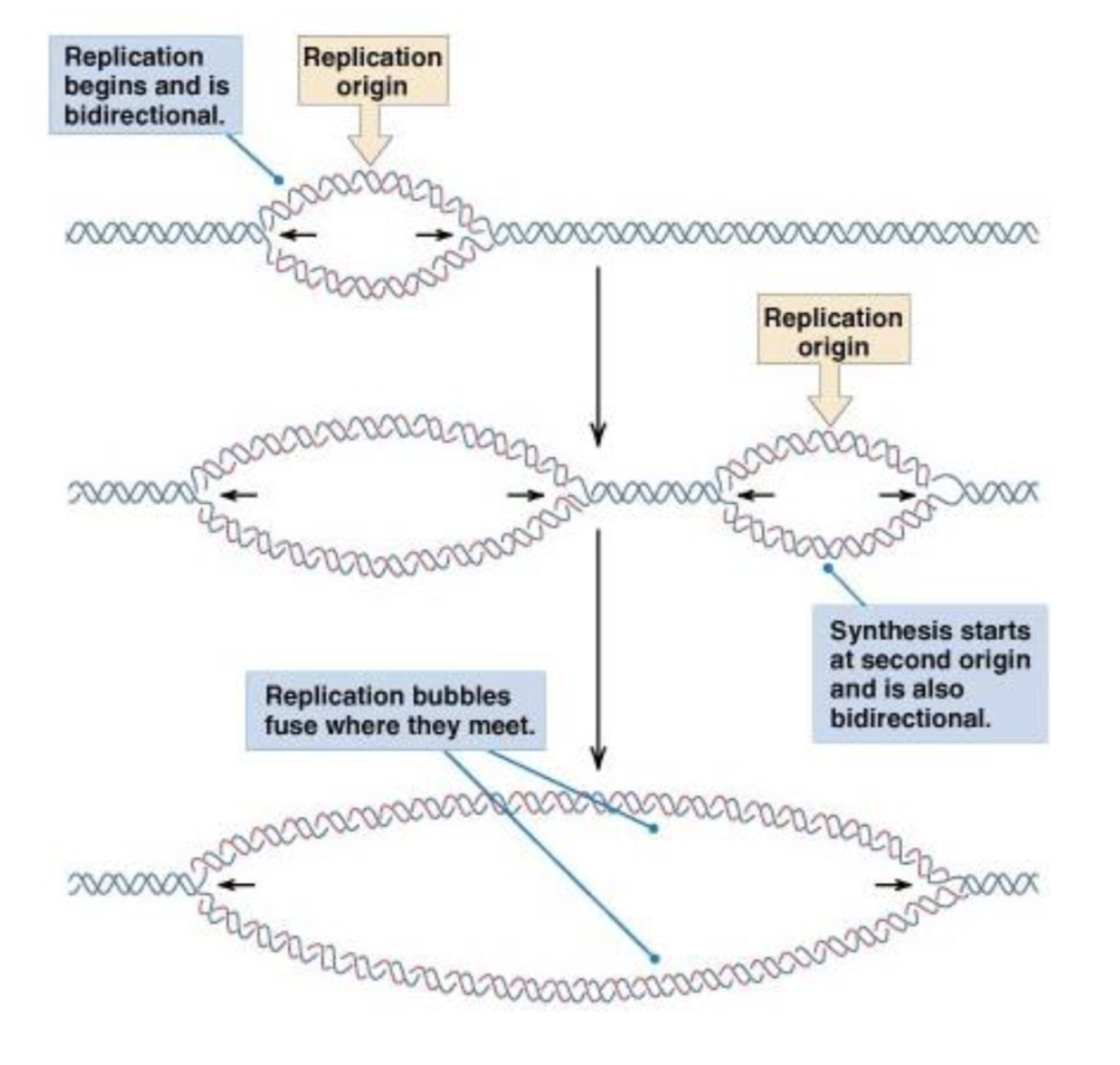
how many replication origins do eukaryotic chromosomes have?
a lot
human genome has >10,000 origins that are 50,000 base pairs apart \
allows for replication to speed up
is DNA replication in both directions at once?
yes
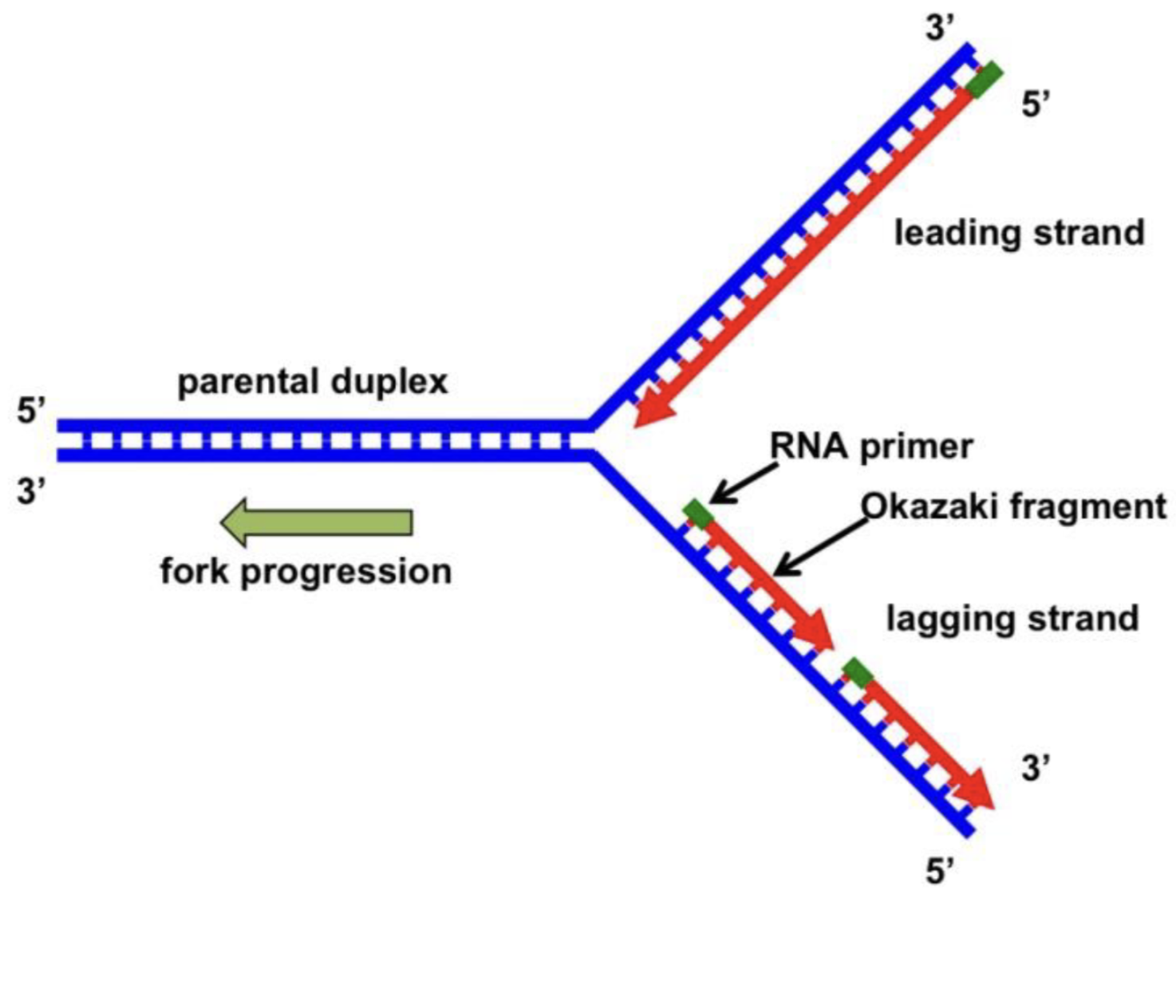
leading vs lagging strand on the replication fork
leading strand is synthesized continuously until it encounters the 5’ end of the adjacent replication bubble
lagging strand is synthesized in shorter pieces and ligated together (phosphodiester bonds via ligase)
DNA replication in cells vs PCR/dideoxy sequencing
Both
need template, primers, DNA polymerase, nucleotides
DNA replication in cells
RNA = primer
enzymes unwind DNA duplex
replication origins, replication forks, leading and lagging strand
multiple enzymes and proteins involved
PCR/dideooxy sequencing
DNA primers
heat unwinds the DNA
no replication origins, forks, leading and lagging strands —> all strands are leading
only DNA polymerase needed
what is the central dogma?
DNA —> transcription —> RNA —> translation —> protein
importance of proteins
give us our phenotype
they are enzymes, structural components of cells, etc.
which ones we have and how much we have of them determine how we function and how we look
DNA controls our phenotype by encoding proteins
what other factors influence phenotype?
non-coding RNAs (tRNAs, rRNAs, microRNAs, IncRNAs)
gene expression
conversion of a gene into a product (protein via an RNA intermediate) —> transcription and translated into protein
how does expression vary among genes?
some genes are expressed all the time and some are only expressed in response to external or integral conditions or signals
do each cell of a multicellular organism contain the same DNA/genes?
yes BUT different cells express different genes and thus different proteins
only a subset of genes are expressed in a given cell type
only a subset of those genes are expressed at a given time
housekeeping genes
expressed in all cells
cell-type specific genes
expressed in certain cells (neurons, muscle cells, blood cells)
transcription
process of RNA being synthesized from DNA template using RNA polymerase
what is the direction of RNA synthesis?
5’ to 3’
what are differences between DNA and RNA?
DNA
composed of dNMPs
synthesized from dNTPs
RNA
composed of NMPs
synthesized from NTPs
uracil used instead of thymine
how does the DNA coding (nontemplate) strand and the mRNA template relate?
they have same polarity and sequence EXCEPT each T in the coding strand appears as a U
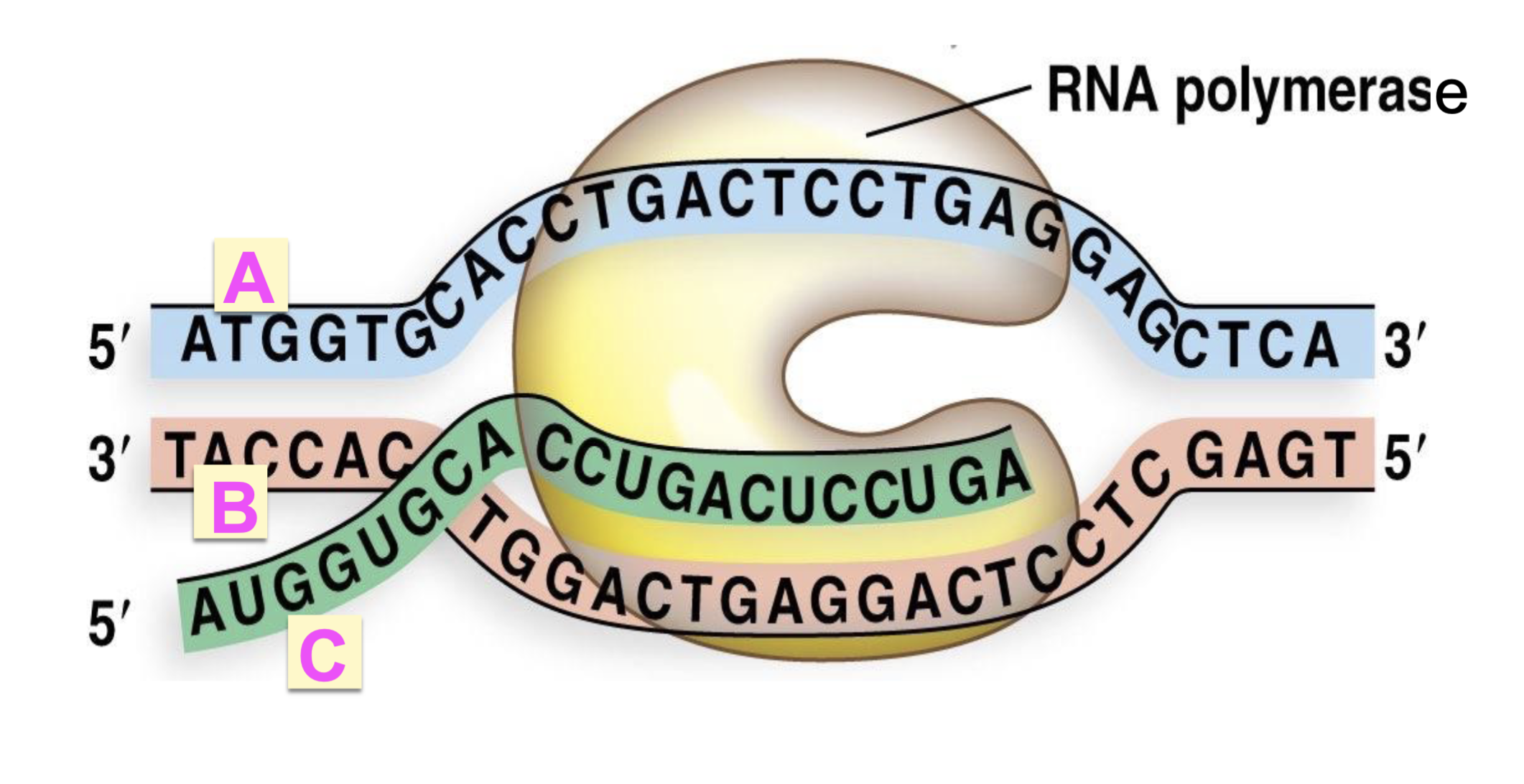
label A, B, C
A = coding/nontemplate sense strand
B = noncoding/template antisense strand
C = mRNA
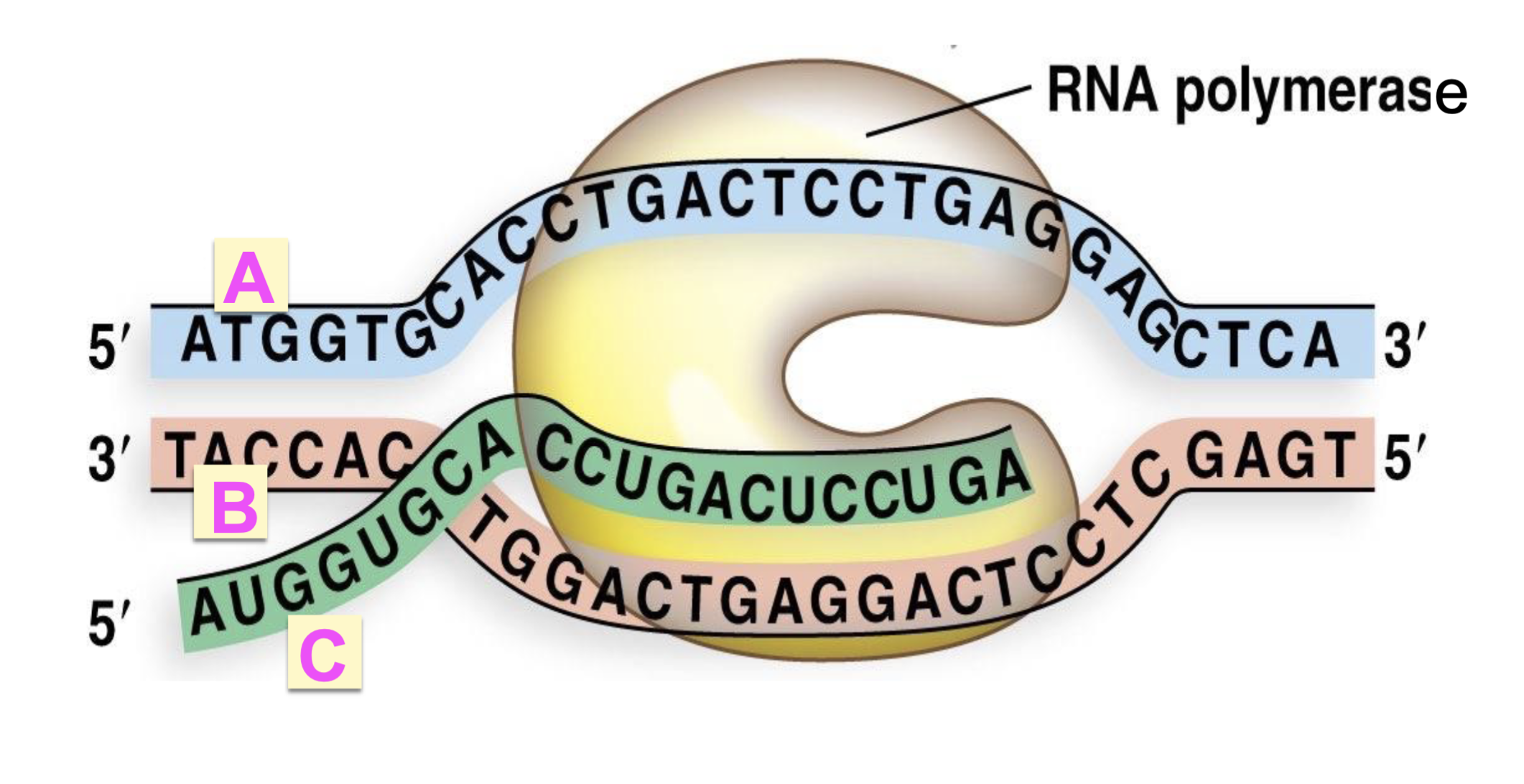
where is the new nucleotide added?
3’ end of the mRNA growing strand (G)
5’ GGUUACAUUC ‘3 is the RNA transcript strand, the coding strand is…
5’ GGTTACATTC 3’
5’ GGUUACAUUC ‘3 is the RNA transcript strand, the template strand is…
5’ GAATGTAACC 3’
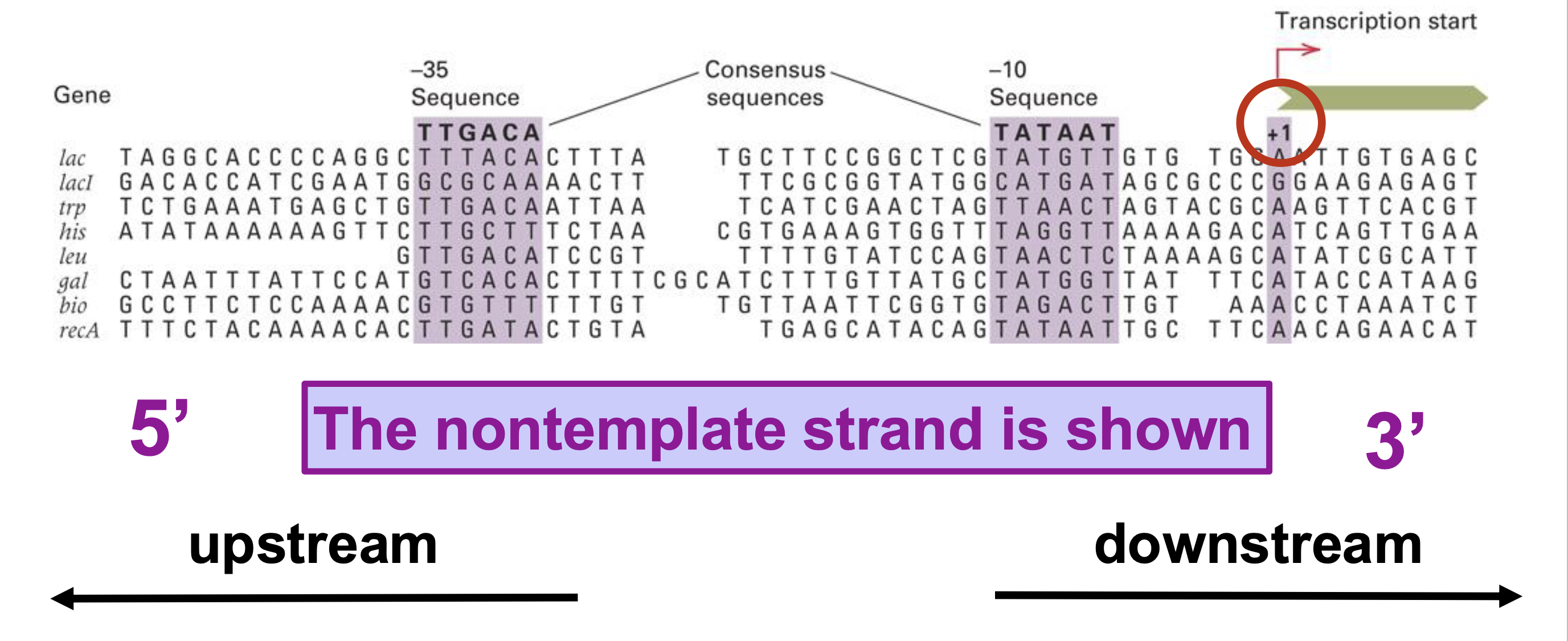
how does RNA polymerase know where to start transcription?
uses promoter
(in prokaryotes) —> promoter consists of -10 sequence and -35 sequence (RNA polymerase binds to these two)
amino acid residues in protein use h-bonds, electrostatic interactions, and hydrophobic interactions to recognize these sequences
determines when transcription starts and direction in which it proceeds
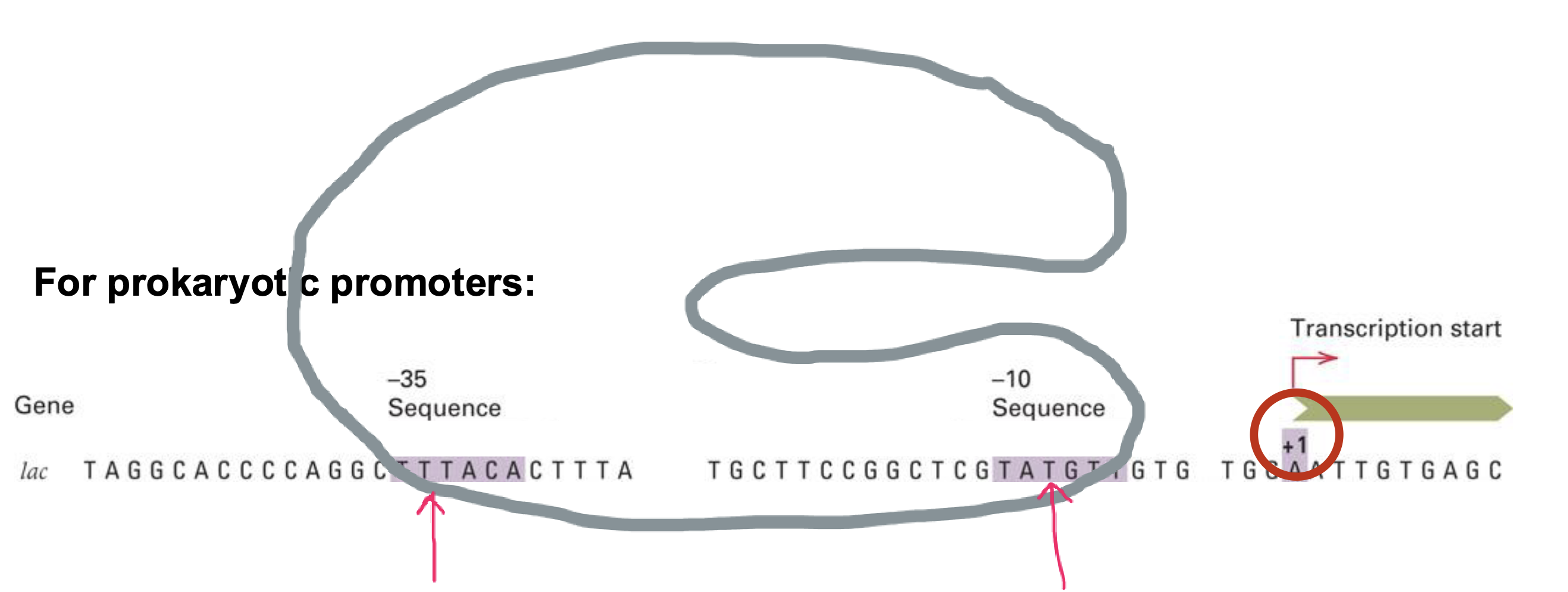
does transcription start upstream or downstream the promoter?
downstream
is the sequence of the -10 and -35 boxes different in different genes?
yes —> consensus sequence of each box can be found by aligning the promoter regions of multiple genes
how else are eukaryotic genes regulated?
by other sequences in the DNA called enhancers and silencers
where are enhancers and silencers located?
varies —> some close to promoter or far upstream or downstream
what do enhancers and silencers contain?
binding sites for specialized proteins called transcription factors
transcription steps
RNA polymerase binds to -10 (TATA box) and -35 promoter consensus sequences
DNA unwinds near the transcription start site to form open promoter sites
RNA polymerase initiates transcription and begins RNA synthesis
continues until it encounters termination sequence —> as RNA synthesis progresses the DNA duplex unwinds to allow the template strand to direct RNA assembly
prokaryote vs eukaryote mRNAs
each eukaryote mRNA encodes for a single protein
prokaryote mRNA can be polycistronic (code for multiple proteins)
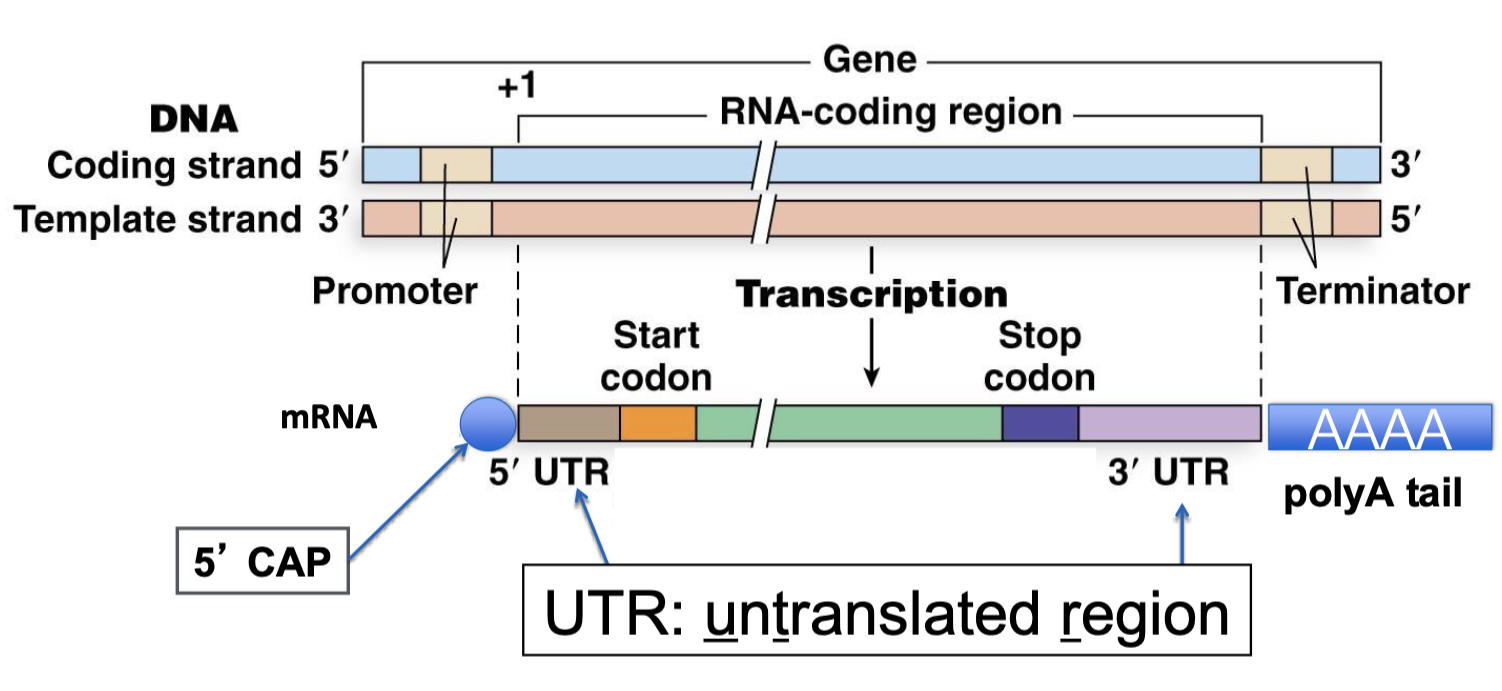
what is added as the primary transcript is processed into mRNA in eukaryotes?
5’ cap and polyA tail added
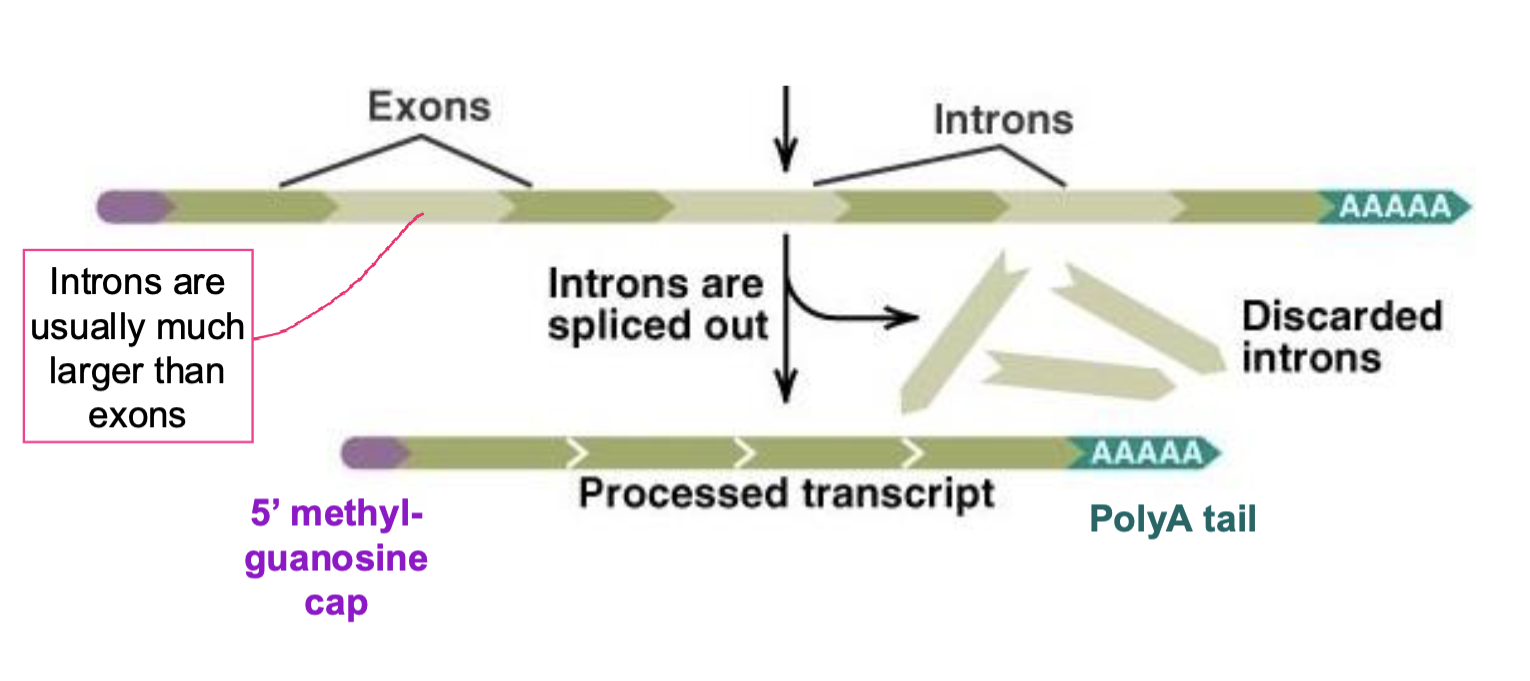
what other modification occurs as primary transcript is processed into mRNA in eukaryotes?
mRNA splicing
introns are splices out
occurs at sites determined by consensus sequences
multiple proteins needed to cut and paste
takes place in nucleus
splices out introns degraded and nucleotides recycled
splicing patterns vary under different conditions (alternative splicing)
introns larger than exons
links up exons from within a given gene, not exons within different genes
re
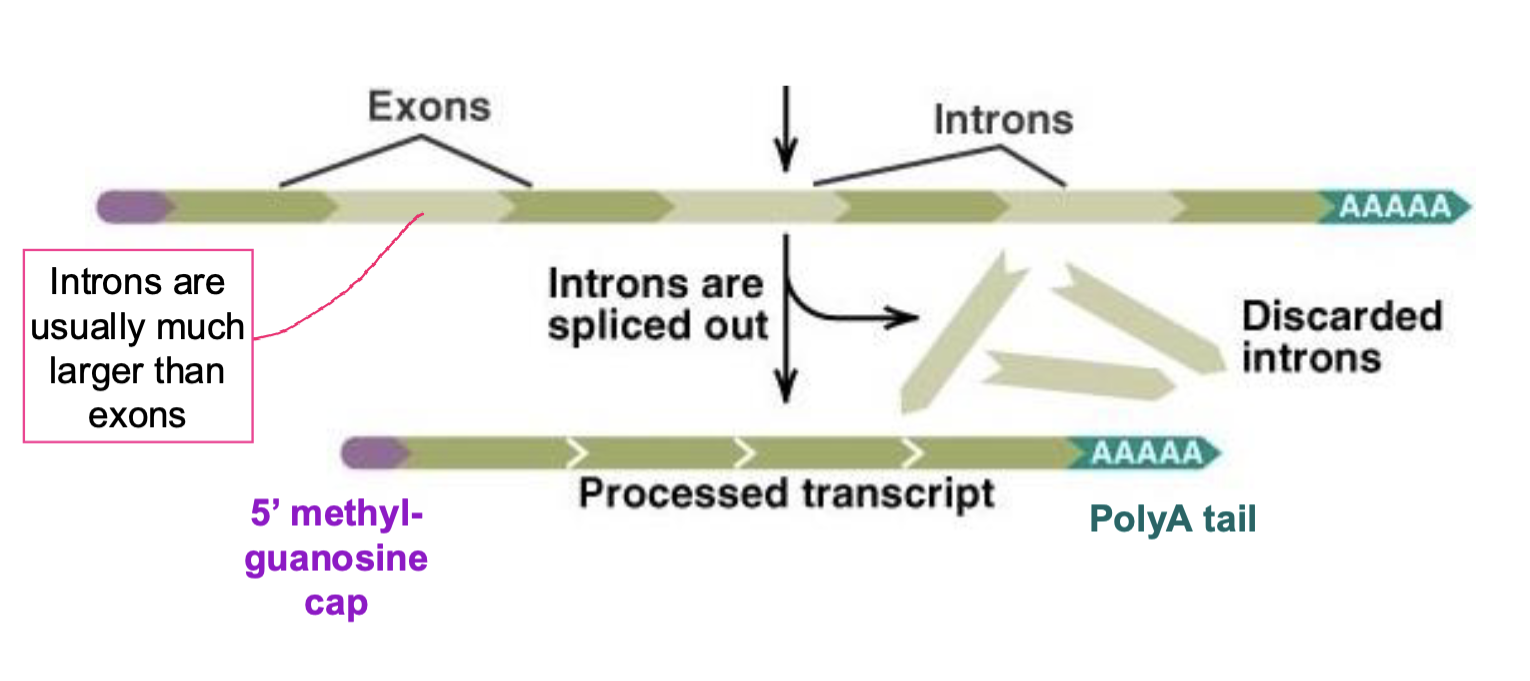
what happens during alternative splicing?
same transcript can be splices differently and give rise to different proteins (order of exons remain the same, results in the inclusion or exclusion an exon) —> forms of proteins can have different functions
does not result in duplication of an exon in the mature mRNA
what doe exons contain/encode?
5’ and 3’ UTR
protein coding sequence (CDS)
translation
take instructions in mRNAs and use to build proteins
building blocks of the fundamental dogma
replication —> DNA nucleotides
transcription —> RNA nucleotides
translation —> amino acids
what are proteins composed of?
amino acids —> 20 different AAs
protein = linked in a chain by peptide bonds —> typical protein is 450 AA long
start as a linear chain of AAs —> will fold into complicated 3-D structures
what are the components of an amino acid?
amino group, carboxyl group, side chain (R) group
what kind of reaction is peptide bond formation?
dehydration synthesis reaction
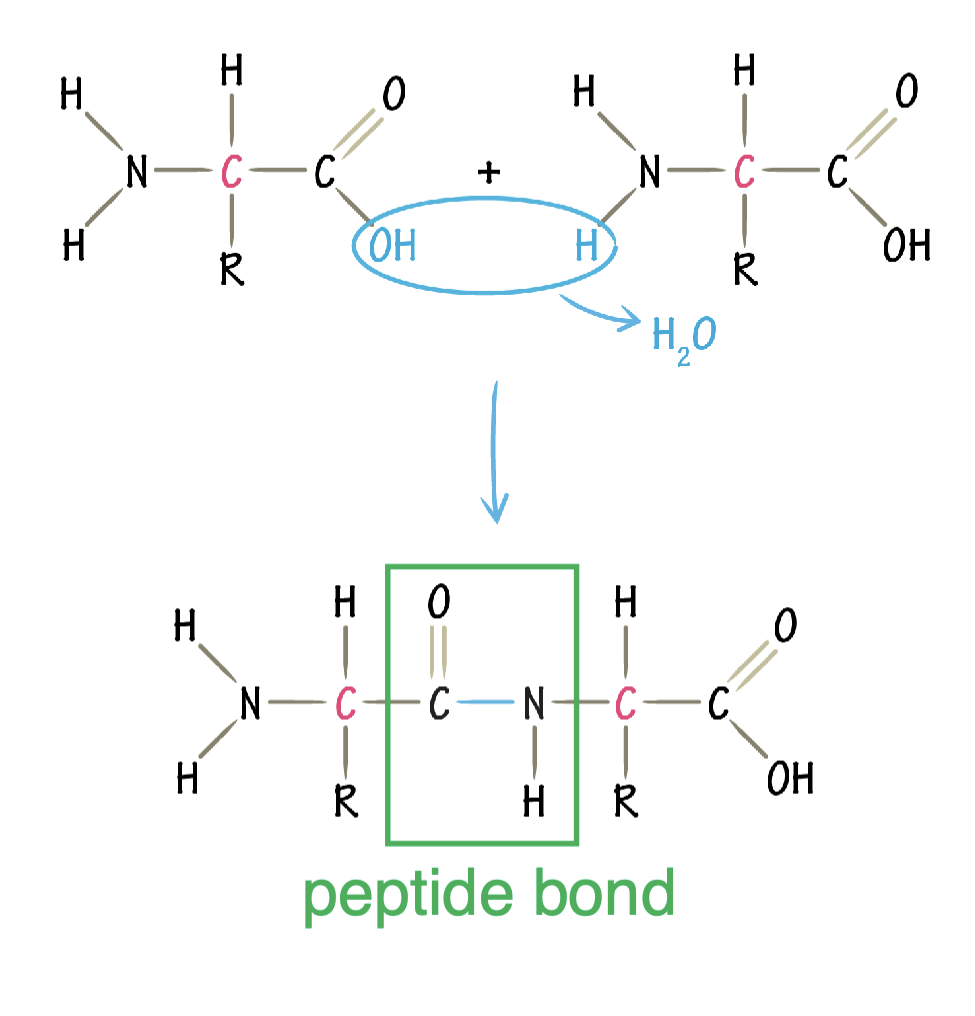
direction of protein synthesis
amino (N) terminus to carboxyl (C) terminus
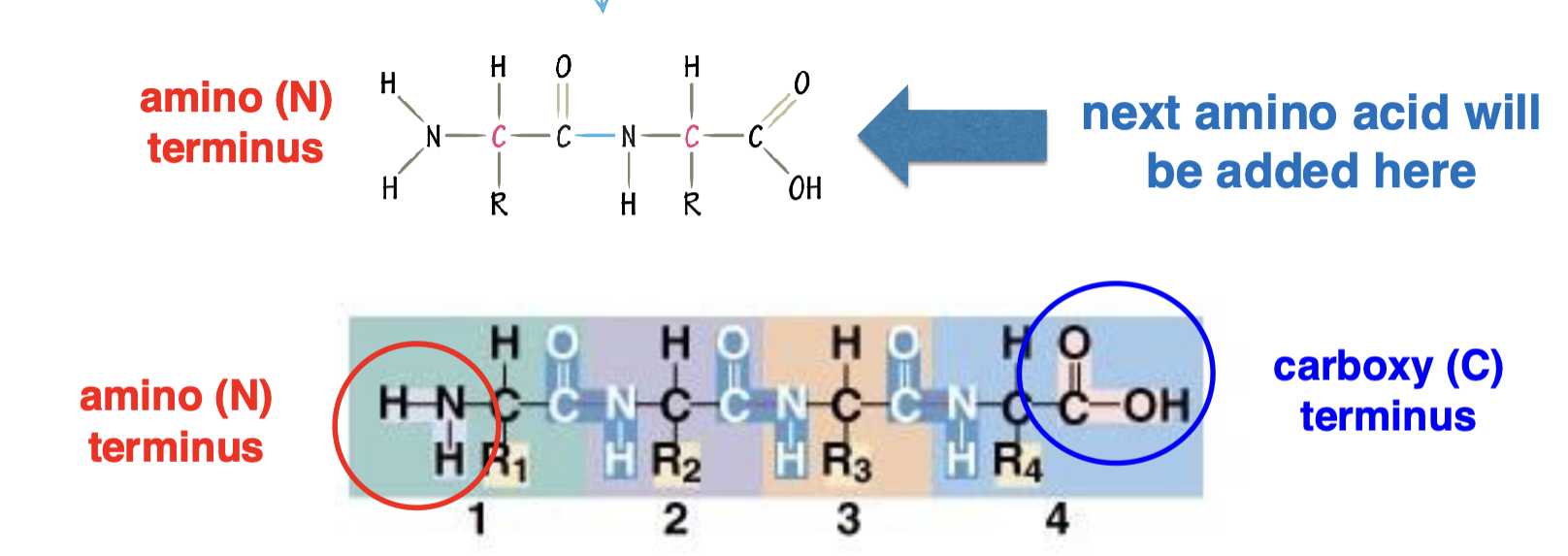
how is protein sequence encoded in DNA/RNA?
protein coding region of an mRNA is made up of non-overlapping nucleotide triplets (codons) which each correspond to an AA
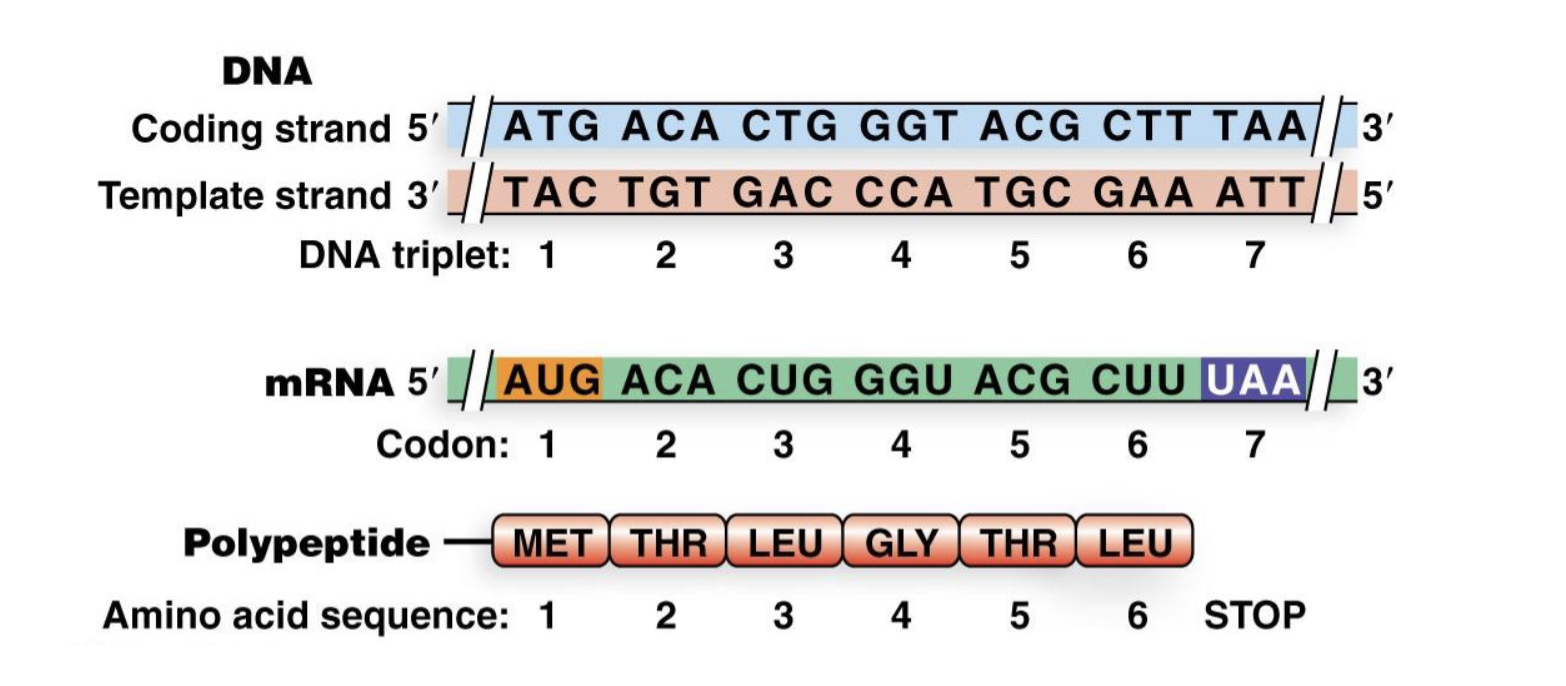
the universal genetic code
64 codons
61 codons for 20 AA (redundancy occurs)
AUG = start codon —> makes methionine (M)
3 stop codons —> signal end of coding sequence reached
what are key players in translation?
ribosomes
tRNAs (transfer RNAs)
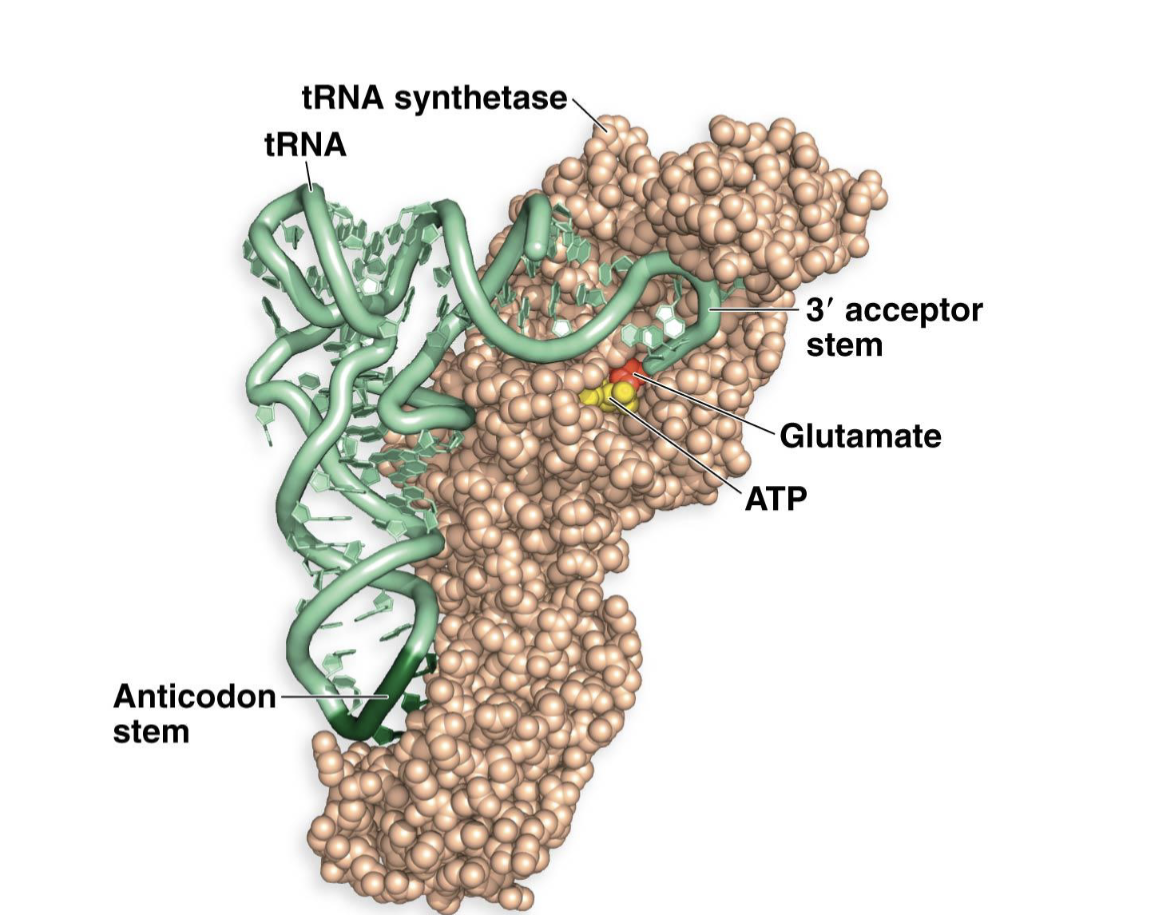
aminoacyl tRNA synthetases
what are ribosomes composed of?
they are machines composed of 3-4 rRNAs (ribosomal RNAs) and over 50 proteins
large and small subunits
have channels that hold RNA and nascent polypeptide
what are the functions of ribosomes?
bind mRNA and identify the start codon for translation (mRNA read from 5’ —> 3’)
help bring about complementary pairing between mRNA codons and tRNA anti-codons
catalyze peptide bond formation between AA
what do tRNAs do?
pair codons with AAs
—> becomes charged when attached to AA
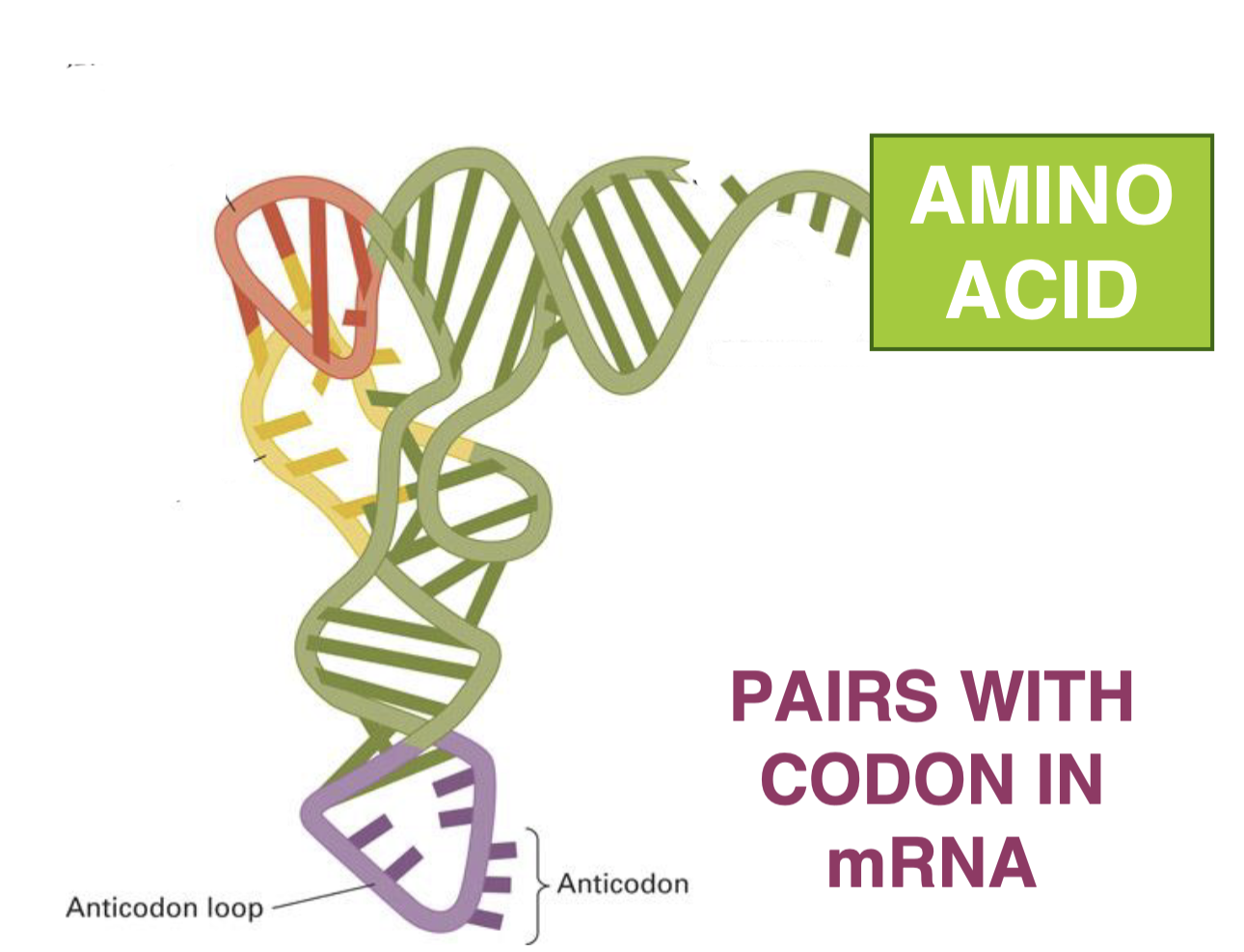
aminoacyl tRNA synthetases
enzymes that recognize the anticodon and correct (cognate) AA and then attach this AA to the tRNA
what are the three phases of translation?
initiation, elongation, termination
step 1 translation (initiation)
initiation complex —> small ribosomal subunit and initiator Met-tRNA binds to 5’ cap on mRNA
initiation complex scans along mRNA 5’ to 3’ until AUG is found
large subunit binds
initiating methionine is tRNA is on the P (peptidyl site)
what are the tRNA binding sites in the ribosome?
peptidyl (P) site —> holds tRNA to which the growing polypeptide chain is attached
acceptor (A) site —> binds tRNA carrying the next AA to be added
exit (E) site —> empty tRNAs leave from here (large subunit shifts) after their AA has been added
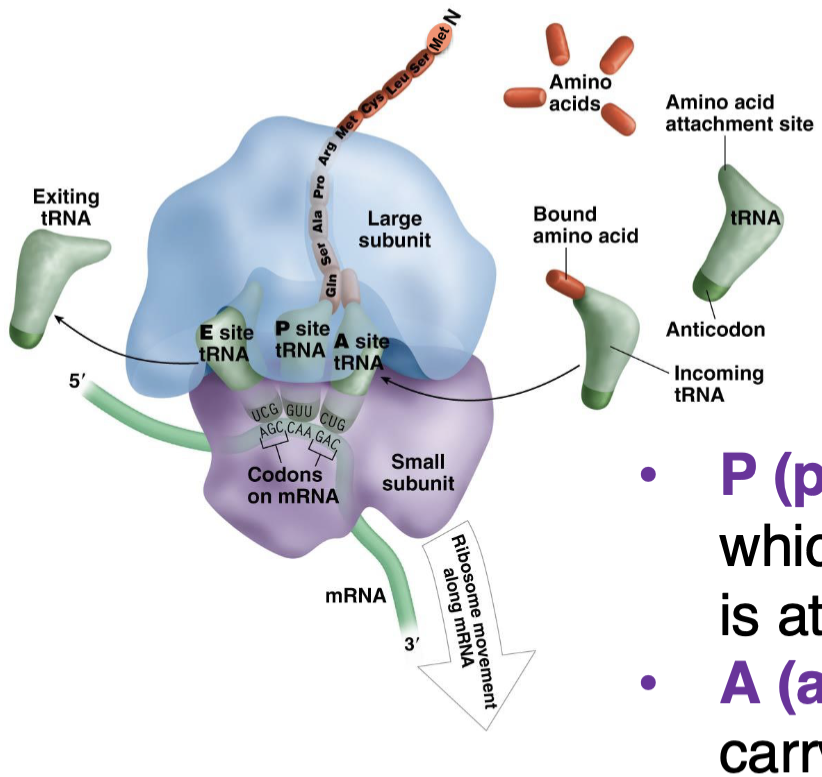
step 2 translation (elongation)
new tRNA enters A site
peptide bond formed between AA in the A and P site
ribosome moves one codon —> tRNA in P site moves to E site and tRNA in A site moves to P site
tRNA in E site exits
next tRNA enters A site —> repeat
step 3 translation (termination)
release factors are recruited when a stop codon occurs at the A site
eRF fills the A site and triggers release of the newly made polypeptide by hydrolysis of GTP
ribosome dissociates and mRNA release
the portion of the mRNA after the stop codon is 3’ UTR
the anticodon is part of the…
tRNA
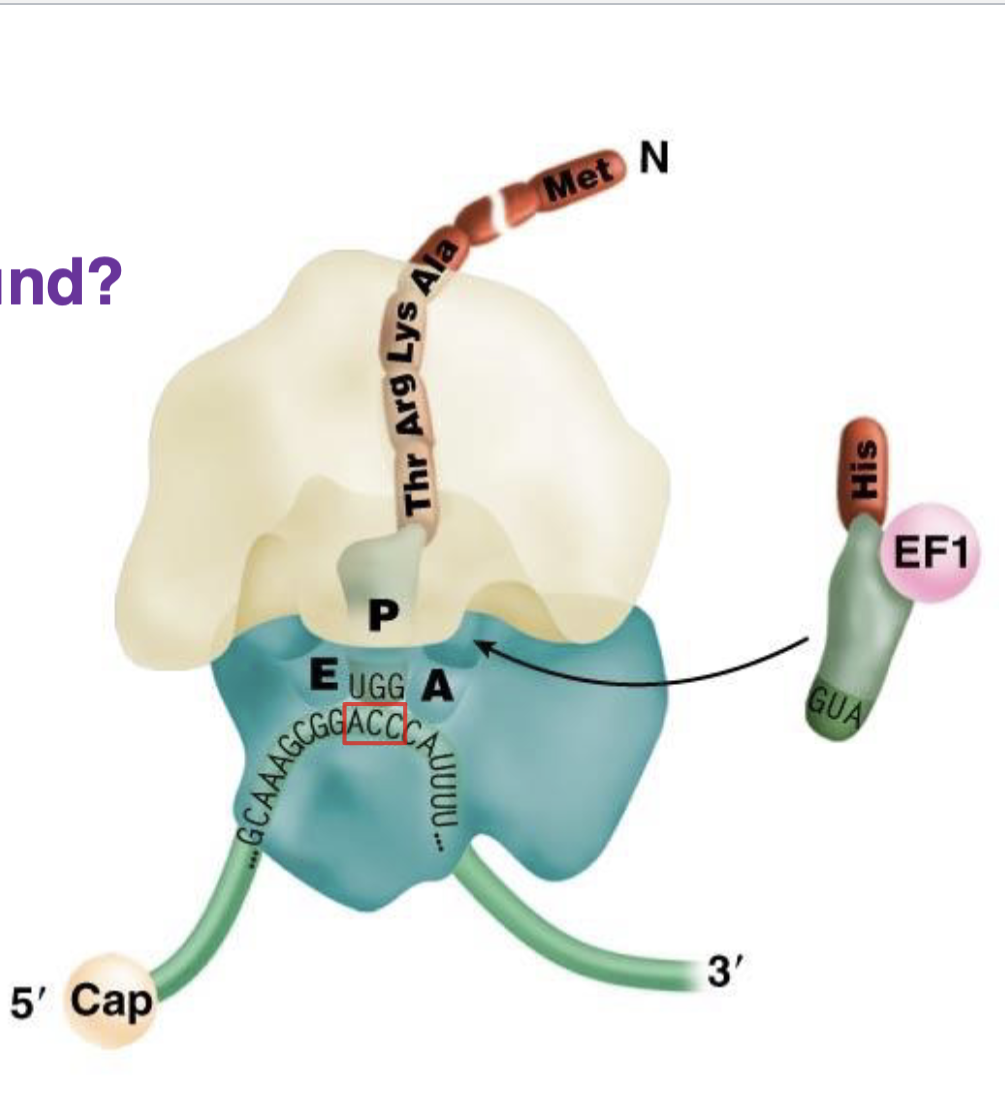
what is the red box?
codon for Thr
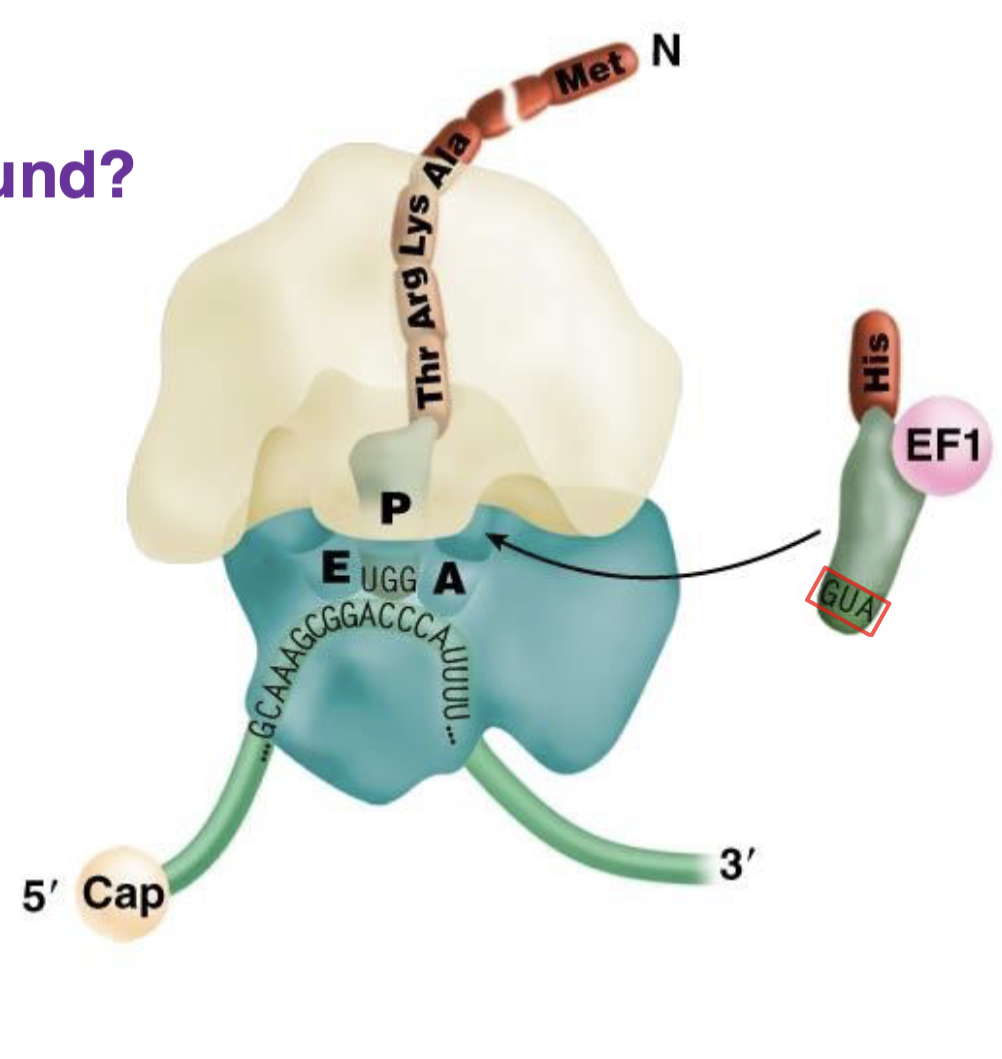
what is the red box around?
anticodon for His (GUA —> CAO (codon) —> His)
what is the anticodon for the Thr codon ACC?
5’ GGU 3’
where are proteins translated in eukaryotes?
mRNA processed in nucleus, then splicing, then exported to cytoplasm where it is translated
where are proteins translated in prokaryotes?
transcription and translation coupled (cuz there is no nucleus in bacteria)
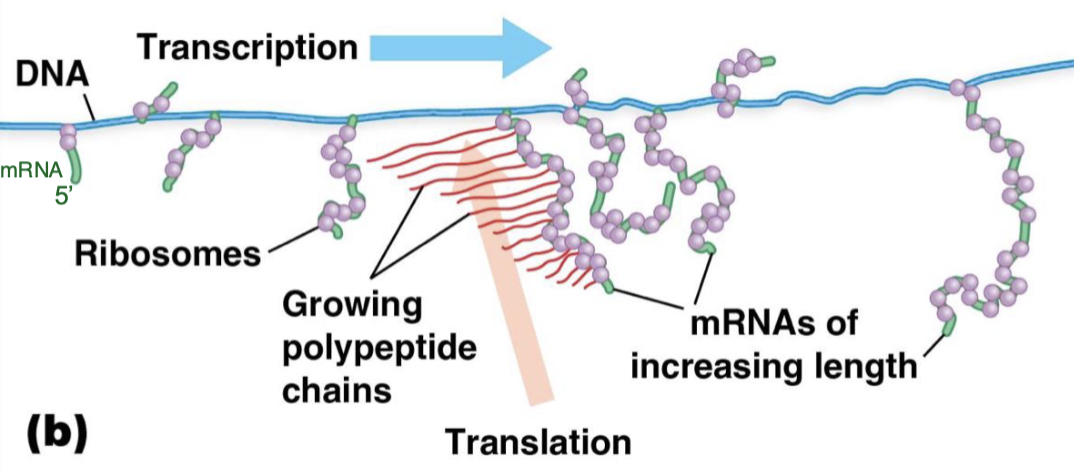
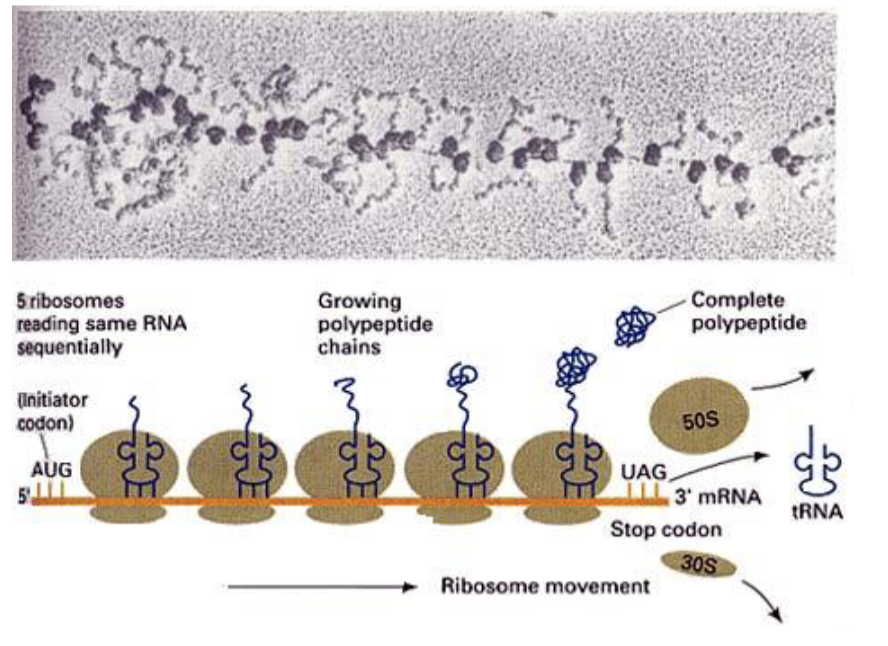
are there many ribosomes on a single mRNA?
yes —> entities are polysomes
train of ribosomes move along mRNA
researchers can isolate polysomes from cells and figure out which mRNA are being actively translated