Non-coding regulatory RNAs
1/11
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
12 Terms
Non-coding RNAs
functional RNA molecules that are trascribed from DNA, but not translated into protein
only 1-3% of the genome is translated into protein (ex. mRNA)
the rest of the transcriptome is made up of non-coding RNAs
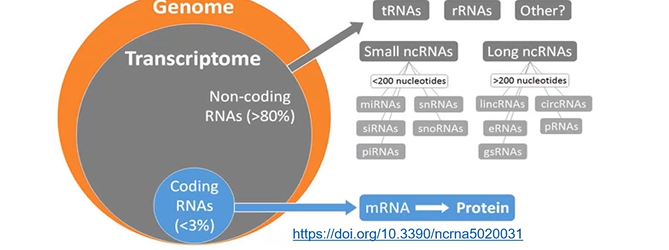
Types of non-coding RNAs
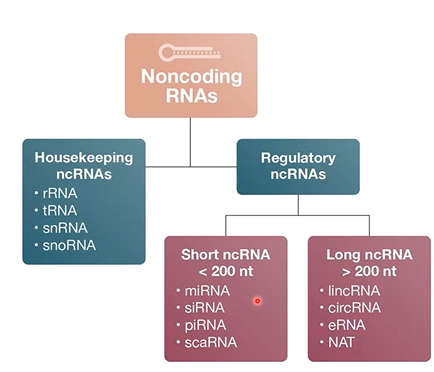
classified on function and size
housekeeping ncRNAs:
sometimes called structural ncRNAs or functional ncRNAs tend to plau functional roles in cellular processes (translation and splicing)
Regulatory ncRNAs:
regukate cellular processes (transcription, gene expression, splicing, translation)
broken into 2 categories based on size
small ncRNA (<200 nt)
long ncRNA(>200 nt)
Housekeeping small non-coding RNAs: rRNA
Ribosomal RNAs (rRNAs): interact with ribosomal proteins to make the ribosome which is critical for protein translation
majority of RNA in the cell (80%)
Biosynthesis:
transcribed in nucleolus from rDNA genes found in tandem repeats throughout the genome
modified and folded into structural RNAs that can interact with other RNAs and proteins
Housekeeping small RNAs: tRNAs
adapters that link the mRNA and the amino acid during protein synthesis (translation)
each tRNA binds a single amino acid via the acceptor stem (amino acid binding arm) and brings it to the ribosome
anticodon arn has 3 bases that are complementary to a codon mRNA
biosythesis
encoded by specific genes scattered throughout the genome
transcribed as pre-tRNAs from their own highly conserved promoters
pre-tRNAs are spliced and processed in the nucleus and then fold into a secondary structure
Housekeeping small non-coding RNAs: snRNAs
Small Nuclear RNAs: structural RNAs that are a major component of the spliceosome
the spliceosome is a large, protein-RNA complex that consists of 5 snRNAs (U1 to U6) and over 150 proteins
aid in the regulation of transcription factors and maintenance of telomeres
biosynthesis:
introns of mRNAs and liberated by splicing or transcribed from their own promoters (BOTH)
Housekeeping small non-coding RNAs: snoRNAs
small nucleolar RNAs: primarily “guide” chemical modificatons (methylations and pseudouridylation) of the other RNAs (rRNAs, tRNAs, and snRNAs)
complexes with proteins to form a complex referred to as a small nucleolar ribonucleoprotein partical (snoRNP)
acts as a guide that binds target RNA through complementary base-pairing
Biosynthesis
as intronts of mRNAs and liberated by splicing or transcribed from their own promoters (BOTH)
Housekeeping small non-coding RNAs: scaRNAs
a class of snoRNAs that localize to the Cajal body
primarily responsible for the modifications of other RNAs
some scaRNAs are precursers to other ncRNAs
Biosynthesis
most as introns of mRNAs liberated by splicing, but sime have their own promoters (BOTH KINDA)
regulatory small ncRNAs: PIWI-interacting RNAs (piRNAs)
silence transposable elements to maintain genomic instability (2 mechanisms)
no secondary structure (completely linear)
pi biogenesis mainly occurs in the germline and some somatic cells (stem cells and neurons)
Biosynthesis
synthesized as precursor piRNAs from piRNA clusters, active transposable elements, or in UTRs of other RNAs (BOTH)
piRNA precursor is cut into piRNA intermediates which are then loaded into Piwi proteins and modified to yeild primary piRNA
a secondary biogenesis pathway exists called ping pong amplification: creates a lot more of the piRNAs
regulatory small ncRNAs: MiRNAs/ siRNAs
MicroRNAs/ small interfering RNAs: regulate gene expression by binding to mRNAs and IncRNAs and either blocking translation or promoting RNA decay
siRNAs use complementary base pairing to recruit RISC to target mRNAs
if base pairing is perfect → mRNA degradation
if imperfect → suppression of translation of mRNA
one miRNA can target many genes bc can be an imperfect fit
miRNA biogeneisis
in canonical pathway, miRNA genes are transcribed from their own promotor, then are modified (capping and poly-A) and folded into hairpin structure-primary miRNA
Pri-miRNA is cleaved by Drosha and DGCR8 into pre-miRNA and then exported from the nucleus
in a non-conical pathway, gene is an intron of mRNA (called mirtron) is spliced out to form the pre-miRNA
in cytoplasm pre-mRNA is processed by Dicer into a miRNA duplex then unwound by Argonaut
one strand is loaded into RISC complex
now called a small infering RNA (siRNA)
Transposable elements (TE) and classes
repetative DNA fragments that can “jump” around the genome
if they land in or near other genes, they can mutate or alter the transcription of that gene → genomic instability
Two main types:
class 1 (retrotransposon): use “copy and paste” - transcribed into RNA → reverse transcribed into DNA → inserted into a new location
sometimes called virus-like transposon
class 2 (DNA transposon): Use “cut and paste” - transposon enzyme cuts out TE and pastes it into new location
transposons contain genes that encode the reverse transcriptase (class 1) or transposase (class 2) necessary for copy and paste or cut and paste
LncRNAs
Long non-coding RNAs: long RNAs (>200 nt) with no obvious open reading frame (ORF)
classified based on where they are made from within the genome
function:
functions both in the nucleus and the cytoplasm. can influence chromosome architecture, transcription, translation, epigenetic regulation, protein localization, and protein function
Biosynthesis
encoded by genes that typically have their own promoters
promoters are simular to protein-coding genes so IncRNAs are expressed in a developmental, cell type-specific, and context-dependent manner
processed like mRNAs
can be alternatively spliced like mRNAs
some evidence that they get translated into proteins
CircRNAs
single stranded RNA that is a closed structure because 5’ and 3’ ends covalently linked
generated by post transcriptional processing of mRNAs or IncRNAs
lack of exposed 3’ or 5’ nucleotides so are resistant to nuclease activity and very stable
Biosynthesis
generated by non-canonical splicing of mRNA and IncRNA (called backsplicing)