Genetics Ch. 12-14🧬
0.0(0)
Card Sorting
1/191
Earn XP
Description and Tags
Last updated 2:51 AM on 11/11/22
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
192 Terms
1
New cards
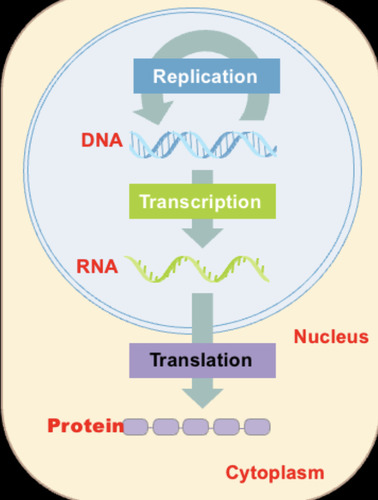
Central Dogma of Biology
DNA -> RNA -> Protein
-DNA is the genetic material. DNA is replicated, transcribed into RNA, and translated into protein
-DNA is the genetic material. DNA is replicated, transcribed into RNA, and translated into protein
2
New cards
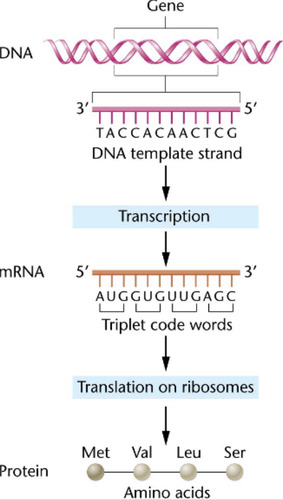
Transcription
DNA is used as a template for the creation of RNA using RNA polymerase.
3
New cards
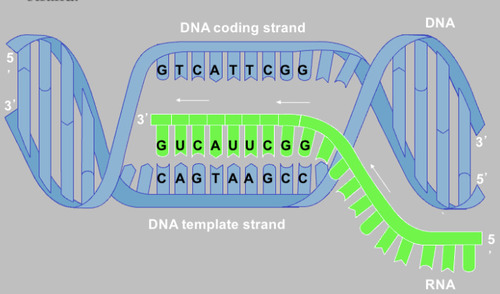
U
The new RNA strand, made during transcription, is formed by incorporating nucleotides that are complementary to the template strand.
So, we the RNA strand is therefore identical to the coding strand, except that the RNA has ________ instead of T.
So, we the RNA strand is therefore identical to the coding strand, except that the RNA has ________ instead of T.
4
New cards
Coding strand
the strand of DNA that is not used for transcription and is identical in sequence to mRNA, except it contains uracil instead of thymine
5
New cards
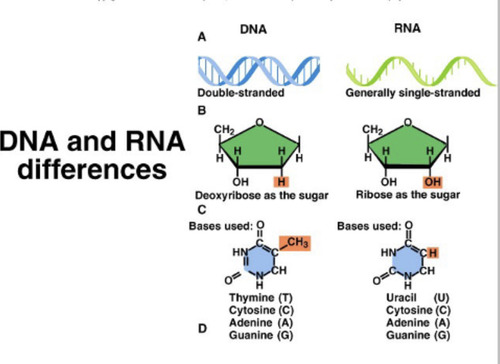
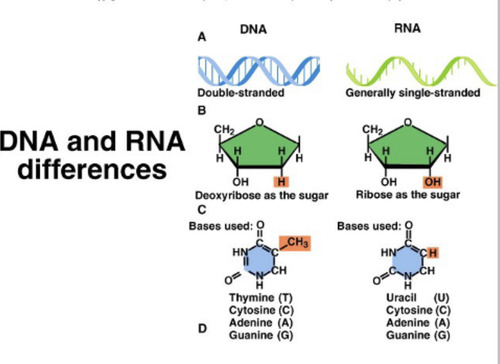
DNA
Double stranded, has deoxyribose as the sugar, bases are ATCG
6
New cards
RNA
Single stranded, has ribose as the sugar, bases used are AUCG
7
New cards
Characteristics of the Genetic Code
-DNA template serves as the template for mRNA
-codon (3 mRNA bases)
-unambiguous: each codon codes for 1 amino acid
-degenerate (more than one codon codes for an amino acid)
-start/ stop codon
-commaless -no breaks
- is non-overlapping
-universality of DNA
-codon (3 mRNA bases)
-unambiguous: each codon codes for 1 amino acid
-degenerate (more than one codon codes for an amino acid)
-start/ stop codon
-commaless -no breaks
- is non-overlapping
-universality of DNA
8
New cards
Triplet Nature of Genetic Code
3 mRNA bases= 1 codon= 1 amino acid
9
New cards
64
What's the correspondence between the mRNA nucleotides and the amino acids of the protein?
-Because DNA consists of 4 different bases and there are 3 bases in a codon... 4^3= ________ possible combinations of codon.
-Proteins are formed from 20 amino acids in humans.
-There are 64 different codons in humans. Of the 64, 61 represent amino acids and the remaining three represent stop codons. (Stop codons don't code for a protein!)
-Because DNA consists of 4 different bases and there are 3 bases in a codon... 4^3= ________ possible combinations of codon.
-Proteins are formed from 20 amino acids in humans.
-There are 64 different codons in humans. Of the 64, 61 represent amino acids and the remaining three represent stop codons. (Stop codons don't code for a protein!)
10
New cards
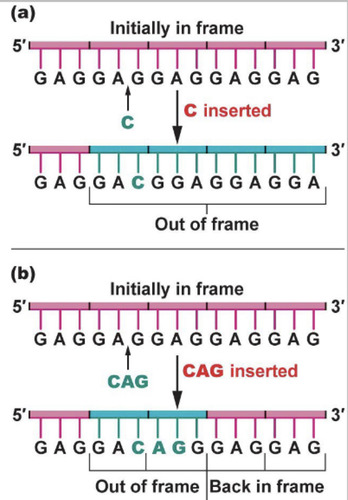
Reading Frame
Every mRNA has this where the DNA sequence/RNA sequence is read 3 base pairs at a time. The start codon determines it.
11
New cards
frameshift mutation
mutation that shifts the "reading" frame of the genetic message by inserting or deleting a nucleotide
12
New cards
The reading frame will not be shifted. A new amino acid will just be added.
A codon of three nucleotides in the reading frame determines the amino acid that's added to the polypeptide chain.
-If 1 or 2 base pairs are added, the reading frame will be messed up.
-If 3 base pairs are added, what will happen?
-If 1 or 2 base pairs are added, the reading frame will be messed up.
-If 3 base pairs are added, what will happen?

13
New cards
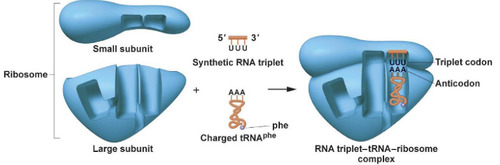
Anticodon
RNA is translated into protein on the ribosome (small subunit and large subunit). tRNA carries the amino acids to the ribosome during translation. Each codon pairs with its complementary sequence: its _____________ on the tRNA
14
New cards
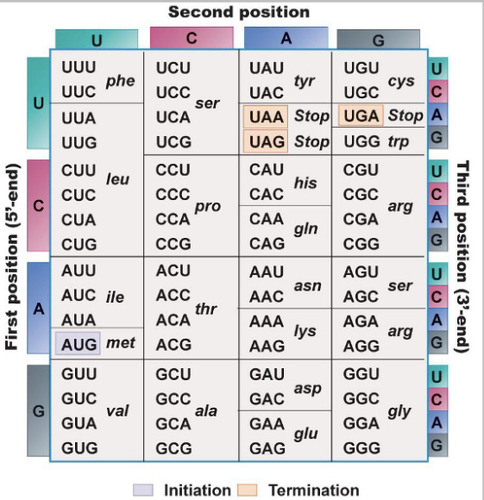
UGA, UAA, UAG
Out of 64 possible codons, 61 code for an amino acid. The other 3 are stop codons. What are the 3 stop codons?
15
New cards
Termination codon
One of the three codons (UAA, UAG, UGA) that signal the termination of translation of a polypeptide.
16
New cards
Initiator codon
The codon for methionine (AUG) that signals the start of translation in a mRNA.
17
New cards
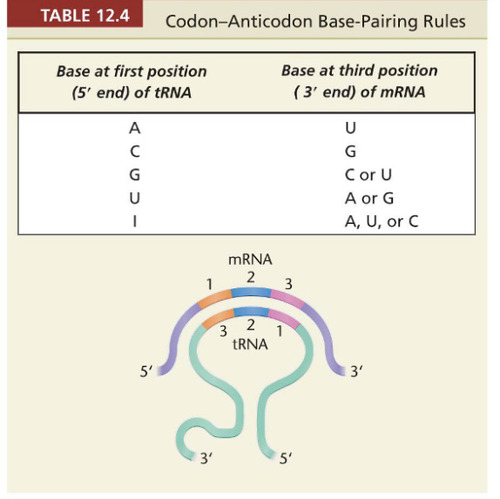
Wobble Hypothesis
the hypothesis that says normal base pairing can occur between nitrogenous bases in positions 1 and 2 of the codon and corresponding bases 2 and 3 in the anticodon. States the degeneracy of the genetic code.
-read in 5' to 3' direction
-complementary base pair rules are not strict with the tRNA
-can be various codons that can pair with an anticodon because the 3rd position of the codon is not strict
-read in 5' to 3' direction
-complementary base pair rules are not strict with the tRNA
-can be various codons that can pair with an anticodon because the 3rd position of the codon is not strict
18
New cards
Nonsense mutation
Mutation that results in a premature stop in translation
19
New cards
Degenerate
The genetic code is __________, meaning different codons can code for the same amino acid.
Ex: GGU, GGC, GGA, and GGG all encode glycine.
-is mostly at the third base of the codon
Ex: GGU, GGC, GGA, and GGG all encode glycine.
-is mostly at the third base of the codon
20
New cards
Commaless
The genetic code is __________, meaning that once the reading frame starts, it does not break until a stop codon is reached.
21
New cards
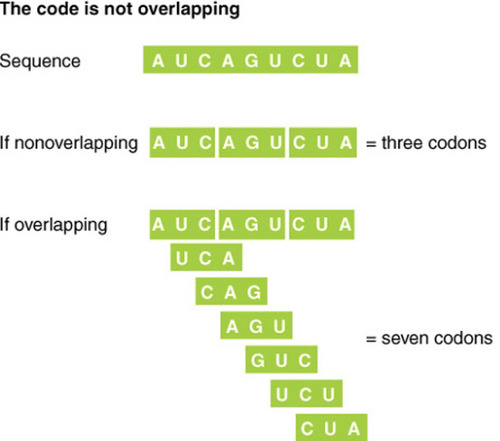
Non-overlapping
Each base appears in only one triplet - each base is only read once in the reading frame 3bp at a time.
22
New cards
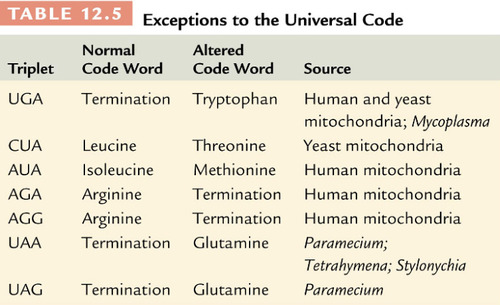
Universal
The genetic code is _____________~ is the same among viruses, bacteria, and eukaryotes (all have the same amino acids and 64 possible codons).
-this is why recombinant DNA technology works (taking DNA from 2 different sources and combining them).
-exceptions to this are found in mitochondria
-this is why recombinant DNA technology works (taking DNA from 2 different sources and combining them).
-exceptions to this are found in mitochondria
23
New cards
Overlapping
There can be one mRNA coding for different genes because it has a different reading frame. Some portions of a gene may be read in various reading frames; common characteristic of viruses.
-this is different than saying the genetic code is nonoverlapping
-this is different than saying the genetic code is nonoverlapping
24
New cards
RNA polymerase
Transcription is mediated by this enzyme. It separates the DNA and creates RNA without any primers needed first. It synthesizes RNA, creating Phosphodiester bonds.
-Requires nucleoside triphosphate and the DNA template.
-it cleaves two of the phosphates to create a nucleoside monophosphate
n(NTP)—> (NMP)n + n(Ppi)
-Requires nucleoside triphosphate and the DNA template.
-it cleaves two of the phosphates to create a nucleoside monophosphate
n(NTP)—> (NMP)n + n(Ppi)
25
New cards
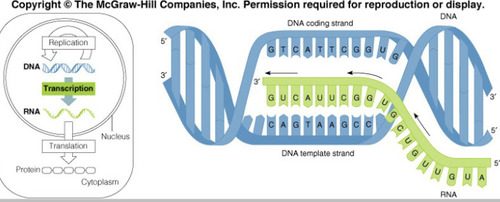
Template strand
This DNA strand is used to synthesize RNA. Other DNA strand is referred to as the DNA coding strand/ partner strand.
26
New cards
3
Prokaryotes use 1 RNA polymerase during translation. How many do eukaryotes use?
27
New cards
Template Binding
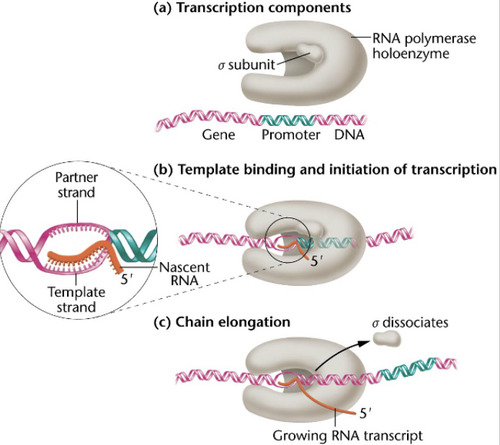
Sigma subunit recognizes and binds RNA polymerase to the promoter region during start of transcription.
-sigma subunit recognizes promoter regions on the DNA that allow RNA polymerase to bind
-After RNA polymerase binds, the sigma subunit can dissociate because it's function is no longer needed
-sigma subunit recognizes promoter regions on the DNA that allow RNA polymerase to bind
-After RNA polymerase binds, the sigma subunit can dissociate because it's function is no longer needed
28
New cards
Promoter sequence
Specific DNA sequence essential for the initiation of RNA transcription.
-is a consensus sequence
-in bacterial genome: there's a sequence called Pribnow box (TATAAT) 10 BP from the start of transcription. There's also a region 35 nucleotides from the end (35 region).
-is a consensus sequence
-in bacterial genome: there's a sequence called Pribnow box (TATAAT) 10 BP from the start of transcription. There's also a region 35 nucleotides from the end (35 region).
29
New cards
cis-acting elements
Sequences or elements that are part of the gene on the same side/ are adjacent parts of the DNA molecule
30
New cards
3 steps of transcription
1. Initiation- control of transcription is regulated by transcription factors -no primer needed
2. Elongation- RNA polymerase adds nucleotides to the growing RNA
3. Termination- sequences in the DNA prompt the RNA polymerase to fall off, ending the transcript
2. Elongation- RNA polymerase adds nucleotides to the growing RNA
3. Termination- sequences in the DNA prompt the RNA polymerase to fall off, ending the transcript
31
New cards
termination factor rho
large hexameric protein that physically interacts with the growing RNA transcript to terminate transcription. It physically stops the chain from growing, and the RNA molecule releases from the DNA template.
32
New cards
Cistrons
The bacterial genome is clustered into genes of similar function, which helps bacterial cell be proficient in making proteins. These genes are called...
33
New cards
polycistronic mRNA
One large RNA molecule in bacteria that encodes for more than one/multiple proteins. It transcribes the whole cluster of genes with a similar function.
34
New cards
Transcription in Eukaryotes
-occurs in the nucleus with 3 forms of RNA polymerases
-promoters and enhancers have more complex interactions
-more processing of mRNA / nNRNA before translation occurs
-After, pre-mRNA is produced. Then, processing (splicing) needs to occur.
-promoters and enhancers have more complex interactions
-more processing of mRNA / nNRNA before translation occurs
-After, pre-mRNA is produced. Then, processing (splicing) needs to occur.
35
New cards
cis-acting elements
can only influence expression of adjacent genes on the same DNA molecule.
Ex: promoters: TATA box (35 nucleotides upstream) and CAATT box (-80)
Enhancers: located upstream, downstream, or within
Ex: promoters: TATA box (35 nucleotides upstream) and CAATT box (-80)
Enhancers: located upstream, downstream, or within
36
New cards
Promoters
region of DNA that indicates to an enzyme where to bind to make RNA. TATA box
37
New cards
Enhancers
A DNA sequence that aides in allowing RNA polymerase to bind to the promoter region and recognize there is a gene to be transcribed.
38
New cards
trans-acting elements
regulatory proteins that bind to such DNA sequences to act on the DNA, like transcription factors.
39
New cards
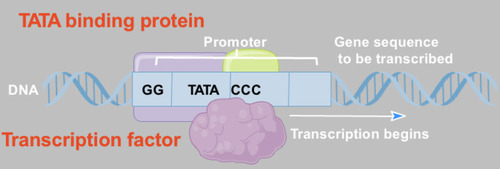
Transcription factors
Proteins that regulate the initiation of transcription. They signal for where RNA polymerase must bind.
Ex: TATA-binding protein (TBP): binds to the TATA box
Ex: TATA-binding protein (TBP): binds to the TATA box
40
New cards
Initiation of Transcription in Eukaryotes
-begins upstream of the gene in a region called the promoter
-the promoter recruits TATA protein , a DNA binding protein, which in turn recruits other proteins
-transcription factors are recruited
-when a complete transcription complex is formed, RNA polymerase binds and transcription begins
-the promoter recruits TATA protein , a DNA binding protein, which in turn recruits other proteins
-transcription factors are recruited
-when a complete transcription complex is formed, RNA polymerase binds and transcription begins
41
New cards
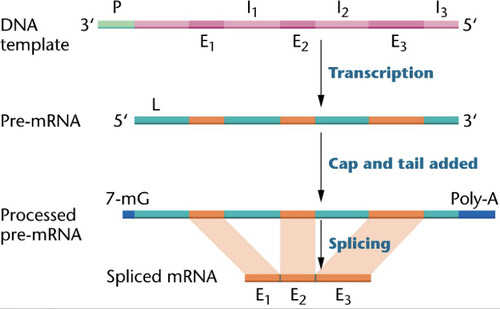
post-transcriptional modification of RNA
This happens in eukaryotes to prevent the degradation of mRNA as it gets sent out of the nucleus and into the cytoplasm for translation. mRNA transcripts are modified before use as a template for translation
-addition of capping nucleotide at the 5' end
-addition of poly A tail to 3' end
-splicing occurs, removing introns
-addition of capping nucleotide at the 5' end
-addition of poly A tail to 3' end
-splicing occurs, removing introns
42
New cards
Addition of capping nucleotide at 5' end
After transcription, a 7-methylguanosine (7mG) is added to the 5' end for protection for nuclease activity and to facilitate transporting the transcript out of the nucleus.
43
New cards
Addition of poly A tail to 3' end
After transcription, this 200 nucleotide long chain is added to the 3' end of mRNA to stabilize against degradation. It cleaves the initial end of the transcript (AAUAAA) and then adds the polyAtail.
44
New cards
Splicing
the process of removing introns and reconnecting exons in a pre-mRNA right after transcription
45
New cards
Introns
sequence of DNA that is not involved in coding for a protein, are removed in processing
46
New cards
Exons
expressed sequence of DNA; codes for a protein
47
New cards
heterogeneous nuclear RNA (hnRNA)
In the nucleus, there's a mix of RNA at different stages of processing until the final mature mRNA is done. This mix is called...
48
New cards
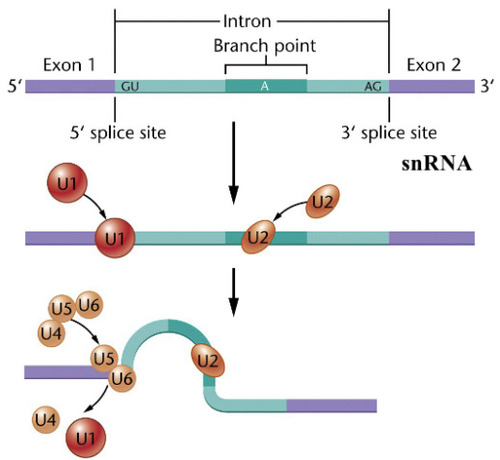
Spliceosome
A large RNA protein complex that catalyzes the removal of introns from nuclear pre-RNA.
-there are nucleotide sequences near the junction of the intron/exon boundary that need to be removed: 5' GU (donor sequence) and 3' AG (acceptor sequence).
-there are nucleotide sequences near the junction of the intron/exon boundary that need to be removed: 5' GU (donor sequence) and 3' AG (acceptor sequence).
49
New cards
Alternative Splicing
Different isoforms/ Related proteins resujt from this process. The combination of exons in genes will lead to different proteins in a gene.
50
New cards
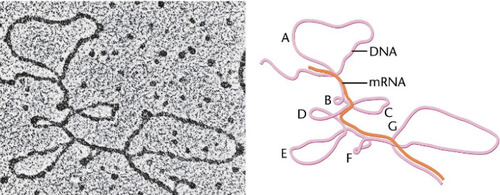
Splicing
Shown here, is the DNA sequence in pink. RNA sequence in orange. Promoter sequences don't get transcribed. The intronic sequences are looped out as they are removed (there are 7 intronic sequences here) . Removal of introns is ...?
51
New cards
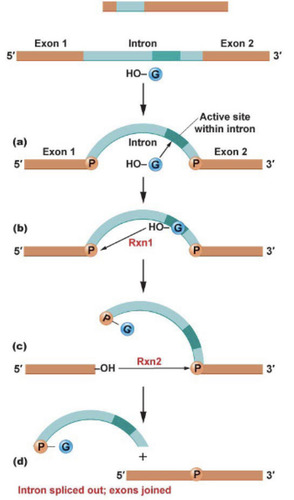
Riboenzymes
RNA capable of splicing themselves; are autocatalytic. Will splice out introns and ligate them back together.
-rRNA is autocatalytic
-protozoans, mitochondria, chloroplasts, and introns are autocatalytic
-nuclear mRNA is not autocatalytic! It requires a number of proteins
-rRNA is autocatalytic
-protozoans, mitochondria, chloroplasts, and introns are autocatalytic
-nuclear mRNA is not autocatalytic! It requires a number of proteins
52
New cards
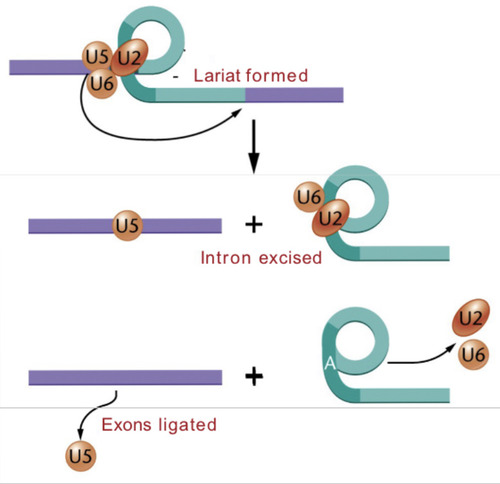
Lariat
A characteristic loop that forms where the intron is excised.
53
New cards
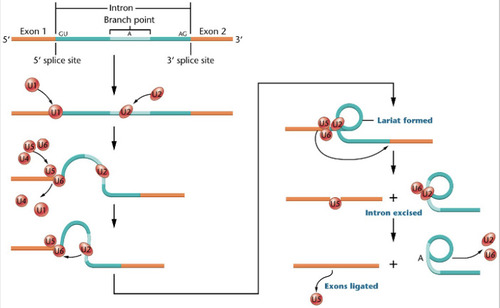
Spliceosome
A model of the splicing mechanism involved during the removal of an intron from from pre-mRNA is shown here. Excision is dependent on various snRNA's (U1, U2,....U6) that combine with proteins to form snRNPs, which are a part of the ____________. The lariat structure in the intermediate stage is characteristic of this splicing mechanism which happens in the nucleus.
54
New cards
TATA box
A promoter DNA sequence crucial in forming the transcription initiation complex. Is rich in thymine and adenine residues.
55
New cards
61
The complete coding dictionary reveals that of the 64 possible triplet codons, __________ encode the 20 amino acids found in proteins, while three triplets terminate translation.
56
New cards
Unambiguous
When we say the genetic code is ___________, we mean each triplet codes for a single amino acid.
57
New cards
Degenerate
When we say the genetic code is ____________, we mean a given amino acid can be specified by more than one triplet codon.
58
New cards
overlapping genes
We know the genetic code is non overlapping-each ribonucleotide in an mRNA is part of only 1 codon. However, this does not rule out that a single mRNA may have multiple initiation points for translation. If so, these points could theoretically create several different reading frames within the same mRNA, thus specifying more than one polypeptide and leading to the concept of ....
59
New cards
No
During transcription, is a primer needed to initiate synthesis by RNA polymerase?
60
New cards
Sigma factor
controls the binding of RNA polymerase to the promoter in initiation of transcription
61
New cards
Introns
The primary transcript in eukaryotes reflects the presence of intervening sequences, or ___________, present in DNA, which must be spliced out to create the mature mRNA.
62
New cards
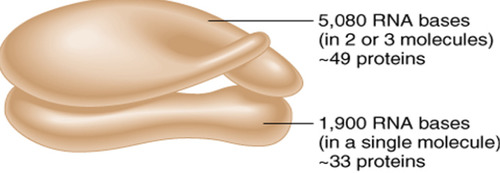
rRNA (ribosomal RNA)
type of RNA that makes up the major part of ribosomes. Helps translate mRNA into protein
63
New cards
Svedberg coefficient
A measure of the density, mass, and shape of a molecule. Is a run on the sucrose gradient, which gives sedimentation with units of S.
-not additive when the large and small subunits come together
-not additive when the large and small subunits come together
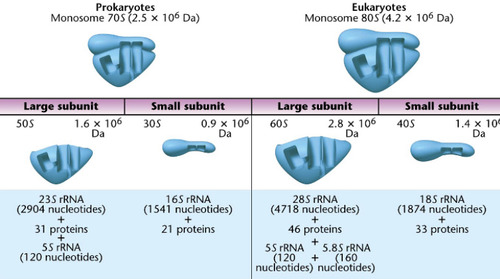
64
New cards
Translation
Synthesis of protein from the mRNA template. Happens in the ribosome (has two subunits). When large and small subunit come together, they can activate this process to occur/ form an active Monosome .
65
New cards
Wobble hypothesis
Post-transcriptional modifications can add rare, odd bases. The _________________told us that modified AUGC in the third position can happen after transcription in the tRNA.
66
New cards
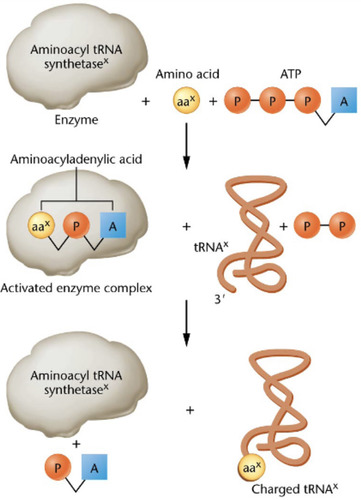
Charging of tRNA
Aminoacyl tRNA synthetase takes an amino acid and attaches it to the 3' end of the tRNA in this process.
-about 31 tRNA (due to Wobble) but 20 synthetases
-about 31 tRNA (due to Wobble) but 20 synthetases
67
New cards
aminoacyl-tRNA synthetase
An enzyme that joins each amino acid to the appropriate tRNA to charge the tRNA.
-is specific for its amino acid
-used energy in the form of ATP
-is called Aminoacylic acid in its active form
-is specific for its amino acid
-used energy in the form of ATP
-is called Aminoacylic acid in its active form
68
New cards
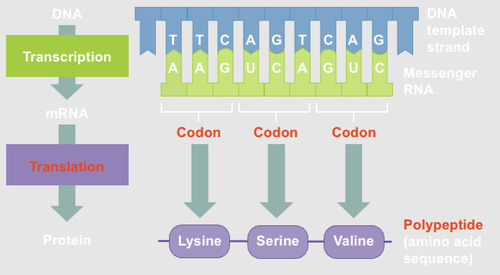
Translation
The process of reading the RNA sequence of an mRNA and creating the amino acid sequence of a protein.
3 stages:
1. Initiation: translation begins at a start codon (AUG= methionine)
2. Elongation= the ribosome uses the tRNA anticodon to match codons to amino acids
3. Termination= translation ends at the stop codon UAA, UAG, or UGA
3 stages:
1. Initiation: translation begins at a start codon (AUG= methionine)
2. Elongation= the ribosome uses the tRNA anticodon to match codons to amino acids
3. Termination= translation ends at the stop codon UAA, UAG, or UGA
69
New cards
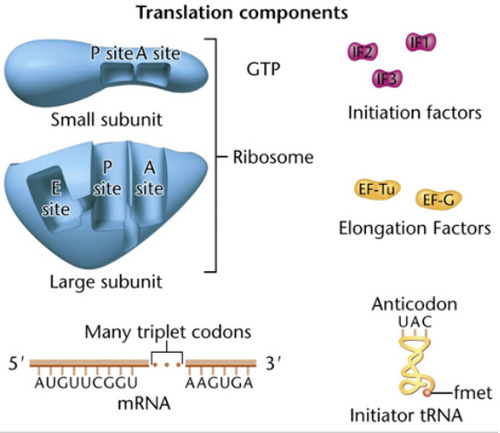
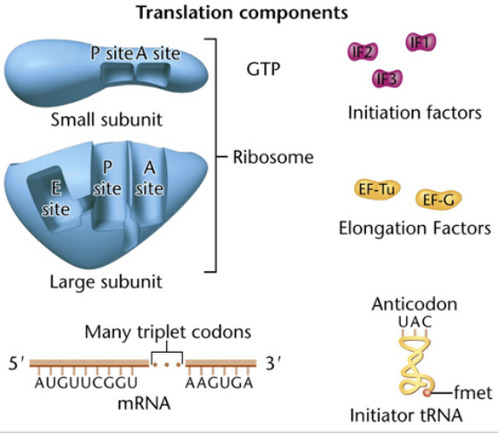
P-site
peptidyl site; where the peptide bond forms during translation in the ribosome
70
New cards
Translation
The process of reading the RNA sequence of an mRNA and creating the amino acid sequence of a protein.
3 stages:
1. Initiation: translation begins at a start codon (AUG= methionine)
2. Elongation= the ribosome uses the tRNA anticodon to match codons to amino acids
3. Termination= translation ends at the stop codon UAA, UAG, or UGA
3 stages:
1. Initiation: translation begins at a start codon (AUG= methionine)
2. Elongation= the ribosome uses the tRNA anticodon to match codons to amino acids
3. Termination= translation ends at the stop codon UAA, UAG, or UGA
71
New cards
snRNA (small nuclear RNA)
Small RNA molecule of 100-200 nucleotides that participates in RNA splicing and is an essential component of the spliceosome.
-are complexed with proteins to form small nuclear ribonucleoproteins (snRNPs or snurps). -which are uridine rich and designated U1, U2, and UG.
-are complexed with proteins to form small nuclear ribonucleoproteins (snRNPs or snurps). -which are uridine rich and designated U1, U2, and UG.
72
New cards
A-site
Amino-acyl tRNA site; the site on a ribosome where the tRNA can enter the ribosome.
73
New cards
E-site
the exit site, where discharged tRNAs leave the ribosome
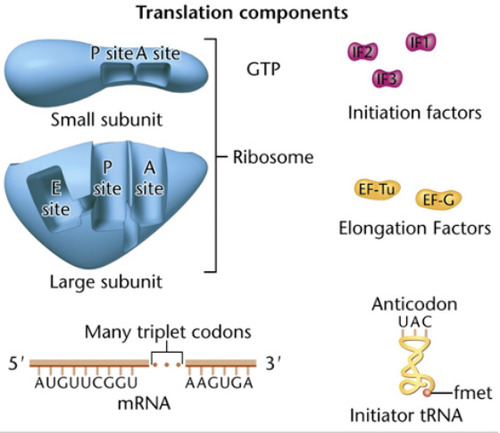
74
New cards
initiation factors (IF)
Proteins that function in binding the small subunit of the ribosome during initiation of translation to start the process.
75
New cards
Elongation factors
Protein that function in bringing the tRNAs into the ribosome through the A-site so the polypeptide chain can be elongated
76
New cards
rRNA genes
repetitive DNA present in clusters throughout the genome at various chromosomal sites (chromosomes 13,14,15,21,22). Are seen as tandem repeats. Are separated by noncoding regions.
77
New cards
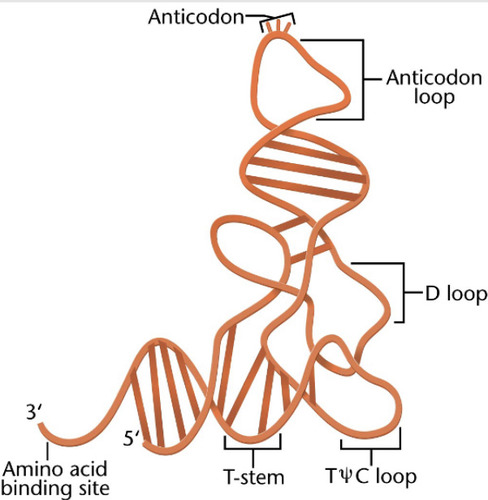
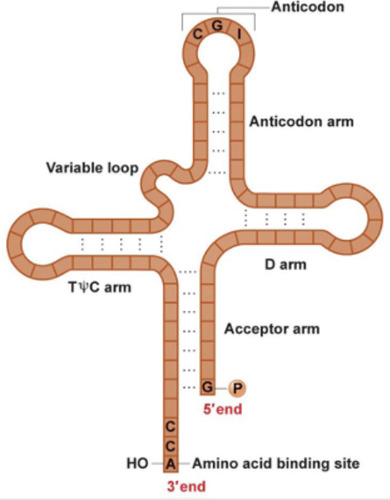
tRNA
carries amino acids to the ribosome during translation process.
-are smaller molecules (75-90 nucleotides long)
-have a clover leaf structure
-base pairing occurs between codon and anticodon
-loops have modified bases in 3rd position
-3' end is where the amino acid will covalently attach to it
-are smaller molecules (75-90 nucleotides long)
-have a clover leaf structure
-base pairing occurs between codon and anticodon
-loops have modified bases in 3rd position
-3' end is where the amino acid will covalently attach to it
78
New cards
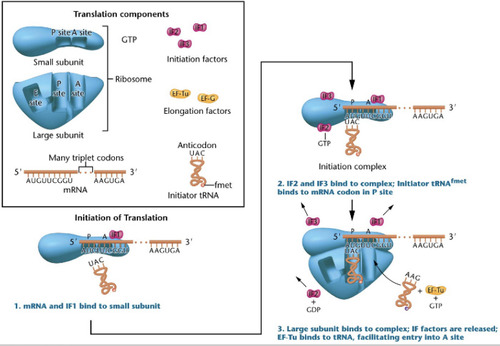
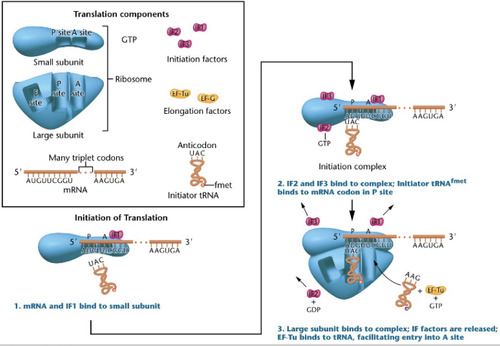
Initiation of Translation
-mRNA binds to the small subunit along with initiation factors
-initiator tRNA binds to mRNA codon in the p-site to form the initiation complex
-large subunit binds to the small subunit and the Monosome complex is formed. tRNA enters through the A-site
-initiator tRNA binds to mRNA codon in the p-site to form the initiation complex
-large subunit binds to the small subunit and the Monosome complex is formed. tRNA enters through the A-site
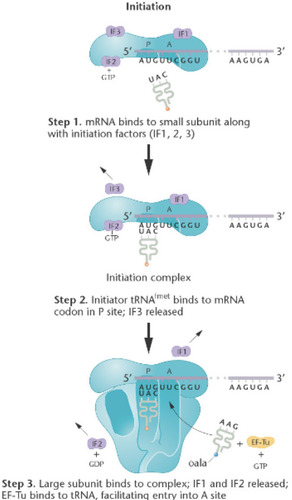
79
New cards
Shine-Dalgarno sequence
Helps the small subunit bind to the mRNA in the first step of initiation of translation.
-in bacterial/ prokaryotes
-in bacterial/ prokaryotes
80
New cards
Elongation of Translation
1. More charged tRNA enters the ribosome through the A-site.
2. Peptide bond forms by an enzyme known as peptidyl transferase. This catalytic rxn is characteristic of 23S rRNA in the large subunit.
The uncharged tRNA then moves to the E-site and exits the ribosome.
3. A shift occurs where the ribosome moves from 5' to 3'. Shifts by 3bp. Requires elongation factors and GTP.
4. More charged tRNA enters the A-site and more elongation occurs until stop codon is reached
2. Peptide bond forms by an enzyme known as peptidyl transferase. This catalytic rxn is characteristic of 23S rRNA in the large subunit.
The uncharged tRNA then moves to the E-site and exits the ribosome.
3. A shift occurs where the ribosome moves from 5' to 3'. Shifts by 3bp. Requires elongation factors and GTP.
4. More charged tRNA enters the A-site and more elongation occurs until stop codon is reached
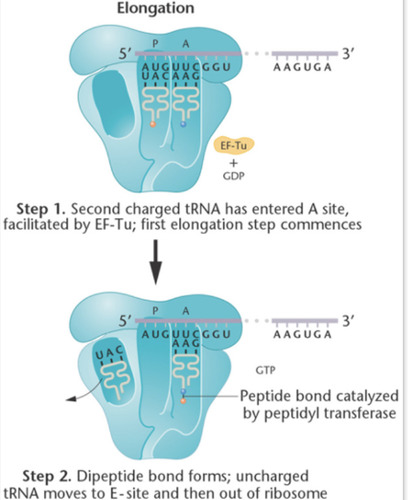
81
New cards
2
At any time during translation, there can only be ________ tRNA bound to the message at any time.
82
New cards
Ribozyme
catalytic RNA
83
New cards
Termination of Translation
occurs when a stop codon/termination codon/ nonsense codon in the mRNA reaches the A site of the ribosome. The polypeptide chain is done being extended. GTP-dependent termination factors stimulate the release of tRNA and dissociation of small and large subunits. The polypeptide folds into a protein.
-polypeptide chain is seen at the P-site
-A-site will be empty
- GTP-dependent release factors cleave the polypeptide bond.
-polypeptide chain is seen at the P-site
-A-site will be empty
- GTP-dependent release factors cleave the polypeptide bond.
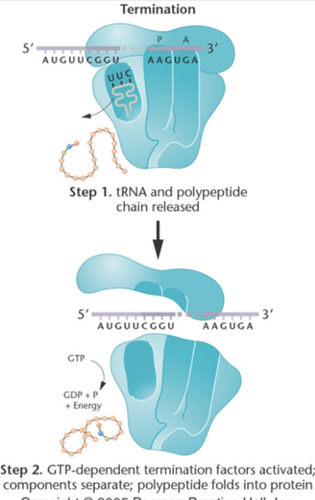
84
New cards
Polyribosomes (polysome)
=2 or more ribosomes actively associated with the mRNA engaged in translation
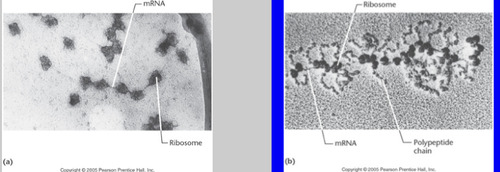
85
New cards
Chaperone proteins
Assist in the process of folding and forming the structure of the polypeptide chain during translation.
During translation we see multiple copies of a protein are made simultaneously. The ribosome moves along the mRNA from 5' to 3'
During translation we see multiple copies of a protein are made simultaneously. The ribosome moves along the mRNA from 5' to 3'
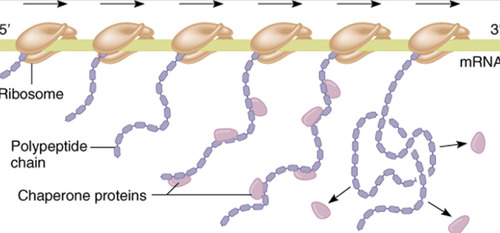
86
New cards
Kozak sequence
plays a role in the initiation of translation (mRNA binding to small subunit ribosomes) in eukaryotes. Is similar to the shine-dalgarno sequence in prokaryotes.
87
New cards
Translation in Eukaryotes
This process is more complex in eukaryotes because transcripts have a longer half-life due to post-translational modifications.
Also, the 5' end cap protects the sequence.
-a recognition sequence around AUG known as the Kozak sequence facilitates binding of the small subunit
Also, the 5' end cap protects the sequence.
-a recognition sequence around AUG known as the Kozak sequence facilitates binding of the small subunit
88
New cards
one gene-one polypeptide hypothesis
there is one gene that codes for one polypeptide
89
New cards
sickle cell anemia
a genetic disorder that causes abnormal hemoglobin, resulting in some red blood cells assuming an abnormal sickle shape and clogging is capillaries in the venous system.
-shows the one gene-one polypeptide hypothesis
-individuals can have crisis during movements/ activities
-are often anemic
-the trait is heterozygous
-shows the one gene-one polypeptide hypothesis
-individuals can have crisis during movements/ activities
-are often anemic
-the trait is heterozygous
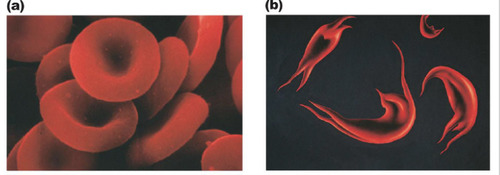
90
New cards
Hemoglobin
Oxygen carrying pigment in red blood cells that is made up of multiple polypeptide chains.
-One amino acid switch results in an entirely different protein.. sickle cell RBC shape
-has 7 different polypeptide chains meaning 7 genes (proves one gene-one polypeptide chain hypothesis)
-HbA protein has 2 alpha, 2 beta chains. Infant has 2 different chains. As individuals age, there's gradual replacement in the chains.
-One amino acid switch results in an entirely different protein.. sickle cell RBC shape
-has 7 different polypeptide chains meaning 7 genes (proves one gene-one polypeptide chain hypothesis)
-HbA protein has 2 alpha, 2 beta chains. Infant has 2 different chains. As individuals age, there's gradual replacement in the chains.
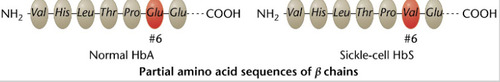
91
New cards
R group of amino acids
gives the amino acid its chemical and physical properties.
Can be categorized based on 4 classes:
1. Nonpolar (hydrophobic)
2. Polar (hydrophilic)
3. Positively charged
4. Negatively charged
Can be categorized based on 4 classes:
1. Nonpolar (hydrophobic)
2. Polar (hydrophilic)
3. Positively charged
4. Negatively charged
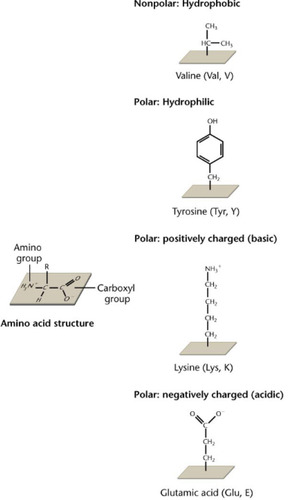
92
New cards
Amino acids
building blocks of proteins.
-has a central carbon, amino group, carboxyl group, and a side chain (R/ radical group)
-the R group gives it unique structures and properties. Can be chemically altered.
-has a central carbon, amino group, carboxyl group, and a side chain (R/ radical group)
-the R group gives it unique structures and properties. Can be chemically altered.
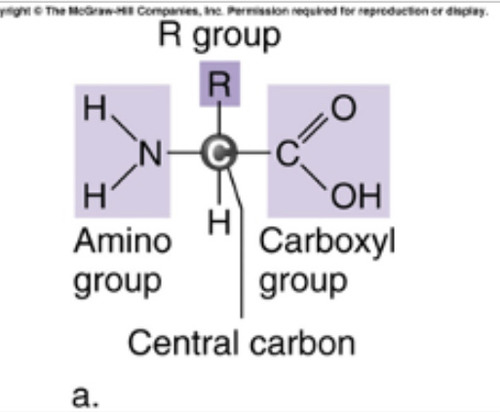
93
New cards
Peptide bond
The chemical bond that forms between the carboxyl group of one amino acid and the amino group of another amino acid. (No side chains involved).
-is always arranged from n-terminus (amino end) to c-terminus (carboxyl end)
-is always arranged from n-terminus (amino end) to c-terminus (carboxyl end)
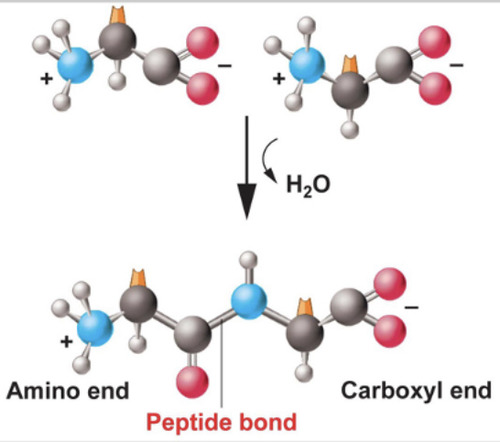
94
New cards
Levels of protein structure
1. Primary
2. Secondary
3. Tertiary
4. Quaternary
2. Secondary
3. Tertiary
4. Quaternary
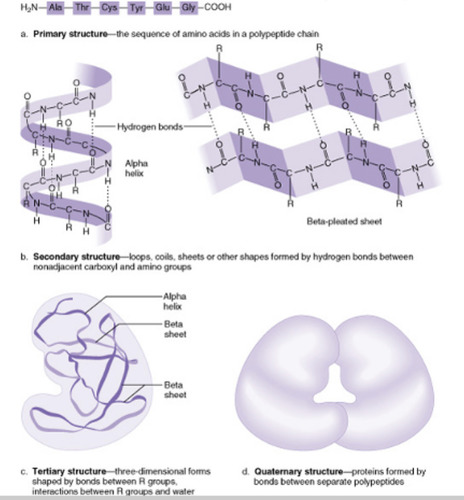
95
New cards
primary protein structure
linear sequence of amino acids
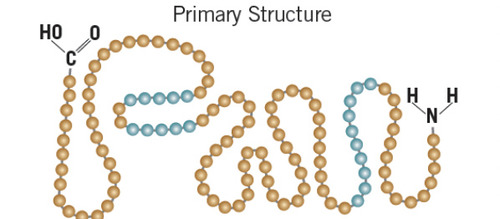
96
New cards
secondary protein structure
occurs when the sequence of amino acids are linked by hydrogen bonds which connect portions of the backbone in a repetitive pattern.
2 types- alpha helix, beta pleated sheet
2 types- alpha helix, beta pleated sheet
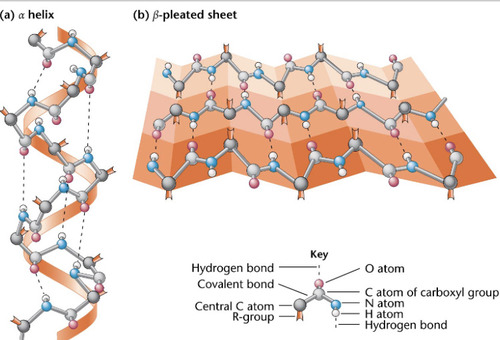
97
New cards
tertiary protein structure
The 3D conformation of a protein.
-has disulfide bonds that reinforce the structure
-ex: in myoglobin
-has disulfide bonds that reinforce the structure
-ex: in myoglobin
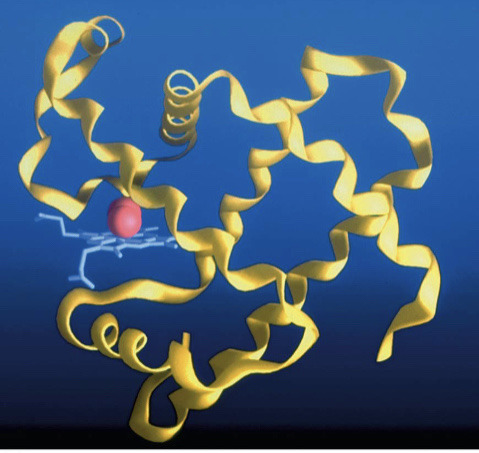
98
New cards
Folding
If the charge of an amino acid is altered, this may affect the ___________ of the polypeptide chain. (Remember.. amino acids are the building blocks of proteins).
99
New cards
quaternary protein structure
protein consisting of more than one amino acid chain. Not all proteins have this.
Ex: hemoglobin has this when all 4 polypeptide chains come together.
Ex: hemoglobin has this when all 4 polypeptide chains come together.
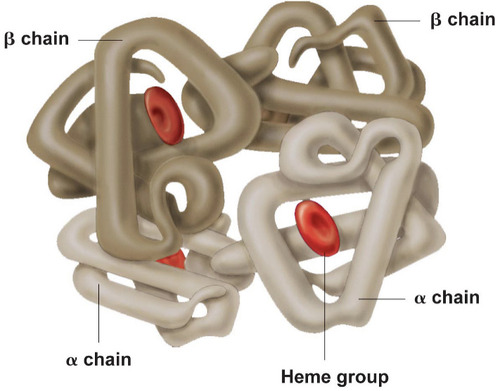
100
New cards
Protein folding
Chaperone proteins aide in this process. Proteins must form different structures depending on their function.