MBIO 2700 / Topic 5a: Proteins
1/41
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
42 Terms
What is a peptide bond?
A special name to the amide bond covalently joining two AAs by condensation of the amino group on one AA and the carboxyl group of another AA.
Differentiate the following: peptides, polypeptides, and proteins.
• Peptides: Short chains of AAs.
• Polypeptides: Long chains of AAs.
• Proteins: Aggregates of polypeptides.
What are the two components of a polypeptide?
• Backbone: Polypeptide chain w/o R groups.
• Side chains: R groups only.
• What are the pKa and pI of the chains and backbone termini similar to?
• Which is the only ionizable stuff in the backbone?
• pKa and pI of free AAs.
• The termini.
+ When covalent peptide bonds are formed, α-amino and α-carboxyl groups of each AA comprising the peptide are now non-ionizable.
• What are the AA units in a peptide or protein also called?
• How is the amino acid sequence of peptides and proteins written?
• AA units are called residues.
• Sequence is written as N-terminus beginning on the left and C-terminus on the right.
Protein folding contributes to an increase in ______ and decrease of __, contributing overall to disorder and spontaneity.
Protein folding contributes to an increase in entropy and decrease of ∆G, contributing overall to disorder and spontaneity.
Polypeptide chains are very _______, so the way they fold to form a 3D structure is essential to understanding how proteins are assembled.
Polypeptide chains are very flexible, so the way they fold to form a 3D structure (that we will soon known as the structure of a protein) is essential to understanding how proteins are made.
What bonds in a polypeptide makes polypeptides so flexible? What bonds do not make it flexible?
Flexible via:
• α-carbon and α-hydrogen.
• α-carbon and R.
• α-carbon and carboxyl.
• α-carbon and amino.
Inflexible via:
• Carbonyl carbon and amine.
Why is the bond between the carbonyl carbon and amine inflexible?
> N lone pair delocalizes into carbonyl.
> Resonance between C=O and C-N.
> C-N peptide bond = shorter, stiffer, and planar → no free rotation like single bond.
Do you expect proteins to be localized to one part of a body?
> Proteins made every single second of day and moves from one place to another in seconds.
> No.
What are the seven primary functions of a protein?
Catalysis, transport, structure, contractile, nutrient, defence, and regulatory.
Are proteins usually small? Large? Light? Heavy?
Can be small to large and light to heavy.
+ Very diverse.
Differentiate the following: glycoproteins, lipoproteins, cofactors, prosthetic groups, and coenzymes.
• Glycoproteins: Proteins conjugated with sugars.
• Lipoproteins: Proteins conjugated with lipids.
• Cofactors: Non-protein molecules that assist in protein activity.
• Prosthetic groups: Cofactors that are permanently bound to the protein.
• Coenzymes: Cofactors for enzymes specifically.
+ Cofactors an be organic (e.g. vitamins) or inorganic (e.g. metal ions). Can be covalently bonded to protein or non-covalently bonded to protein.
Differentiate the four levels of protein structure.
• Primary structure: Linear sequence of AAs.
• Secondary structure: Atoms (on the backbone of a single, same polypeptide) bond via δ+ and δ- in an intermolecular-bonding, intramolecular-appearing fashion, folding the polypeptide.
• Tertiary structure: Atoms (on the side chains of the folded polypeptide) bond in a variety of intramolecular and intermolecular fashions, crumpling the now globular polypeptide.
• Quaternary structure: Two or more tertiary structures form a protein.
When it comes to protein folding, in what way should you take into account sterics and entropy?
You have to find a way to fold a protein that decreases steric hinderance but at the same time increases disorder.
On a 1º structure, where does the folding happen between 1º and 2º? Why does it happen in these places?
Hydrogen bonding. (Shown in picture)
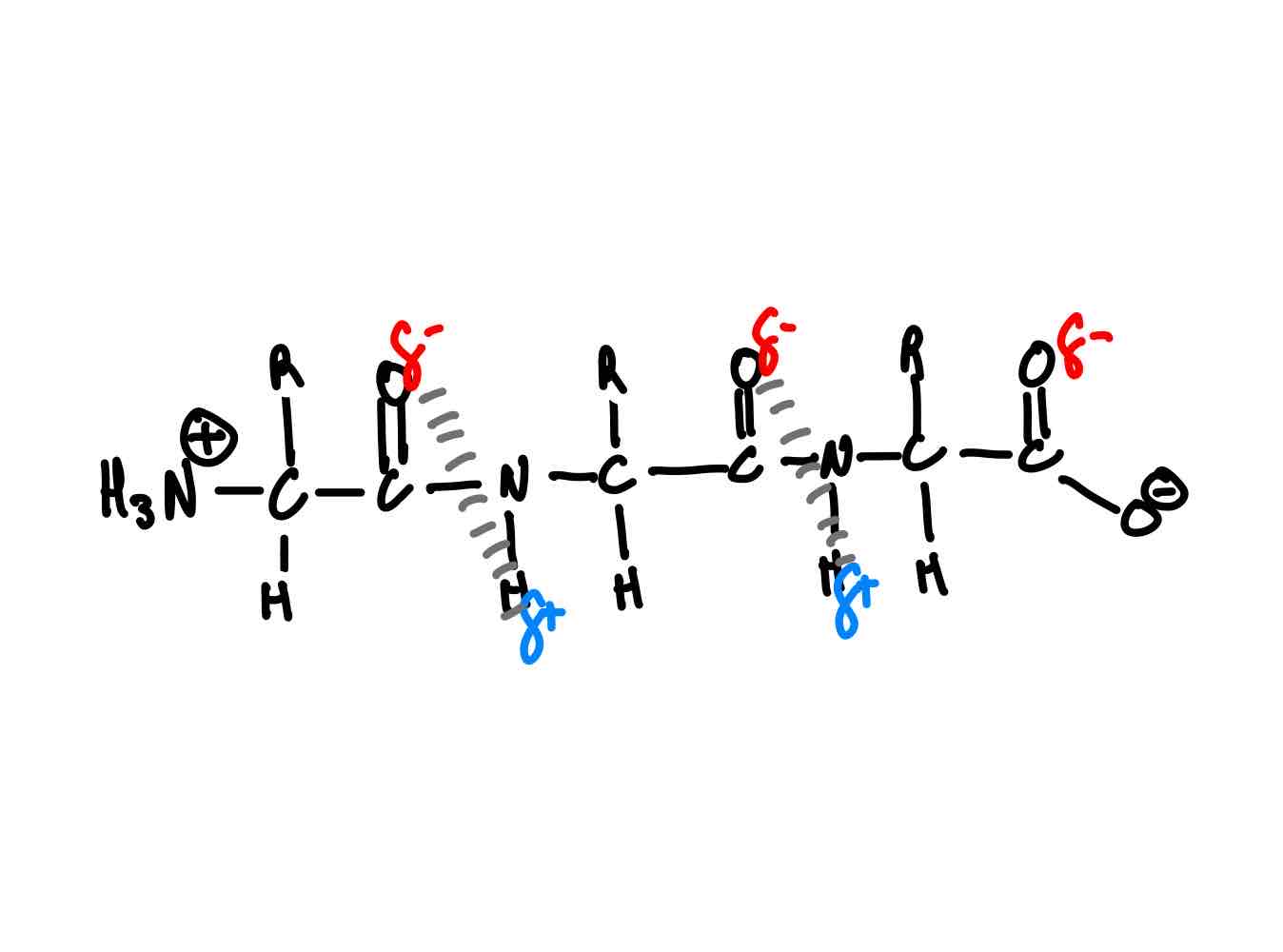
> What are you specifically trying to maximize in secondary folding?
> Why can’t you make too many 2º structure H-bonds?
> You’re trying to max out on H-bonds.
> More H-bonds → more twisting → more strain.
• What are the two main types of 2º structure?
• Can you link these two main types of 2º structures? How so, if ever?
> The two types of 2º are α-helix + β-sheet.
> They are linkable via linkers. α-helix-linker-β-sheet.
What’s the one AA that cannot fold to a 2º?
> Two H-atoms in H2N+ will be used for peptide condensation.
> No H-atoms for H-bonding → no folding to 2º.
In the folding between 1º and 2º, what is the relationship between number of hydrogen bonds, disorder, free energy, and enthalpy?
↑ Hydrogen bonds ↑ Disorder ↓ Free energy (G) ↓ Enthalpy (H)
• In one turn of an α-helix, how many residues are there on average?
• How long is one turn, typically?
• Residues: 3.6 residues.
• Turn: 5.4 Å (= 1E-8cm).
In an α-helix backbone loop, why do the side chains protrude in space?
> If side chains pointed inward/straight down axis → bump into each other → clashes.
> If side chains protrude out in space → minimize spatial interference w/ each other.
If there are high concentrations of charged, bulky, or small residues on an α-helix, what will happen?
Helix disruption.
Across a peptide bond, why must α-carbons be trans to each other?
Minimize steric clashing.
What are the two defining characteristics of an α-helix?
• H-bonds form between every fourth amino acid.
• Side chains tend to point away from the helix.
What are the four disrupting characteristics of an α-helix?
• Electronics clash (e.g. two negatively charged groups).
• Sterics clash (e.g. two bulky groups).
• Small residues wanting to convert to β-conformation.
• Proline due to its (a) inability to form hydrogen bonds + (b) nitrogen’s (comprising the amide) lack of planarity (affecting flexibility).
What is the defining characteristic of a β-sheet?
β-sheets form as overlapping segments of the polypeptide chain when there are side chains spaced out evenly so as to not clash with each other and allow for H-bonds.
Differentiate each way a β-sheet can exist.
• Parallel: Overlapping segments all run in the same direction from C-terminus to N-terminus.
• Antiparallel: Overlapping segments run in different directions, with one sheet going from the C- to the N-terminus, and the adjacent sheet going N- to C- and so on.
Why are H-bonds in one category of β-sheets more stable than in the other?
> Antiparallel sheets have more linear H-bonds → more energy released → lower in energy in the end + stabilized.
> Parallel sheets have crooked H-bonds → higher in energy in the end → weaker + destabilized.
What are the usual side chains that make up β-sheets?
Gly, Ala, and Ser.
What happens when a polypeptide with α-helices is heated then cooled?
> Heating disrupts H-bonds in α-helices → unfolding α-helices.
> Upon cooling, unfolded polypeptide refold into β-sheets.
+ This is temporary. β-sheets will revert back to α-helices The formation of α-helices is a spontaneous and entropically stable reaction. The disruption and conversion of α-helices into β-sheets is the opposite, hence why we need to put in energy - heat.
> What if you had a tertiary structure with a ton of residues containing non-polar side chains? Hint: Where will these residues go within the space of a tertiary structure?
> What if you had a tertiary structure with a ton of residues containing polar side chains? Hint: Where will these residues go within the space of a tertiary structure?
> From lectures past, what is this event called?
> Non-polar side chains buried in protein interior.
> Polar side chains exposed on protein exterior.
> Hydrophobic effect.
What are electrostatic interactions within protein structures?
• Ionic bonds between acidic + basic residues.
• H-bonds between residues w/ partial positive + partial negative ends.
What are the four defining characteristics of a tertiary structure? How does each keep the protein happy?
• Hydrophobic effect: Favourable hydrophilic and hydrophobic interactions.
• Disulphide bonds: Maintenance of globular structure.
• Electrostatic interactions: Maintenance of globular structure.
• Metal chelation: Two functional groups that hate each other together by a mediator - the metal.
• What happens when oxidizing agents are present around cysteine?
• What happens when the pH of the solution is greater than the pKa of cysteine?
• Oxidizes thiol → disulphide bonds are possible.
• pH of solution deprotonates thiols → negatively charged “thiols” → clashing bec of negative charges.
What are the two defining characteristics of 3º structures?
> 3º structures form to minimize unfavourable interactions + maximize favourable ones.
> Compact w/ few cavities + small amt of H2O.
What are the factors that put together a quaternary structure?
The same R-group interactions present in tertiary structures.
On a quaternary structure, do you expect full positives and full negatives to be specifically meant for each other?
Nope.
What are the four different states of a protein? Differentiate each.
• Folding intermediate: Partially folded structure.
• Aggregates: Misfolded, clumpy structure.
• Native proteins: Folded, functional, and correct structure.
• Chaperone proteins: Specialized proteins assisting other proteins in folding correctly, preventing misfolding and aggregation.
+ An aggregate often leads to loss of protein function and diseases.
What is protein denaturation?
Folded/native form → unfolded form.
Do you expect protein denaturation to be costly, energy-wise?
> ∆G between native and unfolded form = ∆G = ~5–10kcal/mol.
> Change in solvent/apply heat → easy denature.
> No, not costly.
• Do you expect denatured proteins to refold spontaneously?
• Do you expect denatured proteins to stay in its denatured appearance? Or does it do something spontaneously?
• What could expect to be great protein denaturants, keeping proteins in denatured form?
• Happens rarely in few, wherein chaperones are usually required.
• Most denatured proteins, other than the rare case of refolding, aggregate together and precipitate.
• High concentrations of solutes that disrupt the H-bonding system of water, e.g. 8M urea, 6M guanidine hydrochloride