Biochem Fu exam 1
1/86
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
87 Terms
DNA Polymerase
enzyme- catalyzes DNA replication
RNA Polymerase
enzyme- Transcription
Hemoglobin
transport protein- transport O2 in blood
Myoglobin
transport protein- storage of oxygen
lactose permease
transport protein- transport lactose across cell membrane
collagen
structural - connective tissue
keratin
structural - hair, nails, horns ; contaminants of MS assay
myosin
protein for motion- muscle tissue ; thicker filament
actin
motion protein- muscle tissue, cell motility ; thinner filament
Luciferase
enzyme that works with cofactor Mg2+ to turn ATP (energy) and Luciferin to oxyluciferin and light. How fireflies light up.
General Structure of an amino acid:
carboxyl group, amino group, chiral center, r group. R group — (side chain) amino acids differ. Glycine is achiral, H for R group, smallest aa.
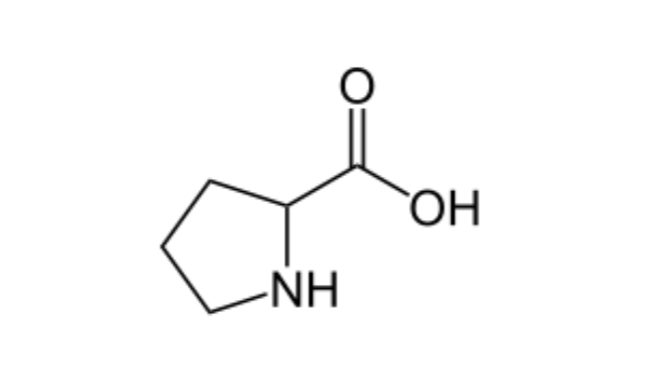
Exception to Amino acid Structure:
Proline- has a cyclic amino acid
Nonpolar Aliphatic R Groups: (7)
Glycine-G, Alanine-A Proline-P Valine-V Leucine-L, Isoleucine-I, Methionine-M
ph: 7
Aromatic R groups: (3)
Phenylalanine- F, Tyrosine-Y, Tryptophan-W
Absorb UV light at 270-280 nm
Non-polar hydrophobic amino acid
Hydrophobic: normally buried inside the protein core.
Y and W absorb UV light more than F
W=4Y
Polar Uncharged R groups (5)
Serine-S, Threonine-T, Cysteine-C, Glutamine-Q, Asparagine-N
Amino acids side chains can form hydrogen bonds.
soluble in water
Cysteine can form disulfide bonds
Positively Charged R Groups (3)
Lysine-K, Arginine-R, Histidine-H
Histidine has imidazole group
Ampholytes
substance with dual acid and base nature
phosphorylation
most common type of regulatory modification- involved in protein activity
Peptide Bonds
small condensation products of amino acids
small compared to proteins
Mw < 10 kDa
Peptide ends (not same)
Amino-terminal end and carboxyl terminal end. Name/Number starting with amino terminus.
Average molecular weight of amino acids
138da
smaller amino acids predominant in proteins: 128da
Protein Extraction
grinding w/wo liquid nitrogen
grinding with sand
bead beater
sonication
french press
buffer w/wo mild detergent
protein precipitation
ammonium sulfate- add saturated solution to sample with buffer and protein will precipitate out
dialysis
selectively permeable membrane- large enzyme molecules cannot pass through pores in membrane
small molecules pass through and equilateral across membrane
SDS Page
SDS- sodium dodecyl phosphate-detergent- binds and unfolds all proteins, gives uniform negative charge no matter shape, rate of movement will depend on size, so smaller molecules will move faster.'
The bound SDS will contributes a negative charge, unmask the intrinsic charge of the protein. Therefore, all protein will have a similar charge to mass ratio. Migration purely based on the molecular weight.
Protein Purification- metal binding
Purification of his-tagged protein.
lyse bacteria cells to release
protein- incubate with nickel NTA agarose beads- wash with salt solution-elute using imidazole competitor- sds page
Fragmenting Polypeptide Chains: Trypsin
trypsin predominantly cleaves proteins at the carboxyl side (or "C-terminal side") of the amino acids lysine and arginine except when either is bound to a C-terminal proline, although large-scale mass spectrometry data suggest cleavage occurs even with proline.
Fragmenting Polypeptide Chains- Submaxillary protease
Cleaves Agr- c terminus
Chymotrypsin
Cleaves Phe,Trp,Tyr- Cterminus
globular protein
S aureus
cleaves Asp, Glu- c terminus
Asp-N-Protease
cleaves Asp, Glu- N terminus
Endoprotease lys C
cleaves lys- c terminus
Cyanogen bromide
cleaves methionine- c terminus
Levels of structure in proteins
Primary, secondary tertiary, quaternary
Primary structure
amino acid residues- sequence, peptide bond, and disulfide bond
polypeptide is made up of a series of linked planes at a carbons
secondary structure
local spatial arrangement of the peptide backbone
two common: a helix, b sheet
irregular arrangement is called random coil
a helix
stabilized by hydrogen bonds between nearby residues.
Helical backbone is held together by hydrogen bonds between the backbone amides of an n and n+4 amino acids
Right-handed helix with 3.6 residues (5.4 Å) per turn
Peptide bonds are aligned roughly parallel with the helical axis
Side chains point out and are roughly perpendicular with the helical axis
too small to fit anything "inside"
happens to fit well in the major groove of dsDNA
Affects on a helix stability
Not all polypeptide sequences adopt a-helical structures
Small hydrophobic residues such as Ala and Leu are strong helix formers
Pro acts as a helix breaker because the rotation around the N-Ca bond is impossible
Gly acts as a helix breaker because the tiny R-group supports other conformations
Attractive or repulsive interactions between side chains 3-4 amino acids apart will affect formation
b sheet
stabilized by hydrogen bonds between adjacent segments that may not be near by.
Parallel or antiparallel orientation of two chains within a sheet are possible
In parallel b sheets the H-bonded strands run in the same direction
Resulting in bent H-bonds (weaker)
In antiparallel b sheets the H-bonded strands run in opposite directions
Resulting in linear H-bonds (stronger)
silk fibroin
main protein in silk for moths and spiders
antiparallel B sheet structure
small side chains (Ala + Gly)= close packing of sheets
structure stabilized by
hydrogen bonding within sheets
and London dispersion interactions between sheets
spider silk is extremly strong, composite material-crystalline and rubber like parts
X-ray crystallography
Measure the locations and intensities of spots produced by a beam of X-ray
Steps needed
Purify the protein
Crystallize the protein
Collect diffraction data
Calculate electron density
Fit residues into density
Pros
No size limits
Well-established
Cons
Difficult for membrane proteins
Cannot see hydrogens
Biomolecular NMR
Nucleic magnetic resonance: manifestation of nuclear spin angular momentum.
Static magnetic field is applied: nuclear spin generates dipoles
Steps needed
Purify the protein
Dissolve the protein
Collect NMR data
Assign NMR signals
Calculate the structure
Pros
No need to crystallize the protein
Can see many hydrogens
Cons
Difficult for insoluble proteins
Works best with small proteins
denaturation
Loss of structural integrity with accompanying loss of activity
Proteins can be denatured by:
heat or cold
pH extremes
organic solvents
chaotropic agents: urea and guanidinium hydrochloride
Ribonuclease refolding experiment
Ribonuclease is a small protein that contains 8 cysteines linked via four disulfide bonds
Urea in the presence of 2-mercaptoethanol fully denatures ribonuclease
When urea and 2-mercaptoethanol are removed How, the protein spontaneously refolds, and the correct disulfide bonds are reformed
The sequence alone determines the native conformation
Quite "simple" experiment, but so important it earned Chris Anfinsen the 1972 Chemistry Nobel Prize
Chaperone Proteins prevent misfolding
1. prevent aggregation of unfolded peptides, Hsp70 bind to regions of unfolded peptide that are rich in hydrophobic residues to protect from denaturation from heat and
2. Prevent new peptide being synthesized
3. Block the folding of certain proteins until they are translocated across the membrane
4. Facilitate quaternary assembly
protein misfolding
basis of numerous human diseases
Amyloidoses: Alzheimer disease, Huntington disease, Parkinson diseases
Proteolytic cleavage of this larger protein leaves the relatively unstable amyloid-β peptide, which loses its α-helical structure. It can then assemble slowly into amyloid fibrils
Formation of disease-causing amyloid fibrils.
The aromatic side chains shown here play a significant role in stabilizing the amyloid structure.
Protein Binding
interaction strength can be expressed as Ka (units M^-1) or Kd (units M) Kd=1/Ka
Strong binding- Kd<10nM
weak binding - Kd> 10uM
Lock and key model
model by Emil fisher- assumes complementary surfaces are preformed
High specificity
proteins typically have it and only bind to certain ligands; explained by the complementary of the binding site and the ligand
Complementary in (high specificity)
size, shape, charge, or hydrophobic/hydrophilic character
Induced fit
Conformational changes may occur upon ligand binding
Induced fit allows for tighter binding of the ligand
Induced fit allows for high affinity for different ligands
Both the ligand and the protein can change their conformations
myoglobin- Heme binding to protein. The bound O2 is hydrogen-bonded to the distal His, His E7 (His64), further facilitating the binding of O2.
Allosteric Protein
Binding of a ligand to one site affects the binding properties of a different site, on the same protein
Can be positive or negative
homotrophic
Normal ligand of the protein is the allosteric regulator
heterotrophic
Different ligand affects binding of the normal ligand
cooperativity
positive homotropic regulation
hemoglobin- is a tetramer of two subunits (a2b2)
Each subunit is similar to myoglobin
hemoglobin T state
T=tense state
more interactions, more stable
lower affinity for oxygen.
oxygen binding triggers a conformational change from T to R
hemoglobin R state
R=relaxed state
fewer interactions, more flexible
higher affinity for O2
T-R conformational change
O2 binding triggers conformational change
involves breaking ion pairs between the α1-b2 interface
pH effect on O2 Binding to hemoglobin(Hb)
Actively metabolizing tissues generate H+, lowering the pH of the blood near the tissues relative to the lungs
Hb Affinity for oxygen depends on the pH
H+ binds to Hb and stabilizes the T state
Protonates His146 which then forms a salt bridge with Asp94
Leads to the release of O2 (in the tissues)
Bohr Effect
The pH difference between lungs and metabolic tissues increases efficiency of the O2 transport
2,3-Bisphosphoglycerate regulates O2 binding
Negative heterotropic regulator of Hb function
Present at mM concentrations in erythrocytes
Produced from an intermediate in glycolysis
Small negatively charged molecule,
binds to the positively charged
central cavity of Hb
Stabilizes the T states
sickle cell anemia
due to a mutation in hemoglobin
Glu6 --Val in the B chain of Hb
The new Valine side chain can bind to a different Hb molecule to form a strand
This sickles the red blood cells
Untreated homozygous individuals generally die in childhood
Heterozygous individuals exhibit a resistance to malaria
Formation of Hb strands
deoxyhemoglobin S has a hydrophobic patch on its surface, which causes the molecules to aggregate into strands that align into insoluble fibers.
Anemia is a condition in which you don't have enough healthy red blood cells to carry adequate oxygen to your tissues.
enzymes
Enzymes are catalysts
Increase reaction rates without being used up
Most enzymes are globular proteins
However, some RNA (ribozymes and ribosomal RNA) also catalyze reactions
Study of enzymatic processes is the oldest field of biochemistry, dating back to late 1700s
Study of enzymes has dominated biochemistry in the past and continues to do so
Holoenzyme:
complete function including coenzyme and metal ions
Apoenzyme:
protein part of the enzyme
Enzyme Substrate complex
enzmyes act by binding substrates
michaelis complex
The noncovalent enzyme substrate complex
Enzymatic Catalysis
Enzymes do not affect equilibrium (ΔG)
Slow reactions face significant activation barriers (ΔG‡) that must be surmounted during the reaction
Enzymes increase reaction rates (k) by decreasing ΔG‡
oxidoreductases
transfer of electrons (hydride ions or H atoms)
transferases
group transfer reactions
hydrolases
hydrolysis reactions (transfer of functional groups to water)
lysases
cleavage of c-o,c,n bonds, or other bonds by elimination, leaving double bonds or rings, or addition of groups to double bonds
isomrases
transfer of groups within molecules to yeild isomeric forms
ligases
formation of c-c,o,n,s bonds via condensation reactions coupled to cleavage of ATP or similar cofactor
Nonlinear Michaelis-Menten plot
should be used to calculate parameters Km and Vmax
Linearized double-reciprocal plot
good for analysis of two-substrate data or inhibition
enzyme inhibitors
compounds that decrease enzyme’s activity
irreversible inhibitors (enzyme inhibition)
React with the enzyme
One inhibitor molecule can permanently shut off one enzyme molecule
They are often powerful toxins but also may be used as drugs
Reversible inhibitors (enzyme inhibition)
bind to and can dissociate from the enzyme
They are often structural analogs of substrates or products
They are often used as drugs to slow down a specific enzyme
Reversible inhibitor binding (enzyme inhibition)
to the free enzyme and prevent the binding of the substrate
to the enzyme-substrate complex and prevent the reaction
competitive inhibition
Competes with substrate for binding
Binds active site
Does not affect catalysis
No change in Vmax; apparent increase in KM
Lineweaver-Burk: lines intersect at the y-axis
uncompetitive inhibition
Uncompetitive inhibitors bind at separate site, but bind only to the ES complex
mixed inhibition
Binds enzyme with or without substrate
Binds to regulatory site
Inhibits both substrate binding and catalysis
Decrease in Vmax; apparent change in KM
Lineweaver-Burk: lines intersect left from the y-axis
Noncompetitive inhibitors are mixed inhibitors such that there is no change in KM
hexokinase
undergoes induced fit on substrate binding
u shape
Catalytic active form
The ends pinch towards each other after binding of D-glucose.
acid base catalysis
Charged intermediates are stabilized by transfer of protons to or from the substrate or intermediate to form a species that breaks down more readily to products.
H3O+ (specific acid catalysis) or HA (general acid catalysis)
covalent catalysis
A transient covalent bond between the enzyme and the substrate
Changes the reaction Pathway
Requires a nucleophile on the enzyme
Can be a reactive serine, thiolate, amine, or carboxylate