Microbio Exam 3
0.0(0)
0.0(0)
Card Sorting
1/156
Earn XP
Description and Tags
Study Analytics
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
157 Terms
1
New cards
Mobile genetic elements
These are different types elements that transfer information.
As genomes are sequenced and annotated, it is apparent that they are rife with genetic elements that move within and between genomes. These elements are referred to as "jumping genes," mobile genetic elements, or transposable elements.
-Transposition refers to the movement of a mobile genetic element.
As genomes are sequenced and annotated, it is apparent that they are rife with genetic elements that move within and between genomes. These elements are referred to as "jumping genes," mobile genetic elements, or transposable elements.
-Transposition refers to the movement of a mobile genetic element.

2
New cards
Insertion sequence
The simplest transposable elements in bacteria are insertion sequences, or IS elements (figure 12.13a). An IS element is a short sequence of DNA (around 750 to 1,600 base pairs [bp] in length). It contains only the gene for the enzyme transposase, and it is bounded at both ends by inverted repeats-identical or very similar sequences of nucleotides in reversed orientation. Inverted repeats are usually about 15 to 25 bp long and vary among IS elements so that each type of IS has a specific nucleotide sequence in its inverted repeats. Transposase recognizes the ends of the IS and catalyzes transposition. IS elements have been observed in many bacteria and some archaea.
![The simplest transposable elements in bacteria are insertion sequences, or IS elements (figure 12.13a). An IS element is a short sequence of DNA (around 750 to 1,600 base pairs [bp] in length). It contains only the gene for the enzyme transposase, and it is bounded at both ends by inverted repeats-identical or very similar sequences of nucleotides in reversed orientation. Inverted repeats are usually about 15 to 25 bp long and vary among IS elements so that each type of IS has a specific nucleotide sequence in its inverted repeats. Transposase recognizes the ends of the IS and catalyzes transposition. IS elements have been observed in many bacteria and some archaea.](https://knowt-user-attachments.s3.amazonaws.com/67daf0bdd16a4468827972750665561d.jpeg)
3
New cards
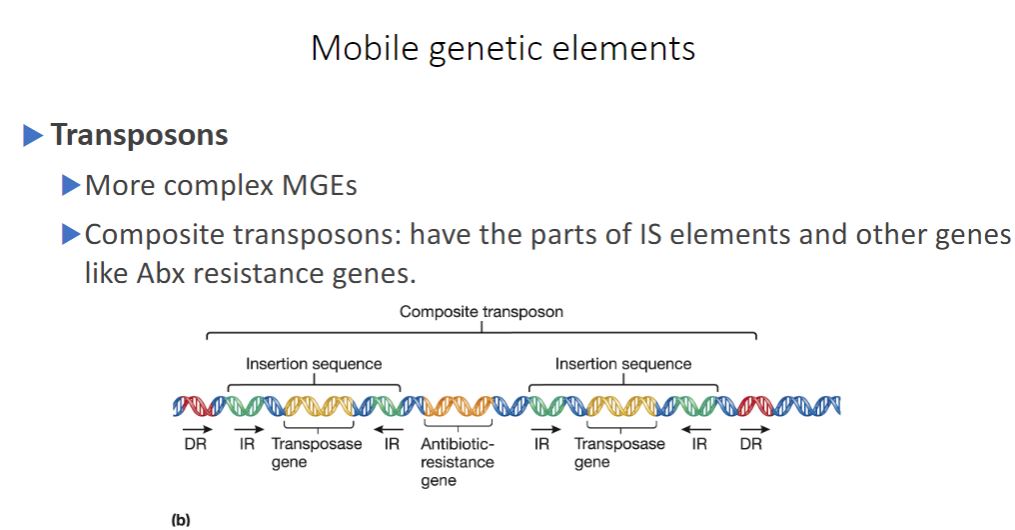
Transposons
Transposons are more complex in structure than IS elements. Some transposons (composite transposons) consist of a central region containing genes unrelated to transposition (e.g., antibiotic-resistance genes) flanked on both sides by IS elements that are identical or very similar in sequence (figure 12.136). The flanking IS elements encode the transposase used by the transposon to move. Other transposons (unit transposons) lack IS elements and encode their own transposition enzymes (
figure 12.13c).
Two major transposition methods have been identified: simple transposition and replicative transposition. Simple transposition is also called cut-and-paste transposition. In this method, transposase catalyzes excision of the transposable element, followed by cleavage of a new target site and ligation of the element into this site (figure 12.14). Target sites are specific sequences about 5 to 9 bp long. When a mobile genetic element inserts at a target site, the target sequence is duplicated so that short, direct-sequence repeats flank the element's terminal inverted repeats. In replicative transposition, the original transposon remains at the parental site on the chromosome and a copy is inserted at the target DNA site. Simple Transposition
figure 12.13c).
Two major transposition methods have been identified: simple transposition and replicative transposition. Simple transposition is also called cut-and-paste transposition. In this method, transposase catalyzes excision of the transposable element, followed by cleavage of a new target site and ligation of the element into this site (figure 12.14). Target sites are specific sequences about 5 to 9 bp long. When a mobile genetic element inserts at a target site, the target sequence is duplicated so that short, direct-sequence repeats flank the element's terminal inverted repeats. In replicative transposition, the original transposon remains at the parental site on the chromosome and a copy is inserted at the target DNA site. Simple Transposition
4
New cards
horizontal gene transfer
-Bacteria and archaea do not reproduce sexually. This suggests that genetic variation in populations of these microbes should be relatively
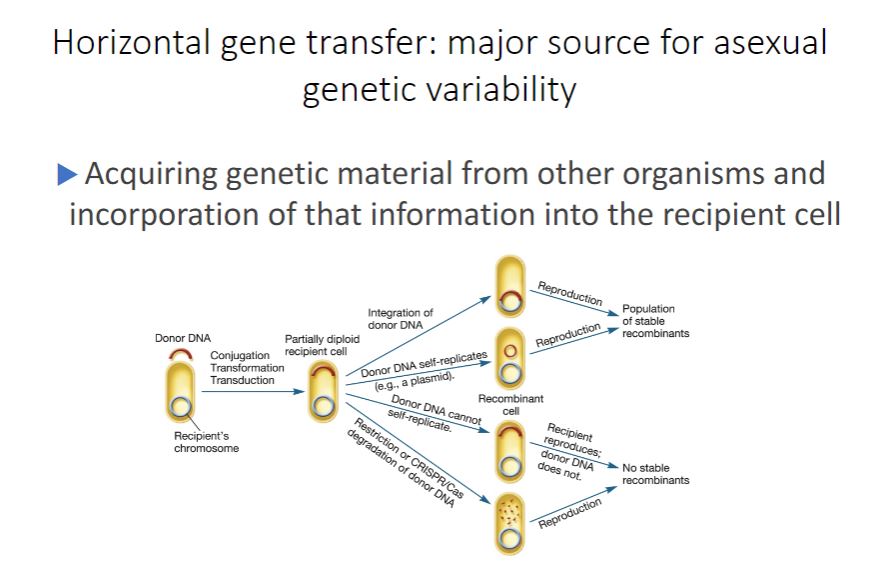
limited, only occurring with the advent of a new mutation and its passage to the next generation by vertical gene transfer. However, this is not the case. Bacteria and archaea have multiple mechanisms for creating recombinants collectively referred to as horizontal (lateral) gene transfer (HGT). HGT is distinctive from vertical gene transfer because genes from one independent, mature organism are transferred to another mature organism, often creating a stable recombinant having characteristics of both donor and recipient. Horizontal Gene Transfer
Image below
-During HGT, a piece of donor DNA, sometimes called the exogenote, enters a recipient. If the donor DNA contains genes already present in the recipient, the recipient will become temporarily diploid for those genes. This partially diploid cell is sometimes called a merozygote (Greek meros, part). The donor DNA has four possible fates in the recipient (figure 12.12). First, when the donor DNA has a sequence homologous to the recipient's chromosome (sometimes called an endogenote), integration may occur. That is, the donor's DNA may pair with the recipient's DNA and recombine. The recombinant then reproduces, yielding a population of stable genetic variants. Second, if the donor DNA is able to replicate (e.g., it is a plasmid), it may persist separate from the recipient's chromosome. When the recipient reproduces, the donor DNA replicates and a population of stable recombinants is formed. Third, the donor DNA remains in the cytoplasm but is unable to replicate. When the recipient divides, the donor DNA is eventually lost from the population. Finally, host restriction or CRISPR/Cas degradation of donor DNA may occur, thereby preventing the formation of a recombinant cell. Responses to viral infection (section 11.6)
limited, only occurring with the advent of a new mutation and its passage to the next generation by vertical gene transfer. However, this is not the case. Bacteria and archaea have multiple mechanisms for creating recombinants collectively referred to as horizontal (lateral) gene transfer (HGT). HGT is distinctive from vertical gene transfer because genes from one independent, mature organism are transferred to another mature organism, often creating a stable recombinant having characteristics of both donor and recipient. Horizontal Gene Transfer
Image below
-During HGT, a piece of donor DNA, sometimes called the exogenote, enters a recipient. If the donor DNA contains genes already present in the recipient, the recipient will become temporarily diploid for those genes. This partially diploid cell is sometimes called a merozygote (Greek meros, part). The donor DNA has four possible fates in the recipient (figure 12.12). First, when the donor DNA has a sequence homologous to the recipient's chromosome (sometimes called an endogenote), integration may occur. That is, the donor's DNA may pair with the recipient's DNA and recombine. The recombinant then reproduces, yielding a population of stable genetic variants. Second, if the donor DNA is able to replicate (e.g., it is a plasmid), it may persist separate from the recipient's chromosome. When the recipient reproduces, the donor DNA replicates and a population of stable recombinants is formed. Third, the donor DNA remains in the cytoplasm but is unable to replicate. When the recipient divides, the donor DNA is eventually lost from the population. Finally, host restriction or CRISPR/Cas degradation of donor DNA may occur, thereby preventing the formation of a recombinant cell. Responses to viral infection (section 11.6)
5
New cards
DNA Sharing sharing conjugation
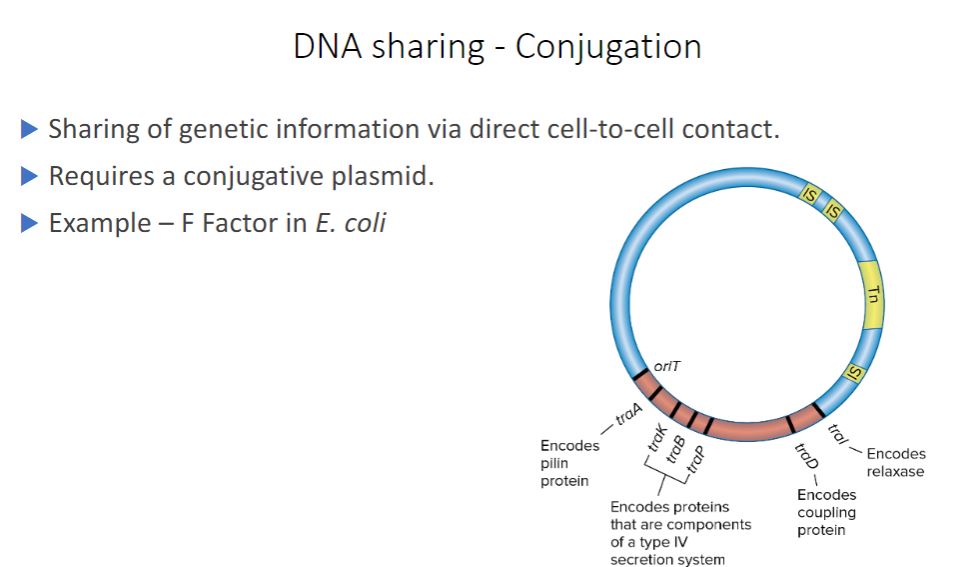
-Conjugation, the transfer of DNA by direct cell-to-cell contact, depends on the presence of a conjugative plasmid. Recall from chapter 3 that plasmids are small, double- stranded DNA molecules that can exist independently of host chromosomes. They have their own replication origins, replicate autonomously, and are stably inherited. Some plasmids are episomes, plasmids that can exist either with or without being integrated into host chromosomes.
6
New cards
F Factor conjugation
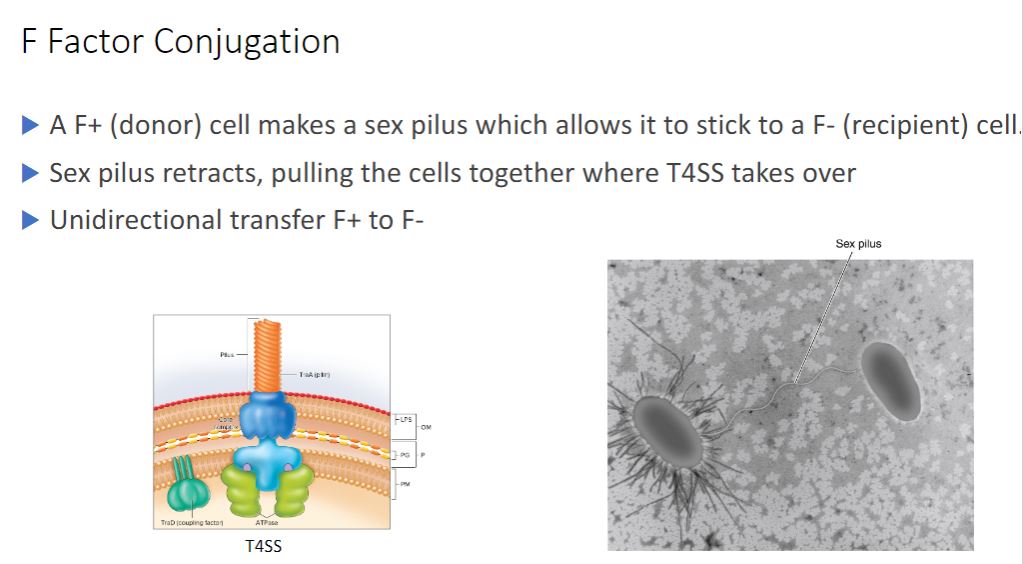
-Perhaps the best-studied conjugative plasmid is the F factor. It plays a major role in conjugation in E. coli, and it was the first conjugative plasmid to be described (figure 12.15). The F factor is about 100,000 bp long and bears genes responsible for cell attachment and plasmid transfer between E. coli cells. Most of the information required for plasmid transfer is located in the tra operon, which contains at least 28 genes. Many of these direct the formation of the sex pilus that attaches the F* cell (the donor cell containing an F plasmid) to an F cell (figure 12.16). Other gene products aid DNA transfer. In addition, the F factor has several IS elements that assist plasmid integration into the host cell's chromosome. Thus the F factor is an episome that can exist outside the bacterial chromosome or be integrated into it.
-Bacterial Conjugation. (image below)
An electron micrograph of two E. coli cells in an early stage of conjugation. The F* cell to the left is covered with fimbriae, and a sex pilus
connects the two cells.
-Bacterial Conjugation. (image below)
An electron micrograph of two E. coli cells in an early stage of conjugation. The F* cell to the left is covered with fimbriae, and a sex pilus
connects the two cells.
7
New cards
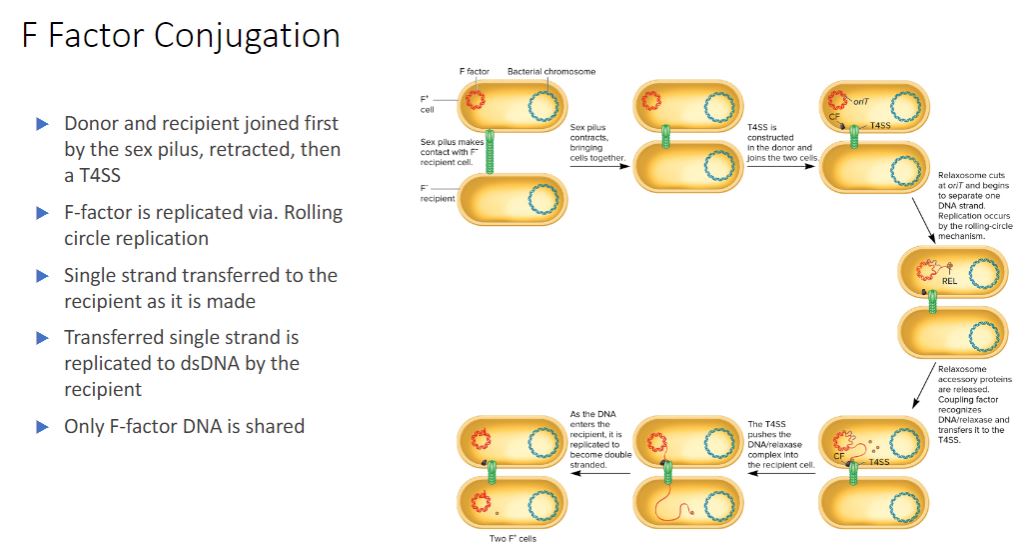
F factor conjugation in steps
In 1952 William Hayes demonstrated that the gene transfer observed by Lederberg and Tatum was unidirectional. That is, there were definite donor (F*, or fertile) and recipient (F", or nonfertile) strains, and gene transfer was nonreciprocal. He also found that in F × F¯ mating, the progeny were only rarely changed with regard to auxotrophy (i.e., chromosomal genes usually were not transferred). However, F" strains frequently became
-These results are now understood and readily explained in the following way. An F* strain contains an extrachromosomal F factor carrying the genes for sex pilus formation and plasmid transfer. The sex pilus is used to establish contact between the F* and F cells (figure 12.18). Once contact is made, the pilus retracts, bringing the cells into close physical contact. The F* cell prepares for DNA transfer by assembling a type IV secretion system, using many of the same proteins in the sex pilus membrane-spanning structure (figure 12.19). The term sex pilus refers to the extracellular structure, while the membrane-bound components are termed the type IV secretion system (T4SS). Although the two structures share polypeptide components and have complementary
roles in conjugation, pilus formation and DNA transfer are independent processes.
- (image below)
F Factor-Mediated Conjugation. The F factor encodes proteins for building the
sex pilus and the type IV secretion (T4SS) system that anchors the donor to the F recipient and transfers DNA. One protein, the coupling factor (CF), escorts the DNA to the T4SS. During
F* * F conjugation, only the F factor is transferred because the plasmid is extrachromosomal.
The recipient cell becomes F
-These results are now understood and readily explained in the following way. An F* strain contains an extrachromosomal F factor carrying the genes for sex pilus formation and plasmid transfer. The sex pilus is used to establish contact between the F* and F cells (figure 12.18). Once contact is made, the pilus retracts, bringing the cells into close physical contact. The F* cell prepares for DNA transfer by assembling a type IV secretion system, using many of the same proteins in the sex pilus membrane-spanning structure (figure 12.19). The term sex pilus refers to the extracellular structure, while the membrane-bound components are termed the type IV secretion system (T4SS). Although the two structures share polypeptide components and have complementary
roles in conjugation, pilus formation and DNA transfer are independent processes.
- (image below)
F Factor-Mediated Conjugation. The F factor encodes proteins for building the
sex pilus and the type IV secretion (T4SS) system that anchors the donor to the F recipient and transfers DNA. One protein, the coupling factor (CF), escorts the DNA to the T4SS. During
F* * F conjugation, only the F factor is transferred because the plasmid is extrachromosomal.
The recipient cell becomes F
8
New cards
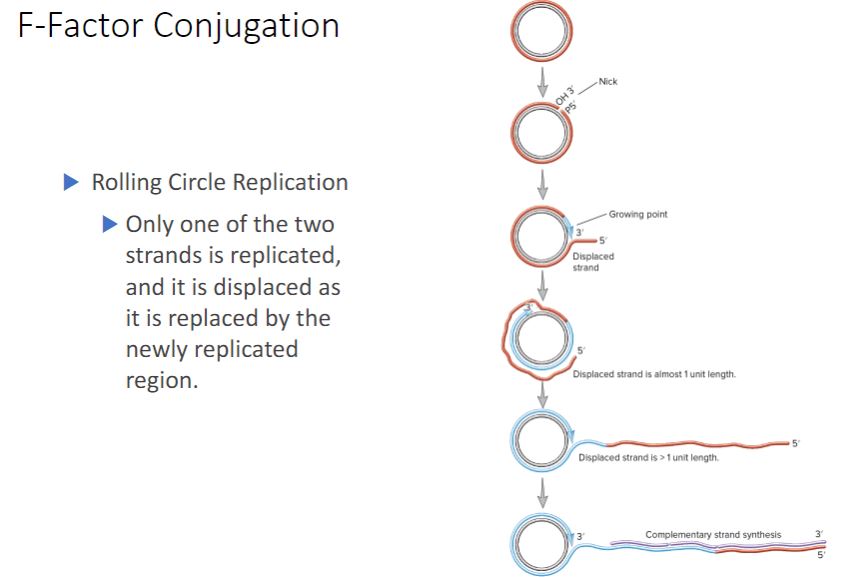
F-Factor conjugation: rolling circle replication
-As the F plasmid is being transferred, it is also copied using a process called rolling-circle replication. One strand of the circular DNA is nicked, and the free 3'-hydroxyl end is extended by replication enzymes (figure 12.20). The 3′ end is lengthened while the growing point rolls around the circular template and the 5′ end of the strand is displaced to form an ever-lengthening tail, much like the peel of an apple is displaced by a knife as it is pared.
-During conjugation, rolling-circle replication is initiated by the relaxosome, a complex of proteins encoded by the F factor (figure 12.15). The relaxosome nicks one strand of the F factor at a site called oriT (for origin of transfer). The major component of the relaxosome is a protein called Tral. It has relaxase activity and remains attached to the 5' end of the nicked strand. As the F factor is replicated, Tral guides the displaced strand through the T4SS to the recipient cell. It is believed that both ATP hydrolysis and the proton motive force provide the energy for DNA translocation. During plasmid transfer, the entering strand is copied to produce double-stranded DNA. When this is complete,
the F- recipient cell becomes F+.
-(image below)
Rolling-Circle Replication. A single-stranded tail, often composed of more than one genome copy, is generated and can be converted to the double-stranded form by synthesis of a complementary strand. The "free end" of the rolling-circle strand is bound to the relaxosome. OH 3' is the 3'-hydroxyl and P 5' is the 5'-phosphate created when the DNA strand is nicked.
-During conjugation, rolling-circle replication is initiated by the relaxosome, a complex of proteins encoded by the F factor (figure 12.15). The relaxosome nicks one strand of the F factor at a site called oriT (for origin of transfer). The major component of the relaxosome is a protein called Tral. It has relaxase activity and remains attached to the 5' end of the nicked strand. As the F factor is replicated, Tral guides the displaced strand through the T4SS to the recipient cell. It is believed that both ATP hydrolysis and the proton motive force provide the energy for DNA translocation. During plasmid transfer, the entering strand is copied to produce double-stranded DNA. When this is complete,
the F- recipient cell becomes F+.
-(image below)
Rolling-Circle Replication. A single-stranded tail, often composed of more than one genome copy, is generated and can be converted to the double-stranded form by synthesis of a complementary strand. The "free end" of the rolling-circle strand is bound to the relaxosome. OH 3' is the 3'-hydroxyl and P 5' is the 5'-phosphate created when the DNA strand is nicked.
9
New cards
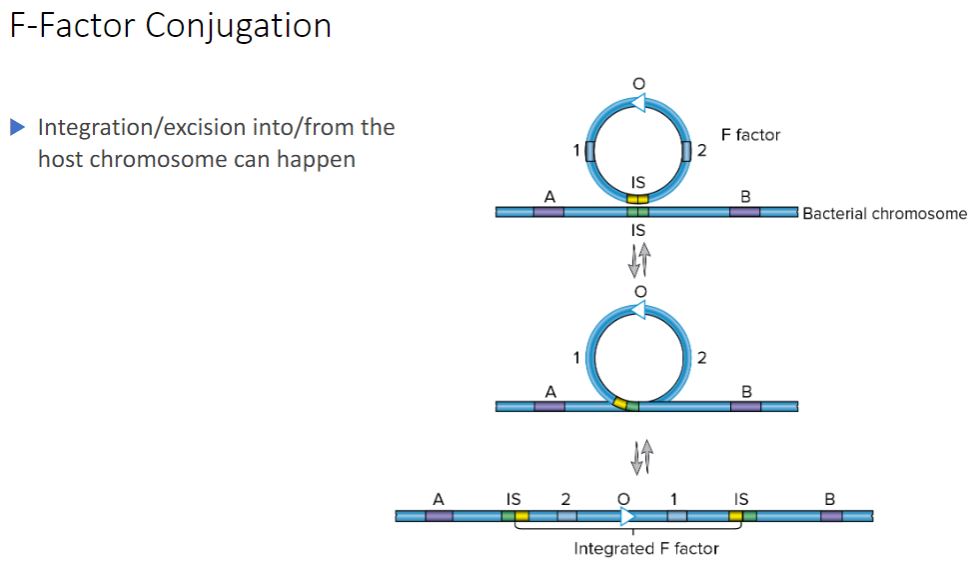
Integration/excision into
-By definition, an F* cell has the F factor separate from the chromosome, so in an F* x
F mating, chromosomal DNA is not transferred. However, within this population, a few cells have the F plasmid integrated (i.e., recombined) into their chromosomes. This
explains why not long after the discovery of F* × F mating, a second type of F factor- mediated conjugation was discovered. In this type of conjugation, the donor transfers chromosomal genes with great efficiency but does not change the recipient bacteria into
F* cells. Because of the high frequency of recombinants produced by this mating, it is referred to as Hfr conjugation and the donor is called an Hfr strain.
F mating, chromosomal DNA is not transferred. However, within this population, a few cells have the F plasmid integrated (i.e., recombined) into their chromosomes. This
explains why not long after the discovery of F* × F mating, a second type of F factor- mediated conjugation was discovered. In this type of conjugation, the donor transfers chromosomal genes with great efficiency but does not change the recipient bacteria into
F* cells. Because of the high frequency of recombinants produced by this mating, it is referred to as Hfr conjugation and the donor is called an Hfr strain.
10
New cards
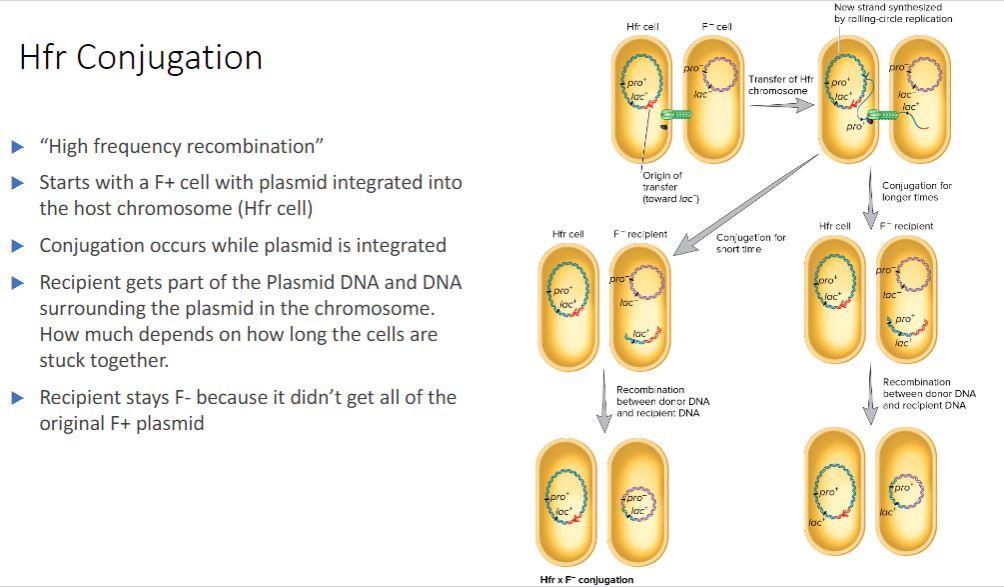
Hfr Conjugation
-Hfr conjugation- Conjugation involving an Hfr strain and an F− strain.
-Hfr strains contain the F factor integrated into their chromosome, rather than as a plasmid (figure 12.21). When integrated, the F factor's tra operon is still functional; it can direct the synthesis of pili, carry out rolling-circle replication, and transfer genetic material to an F recipient. However, rather than transferring just itself, the F factor also directs the transfer of the host chromosome. DNA transfer begins when the integrated F factor is nicked at oriT. As it is replicated, the F factor begins to move into the recipient ( figure 12.22). Initially only part of the F factor is transferred, followed by the donor's chromosome. If the cells remain connected, the entire chromosome with the rest of the integrated F factor will be transferred; this takes about 100 minutes to accomplish. However, the connection between the cells usually breaks before this process is finished. Thus a complete F factor is rarely transferred, and the recipient remains F.
When an Hfr strain participates in conjugation, bacterial genes are transferred to the recipient in either a clockwise or a counterclockwise direction around a circular chromosome, depending on the orientation of the integrated F factor. After the replicated donor chromosome enters the recipient cell, it may be degraded or incorporated into the F genome by recombination.
- (image below)
Hfr * F Conjugation. Shown are the two cells after initial contact and elaboration of the T4SS. As illustrated, during Hfr
F conjugation, some plasmid genes and some chromosomal genes are transferred to the recipient. Note that only a portion of the F factor moves into the recipient. Because the entire plasmid is not transferred,
the recipient remains F. In addition, the incoming DNA must recombine with the recipient's chromosome if it is to be stably maintained.
-Hfr strains contain the F factor integrated into their chromosome, rather than as a plasmid (figure 12.21). When integrated, the F factor's tra operon is still functional; it can direct the synthesis of pili, carry out rolling-circle replication, and transfer genetic material to an F recipient. However, rather than transferring just itself, the F factor also directs the transfer of the host chromosome. DNA transfer begins when the integrated F factor is nicked at oriT. As it is replicated, the F factor begins to move into the recipient ( figure 12.22). Initially only part of the F factor is transferred, followed by the donor's chromosome. If the cells remain connected, the entire chromosome with the rest of the integrated F factor will be transferred; this takes about 100 minutes to accomplish. However, the connection between the cells usually breaks before this process is finished. Thus a complete F factor is rarely transferred, and the recipient remains F.
When an Hfr strain participates in conjugation, bacterial genes are transferred to the recipient in either a clockwise or a counterclockwise direction around a circular chromosome, depending on the orientation of the integrated F factor. After the replicated donor chromosome enters the recipient cell, it may be degraded or incorporated into the F genome by recombination.
- (image below)
Hfr * F Conjugation. Shown are the two cells after initial contact and elaboration of the T4SS. As illustrated, during Hfr
F conjugation, some plasmid genes and some chromosomal genes are transferred to the recipient. Note that only a portion of the F factor moves into the recipient. Because the entire plasmid is not transferred,
the recipient remains F. In addition, the incoming DNA must recombine with the recipient's chromosome if it is to be stably maintained.
11
New cards
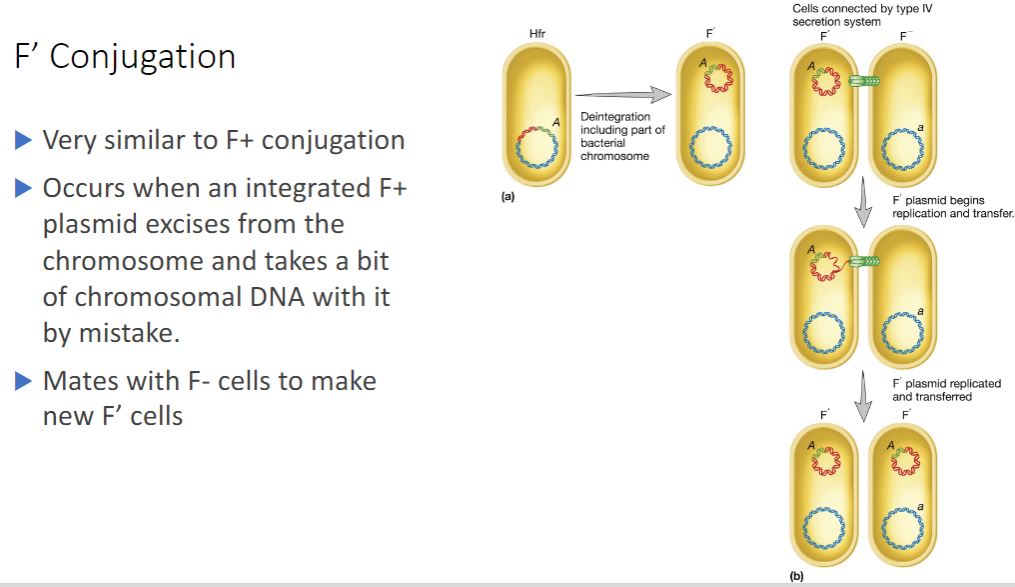
F' Conjugation
Because the F factor is an episome, it can leave the bacterial chromosome and resume status as an autonomous plasmid. Sometimes during excision an error occurs and a portion of the chromosome is excised, becoming part of the F plasmid. Because this erroneously excised plasmid is larger and genotypically distinct from the original F factor, it is called an F' plasmid (figure 12.23a). A cell containing an F' plasmid retains all of its genes, although some of them are on the plasmid. It mates only with an
F recipient, and F' x F" conjugation is similar to an F* x F mating. Once again, the plasmid is transferred as it is copied by rolling-circle replication. Bacterial genes on the chromosome are not transferred (figure 12.236), but bacterial genes on the F' plasmid are transferred. These genes need not be incorporated into the recipient chromosome to be expressed. The recipient becomes F' and is partially diploid because the same bacterial genes present on the F' plasmid are also found on the recipient's chromosome. In this way, specific bacterial genes may spread rapidly throughout a bacterial population.
-(image below)
F' Conjugation. (a) Due to an
error in excision, the A gene of an Hfr cell is incorporated into the F factor. (b) During conjugation, the A gene is transferred to a recipient, which becomes diploid for that gene
F recipient, and F' x F" conjugation is similar to an F* x F mating. Once again, the plasmid is transferred as it is copied by rolling-circle replication. Bacterial genes on the chromosome are not transferred (figure 12.236), but bacterial genes on the F' plasmid are transferred. These genes need not be incorporated into the recipient chromosome to be expressed. The recipient becomes F' and is partially diploid because the same bacterial genes present on the F' plasmid are also found on the recipient's chromosome. In this way, specific bacterial genes may spread rapidly throughout a bacterial population.
-(image below)
F' Conjugation. (a) Due to an
error in excision, the A gene of an Hfr cell is incorporated into the F factor. (b) During conjugation, the A gene is transferred to a recipient, which becomes diploid for that gene
12
New cards
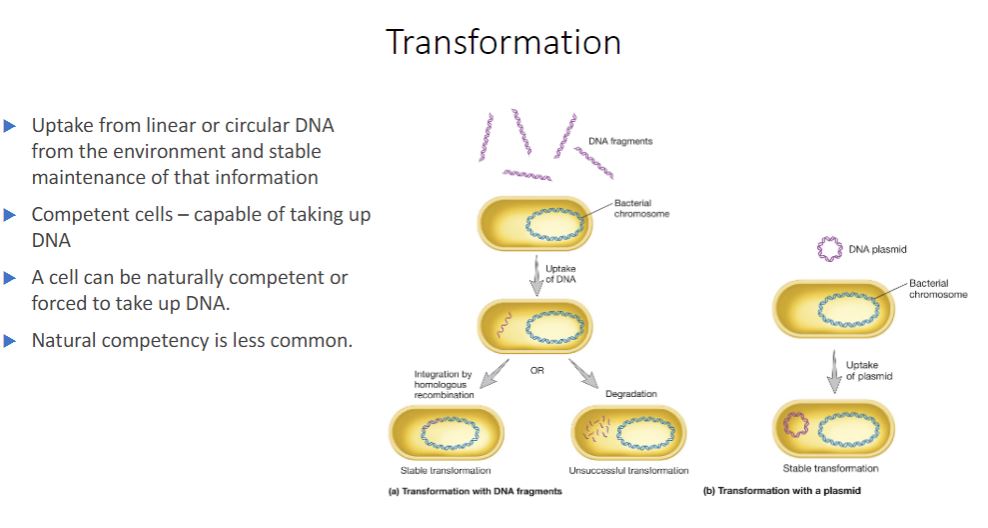
Transformation
Another HGT mechanism is transformation, discovered by Fred Griffith in 1928. Transformation is the uptake of circular or linear DNA from the environment outside the cell and maintenance of the DNA in the recipient cell in a heritable form. Natural transformation is distinct from artificial transformation, a laboratory technique that induces cells to take up DNA. Natural transformation has been observed in some archaea and in members of several bacterial phyla. It occurs in soil and aquatic ecosystems, in vivo during infection, and in biofilm and other microbial communities. Introducing recombinant DNA to host cells (section 31.1)
Natural transformation occurs when bacteria lyse and release their DNA into the surrounding environment. These fragments may be relatively large and contain multiple genes. If a fragment contacts a competent cell-a cell that is able to take up DNA and be transformed-the DNA is bound to the cell and imported (figure 12.24a). The transformation frequency of competent cells is around 103 for most genera when an excess of DNA is used. That is, about one cell in every thousand will take up and integrate the gene.
-image below
Bacterial Transformation. The transforming DNA is in purple. (a) Integration of linear DNA is at a homologous region of the genome. (b) Transformation with a plasmid often is induced artificially in the laboratory.
Natural transformation occurs when bacteria lyse and release their DNA into the surrounding environment. These fragments may be relatively large and contain multiple genes. If a fragment contacts a competent cell-a cell that is able to take up DNA and be transformed-the DNA is bound to the cell and imported (figure 12.24a). The transformation frequency of competent cells is around 103 for most genera when an excess of DNA is used. That is, about one cell in every thousand will take up and integrate the gene.
-image below
Bacterial Transformation. The transforming DNA is in purple. (a) Integration of linear DNA is at a homologous region of the genome. (b) Transformation with a plasmid often is induced artificially in the laboratory.
13
New cards
Transduction
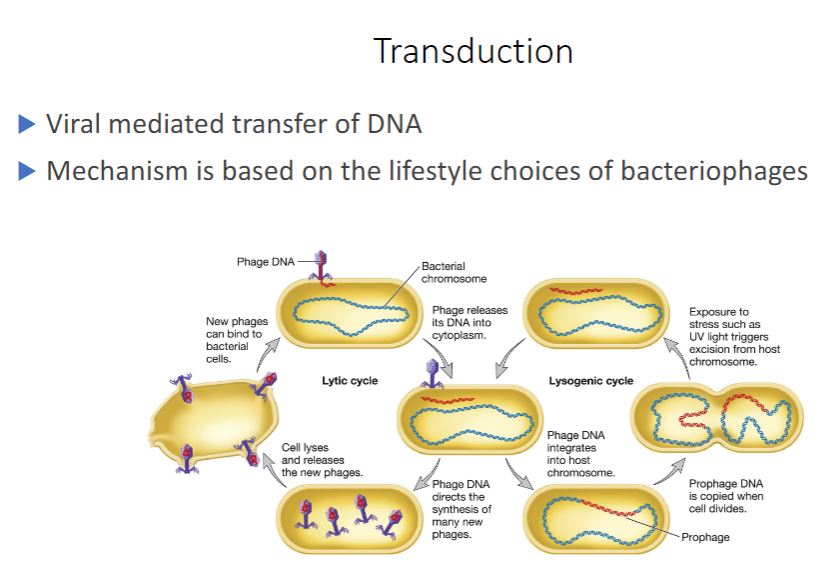
-Transduction is mediated by viruses. It is a frequent mode of HGT in nature. Indeed evidence suggests that the number of genes moved by marine viruses from one host cell to another is huge (perhaps 1024 per year). Furthermore, viruses in marine environments and hot springs move genes between organisms in all three domains of life. Marine viruses: mortality at sea (section 21.2)
Virus particles are structurally simple, often composed of just a nucleic acid genome protected by a protein coat called the capsid. Viruses are unable to multiply autonomously. Instead, they infect and take control of a host cell, forcing it to make many copies of the virus. Viruses that infect bacteria are called bacteriophages, or phages for short. Virulent bacteriophages multiply in their bacterial host immediately after entry. After the progeny phage particles reach a certain number, they cause the host to lyse, so they are released to infect new host cells (see figure 18.14). Thus this process is called the lytic cycle. Temperate bacteriophages, on the other hand, do not immediately kill their host. Instead, the phage establishes a relationship with their host called lysogeny, and bacteria that have been lysogenized are called lysogens. Many temperate phages establish lysogeny by inserting their genomes into the bacterial chromosome. The inserted viral genome is called a prophage. The host bacterium is unharmed by this, and the phage genome is replicated along with the host cell's genome. Temperate phages may remain inactive in their hosts for many generations. However, they can be induced to switch to a lytic cycle under certain conditions, including UV irradiation. When this occurs, the prophage is excised from the bacterial genome and the lytic cycle proceeds. There are several types of viral infections (section 18.4)
Transduction is the transfer of bacterial or archaeal genes by virus particles. It is important to understand that host genes are packaged in the virus particle because of errors made during the virus's life cycle. The virion containing these genes then transfers them to a new cell. Two kinds of transduction have been described: generalized and specialized.
- Most bacteriophages are either virulent or temperate. Virulent phages have only one option: to begin multiplying immediately upon entering its bacterial host, followed by release from the host by lysis. T4 is an example of a virulent phage. Temperate phages have two options: upon entry into the host, they can multiply like virulent phages and
lyse the host cell, or they can remain within the host without destroying it (figure 18.14). Bacteriophage lambda is an example of this type of phages.
Virus particles are structurally simple, often composed of just a nucleic acid genome protected by a protein coat called the capsid. Viruses are unable to multiply autonomously. Instead, they infect and take control of a host cell, forcing it to make many copies of the virus. Viruses that infect bacteria are called bacteriophages, or phages for short. Virulent bacteriophages multiply in their bacterial host immediately after entry. After the progeny phage particles reach a certain number, they cause the host to lyse, so they are released to infect new host cells (see figure 18.14). Thus this process is called the lytic cycle. Temperate bacteriophages, on the other hand, do not immediately kill their host. Instead, the phage establishes a relationship with their host called lysogeny, and bacteria that have been lysogenized are called lysogens. Many temperate phages establish lysogeny by inserting their genomes into the bacterial chromosome. The inserted viral genome is called a prophage. The host bacterium is unharmed by this, and the phage genome is replicated along with the host cell's genome. Temperate phages may remain inactive in their hosts for many generations. However, they can be induced to switch to a lytic cycle under certain conditions, including UV irradiation. When this occurs, the prophage is excised from the bacterial genome and the lytic cycle proceeds. There are several types of viral infections (section 18.4)
Transduction is the transfer of bacterial or archaeal genes by virus particles. It is important to understand that host genes are packaged in the virus particle because of errors made during the virus's life cycle. The virion containing these genes then transfers them to a new cell. Two kinds of transduction have been described: generalized and specialized.
- Most bacteriophages are either virulent or temperate. Virulent phages have only one option: to begin multiplying immediately upon entering its bacterial host, followed by release from the host by lysis. T4 is an example of a virulent phage. Temperate phages have two options: upon entry into the host, they can multiply like virulent phages and
lyse the host cell, or they can remain within the host without destroying it (figure 18.14). Bacteriophage lambda is an example of this type of phages.
14
New cards
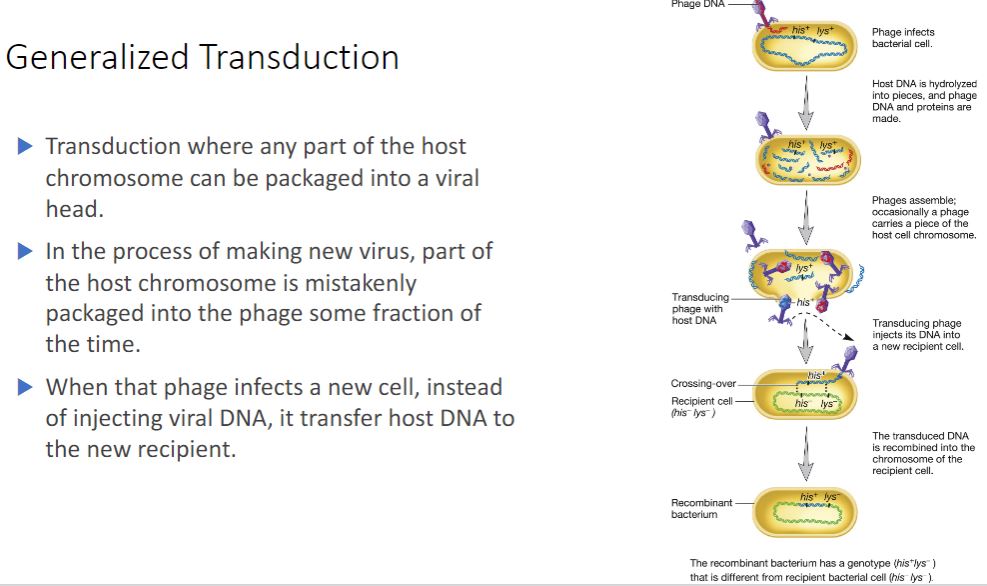
Generalized transduction
-Generalized transduction most often occurs during the lytic cycle of virulent phages but
sometimes happens during the lytic cycle of temperate phages. Any part of the bacterial genome can be transferred after being partially degraded as the virus takes control of its host (figure 12.26). During the assembly stage, viral genomes are packaged by the "headful mechanism"; that is, only genomes of a certain length are packaged. During generalized transduction, a fragment of the host genome that happens to be about the same size as the phage genome is mistakenly packaged. Such a phage is called a generalized transducing particle, because once it is released, it may encounter a susceptible host cell and eject the bacterial DNA it carries into that cell. However, because it lacks viral genes this does not initiate a lytic cycle. As in transformation, once the DNA fragment has been released into the recipient cell, it must be incorporated into the recipient cell's chromosome to preserve the transferred genes. The DNA remains double stranded during transfer, and both strands are integrated into the recipient's chromosome. About 70 to 90% of the transferred DNA is not integrated but often is able to remain intact temporarily and be expressed. Abortive transductants are bacteria that contain this nonintegrated, transduced DNA and are partial diploids.
sometimes happens during the lytic cycle of temperate phages. Any part of the bacterial genome can be transferred after being partially degraded as the virus takes control of its host (figure 12.26). During the assembly stage, viral genomes are packaged by the "headful mechanism"; that is, only genomes of a certain length are packaged. During generalized transduction, a fragment of the host genome that happens to be about the same size as the phage genome is mistakenly packaged. Such a phage is called a generalized transducing particle, because once it is released, it may encounter a susceptible host cell and eject the bacterial DNA it carries into that cell. However, because it lacks viral genes this does not initiate a lytic cycle. As in transformation, once the DNA fragment has been released into the recipient cell, it must be incorporated into the recipient cell's chromosome to preserve the transferred genes. The DNA remains double stranded during transfer, and both strands are integrated into the recipient's chromosome. About 70 to 90% of the transferred DNA is not integrated but often is able to remain intact temporarily and be expressed. Abortive transductants are bacteria that contain this nonintegrated, transduced DNA and are partial diploids.
15
New cards
specialized transduction
-In specialized transduction, only specific portions of the bacterial genome are carried by transducing particles. Specialized transduction is made possible by an error in the lysogenic life cycle of temperate phages that insert their genomes into a specific site in the host chromosome. When a prophage is induced to leave the host chromosome, excision is sometimes carried out improperly. The resulting phage genome contains portions of the bacterial chromosome (about 5 to 10% of the bacterial DNA) next to the integration site, much like the situation with F plasmids (figure 12.27). However, the transducing particle is defective because it lacks some viral genes and cannot reproduce without assistance. In spite of this, it will inject the remaining viral genome and any bacterial genes it carries into another bacterium. The bacterial genes may become stably incorporated under the proper circumstances.
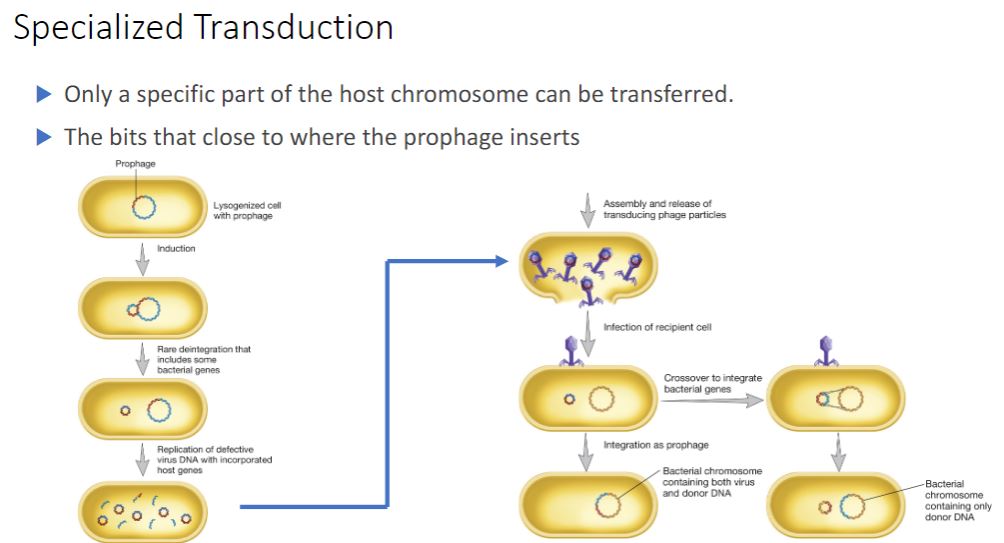
16
New cards
Continued of specialized transduction
-The best-studied example of specialized transduction is carried out by the E. coli phage lambda. The lambda genome inserts into the host chromosome at specific locations known as attachment or att sites (figure 12.28). The att site for lambda is between the gal and bio genes on the E. coli chromosome; consequently when lambda excises incorrectly to generate a specialized transducing particle, these bacterial genes are most often present. Bacteriophage lambda: a temperate bacteriophage.
-The Mechanism of Transduction for Phage Lambda and E. coli. Integrated lambda phage lies between the gal and bio genes in the E. coli chromosome. When it excises normally (top left), the new phage is complete and contains no bacterial genes. Rarely, excision occurs asymmetrically (top right), and either the gal or bio genes are picked up and some phage genes are lost (only aberrant excision involving the gal genes is shown). The result is a defective lambda phage that carries bacterial genes and can transfer them to a new recipient.
-The Mechanism of Transduction for Phage Lambda and E. coli. Integrated lambda phage lies between the gal and bio genes in the E. coli chromosome. When it excises normally (top left), the new phage is complete and contains no bacterial genes. Rarely, excision occurs asymmetrically (top right), and either the gal or bio genes are picked up and some phage genes are lost (only aberrant excision involving the gal genes is shown). The result is a defective lambda phage that carries bacterial genes and can transfer them to a new recipient.
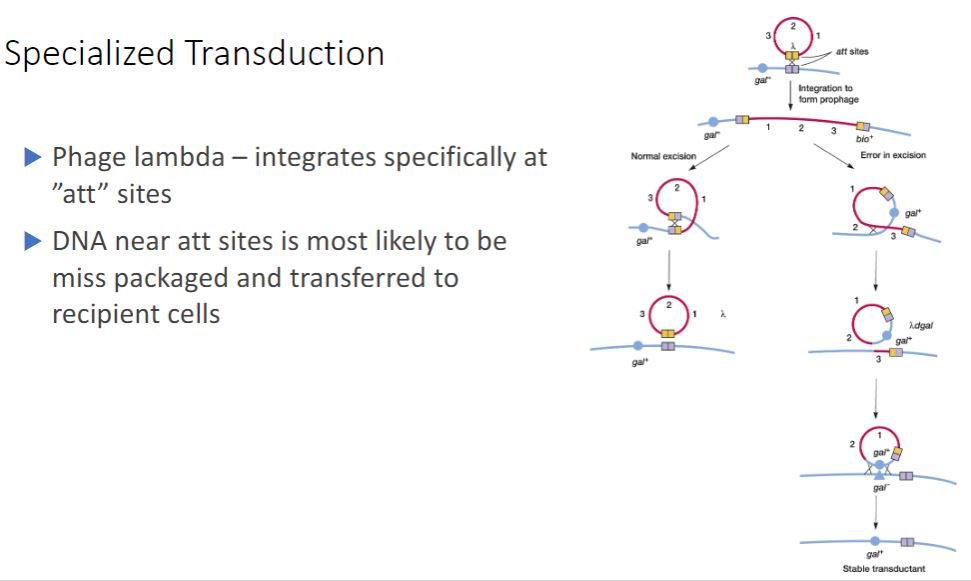
17
New cards
Viruses
- Based on textbooks they are not a cell and not alive. Has no membrane, cannot replicate.
But virologist disagree as they have observed, they are alive and that they are just very complex
But virologist disagree as they have observed, they are alive and that they are just very complex

18
New cards
General structural properties
-Virions can be extraordinarily tiny (about 20 nm in diameter) to about the same size as a rod-shaped bacterial cell (1.5 × 0.5 µm) (figure 18.1). The simplest virions consist only of a nucleocapsid, which is composed of a nucleic acid, either DNA or RNA, and a protein coat called a capsid (figure 18.2). The capsid surrounds the viral nucleic acid, protects the viral genome, and often aids in its transfer between host cells. Some virions are covered by a lipid membrane, and these are termed enveloped viruses, whereas those lacking a membrane are called nonenveloped or naked viruses. Notice what is missing from viruses: ribosomes for protein synthesis and a mechanism for generating ATP. Cytoplasm is absent, and while a few enzymes may be found, there are not enough to sustain cellular processes.
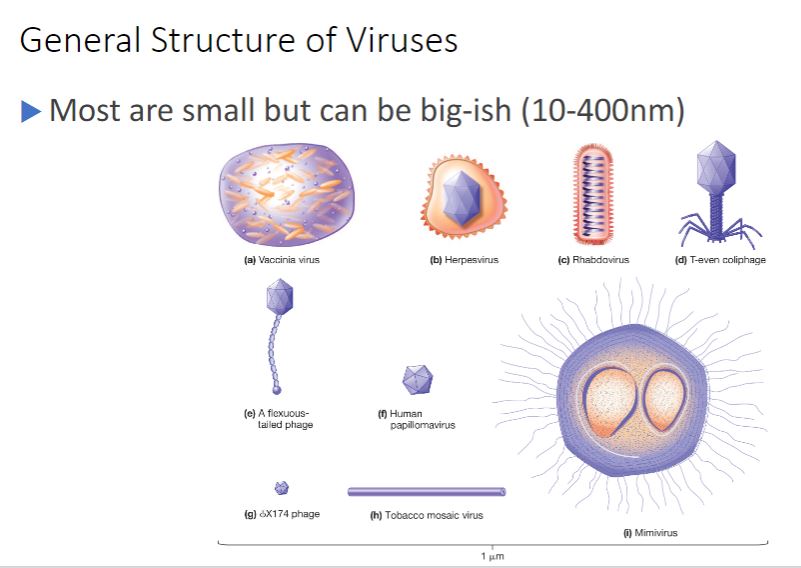
19
New cards
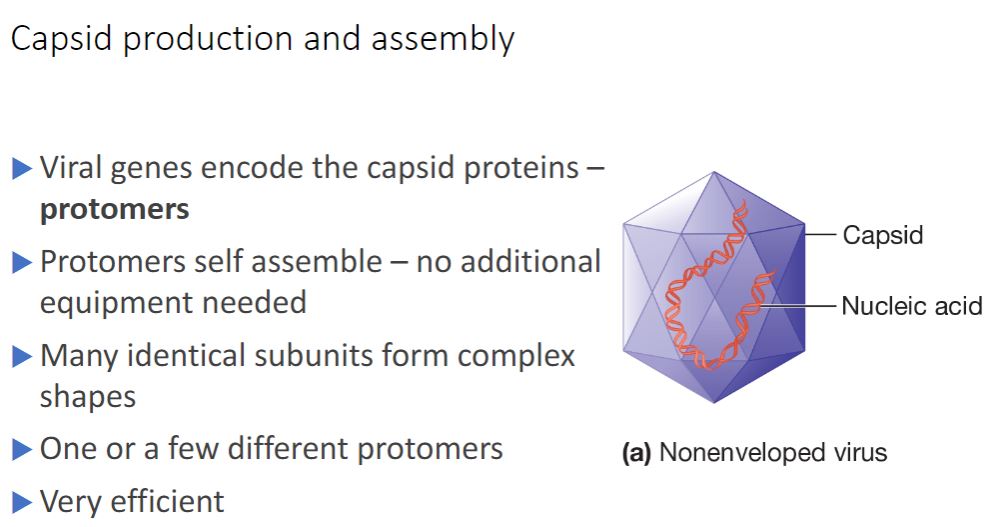
Non-enveloped
Nonenveloped viruses construct a capsid from many copies of one protein and a few minor proteins. Each subunit is termed a protomer, and thousands of protomers self- assemble to form the capsid (figure 18.3). In contrast, enveloped viruses require both nucleocapsid proteins and additional proteins to anchor the membrane. Some viruses use noncapsid proteins as scaffolding upon which the capsids are assembled. Probably the most important advantage of this design strategy is that the viral genome is used with maximum efficiency. For example, the tobacco mosaic virus (TMV) capsid is constructed using a single type of protomer. Recall that the building blocks of proteins are amino acids and that each amino acid is encoded by three nucleotides. The TMV protomer is 158 amino acids in length. Therefore only about 474 nucleotides are required to code for the coat protein. The entire TMV genome consists of only 6,400 nucleotides. Thus only a small fraction of the genome is used to code for the capsid.
- (image below)
Generalized Structure of Virions. (a) A nonenveloped
virus consists of a capsid assembled around its nucleic acid
(nucleocapsid).
- (image below)
Generalized Structure of Virions. (a) A nonenveloped
virus consists of a capsid assembled around its nucleic acid
(nucleocapsid).
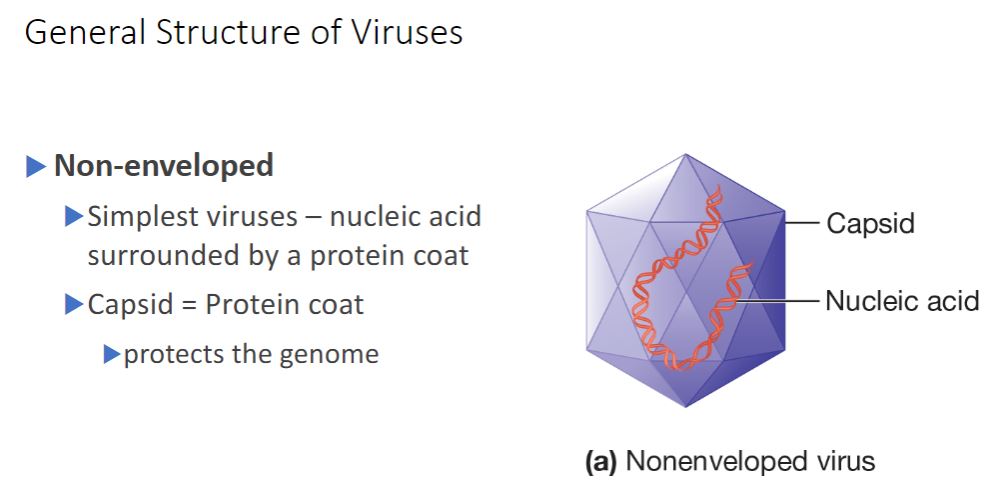
20
New cards
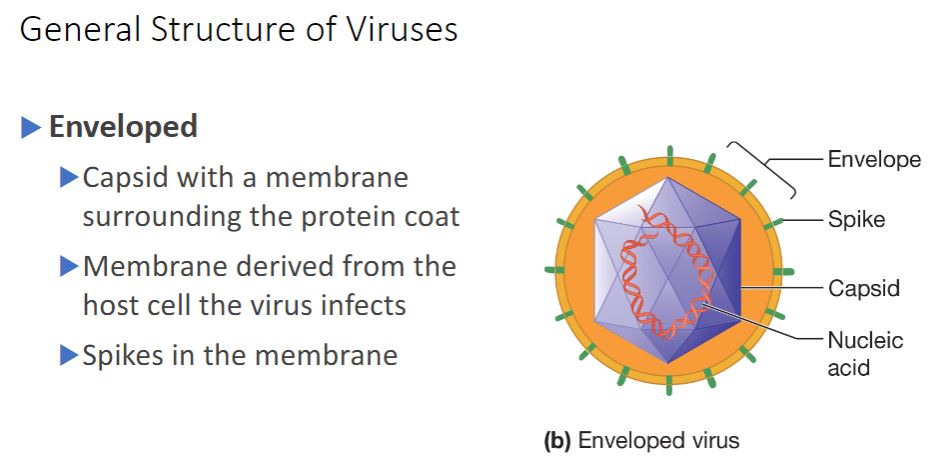
Enveloped
- (Image below)
(b) An enveloped virus is composed of a nucleocapsid has viral proteins called spikes inserted into it.
surrounded by a membrane called an envelope. The envelope usually
(b) An enveloped virus is composed of a nucleocapsid has viral proteins called spikes inserted into it.
surrounded by a membrane called an envelope. The envelope usually
21
New cards
Capsid production and assembly
refer to the previous card, as it is very brief
22
New cards
Tobacco Mosaic Virus ( Helical Capsids)
-Helical capsids are shaped like hollow tubes with protein walls. Tobacco mosaic virus is a well-studied example of helical capsid structure (figure 18.3). The self-assembly of TMV protomers into a helical arrangement produces a rigid tube. The capsid encloses an RNA genome, which is wound in a spiral and lies within a groove formed by the protein subunits.
The size of a helical capsid is influenced by both its protomers and the viral genome. The diameter of the capsid is a function of the size, shape, and interactions of the protomers. The length of the capsid appears to be determined by the size of the viral genome.
The size of a helical capsid is influenced by both its protomers and the viral genome. The diameter of the capsid is a function of the size, shape, and interactions of the protomers. The length of the capsid appears to be determined by the size of the viral genome.

23
New cards
Viral classification is complicated
a

24
New cards
Viral classification 2
Just know that there is a lot of diversity and not to memorize the image below.
25
New cards
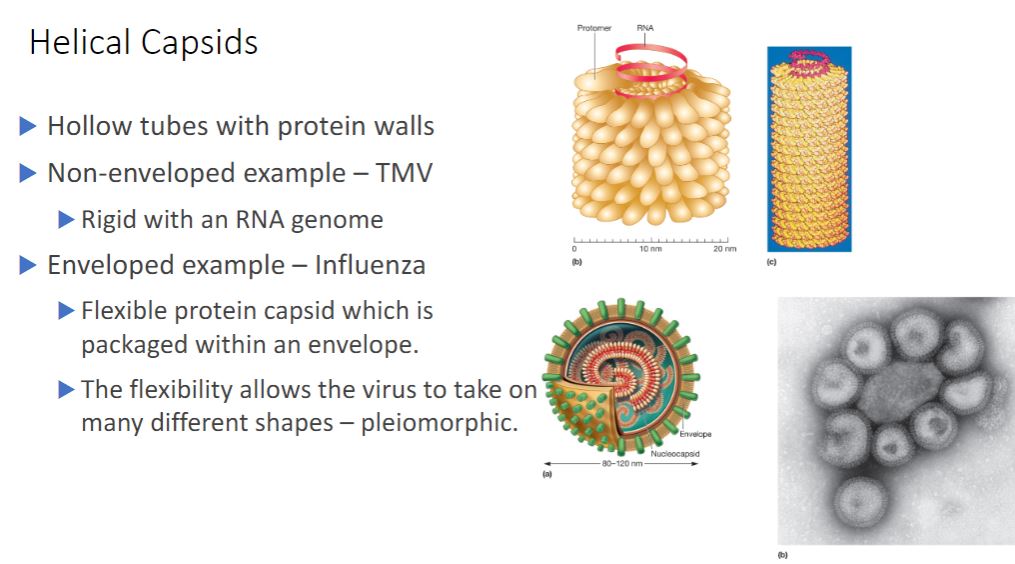
Helical Capsids
-Helical capsids are shaped like hollow tubes with protein walls. Tobacco mosaic virus is a well-studied example of helical capsid structure (figure 18.3). The self-assembly of TMV protomers into a helical arrangement produces a rigid tube. The capsid encloses an RNA genome, which is wound in a spiral and lies within a groove formed by the protein subunits.
The size of a helical capsid is influenced by both its protomers and the viral genome. The diameter of the capsid is a function of the size, shape, and interactions of the protomers. The length of the capsid appears to be determined by the size of the viral genome.
The size of a helical capsid is influenced by both its protomers and the viral genome. The diameter of the capsid is a function of the size, shape, and interactions of the protomers. The length of the capsid appears to be determined by the size of the viral genome.
26
New cards
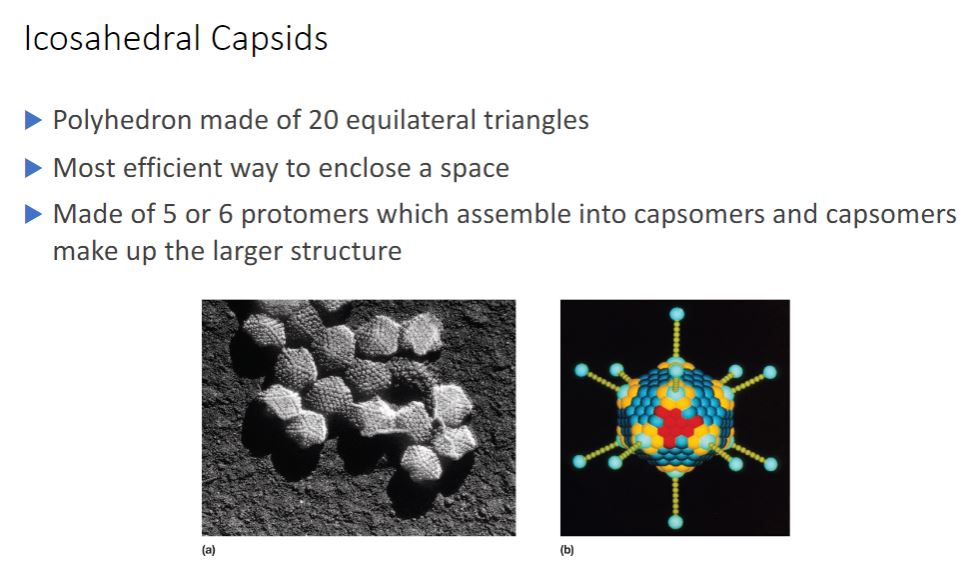
Icosahedral Capsids
-An icosahedron is a regular polyhedron with 20 equilateral triangular faces and 12 vertices (figure 18.1e). Icosahedral capsids are the most efficient way to enclose a space. They are constructed from assemblages of five or six protomers; the assemblages are called capsomers (figure 18.4). Capsomers composed of five protomers are called pentamers (pentons); hexamers (hexons) are capsomers that possess six protomers. Although many icosahedral capsids contain both pentamers and hexamers, some have only pentamers.
27
New cards
Complex symmetry capsids
-Most viruses have either icosahedral or helical capsids, but some viruses do not fit into either category. Poxviruses and large bacteriophages are two important examples.
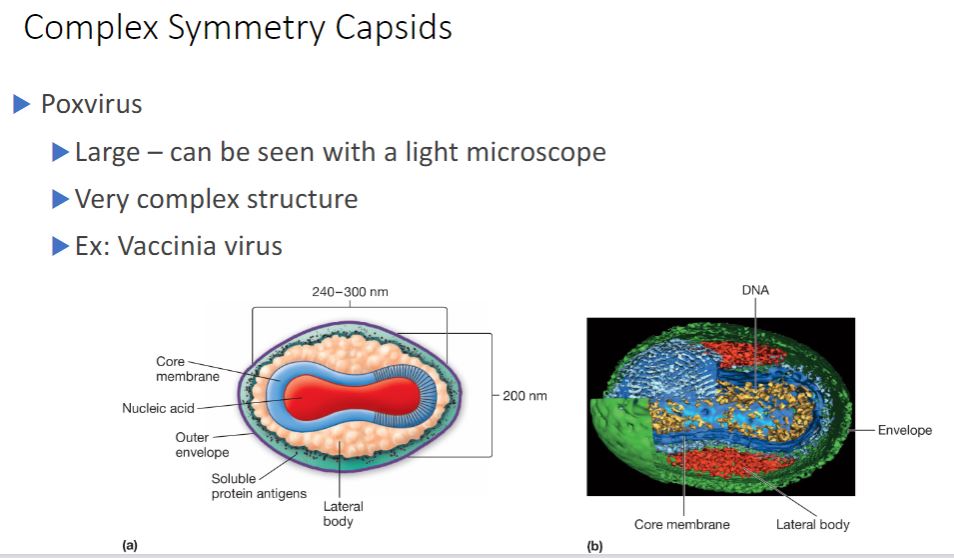
Poxvirus virions are animal viruses that measure about 400 x 240 x 200 nm in size and can be seen with a light microscope. They possess an exceptionally complex internal structure with an ovoid- to brick-shaped exterior. Figure 18.5 shows the virion morphology of the vaccinia virus.
Poxvirus virions are animal viruses that measure about 400 x 240 x 200 nm in size and can be seen with a light microscope. They possess an exceptionally complex internal structure with an ovoid- to brick-shaped exterior. Figure 18.5 shows the virion morphology of the vaccinia virus.
28
New cards
Bacteriophage
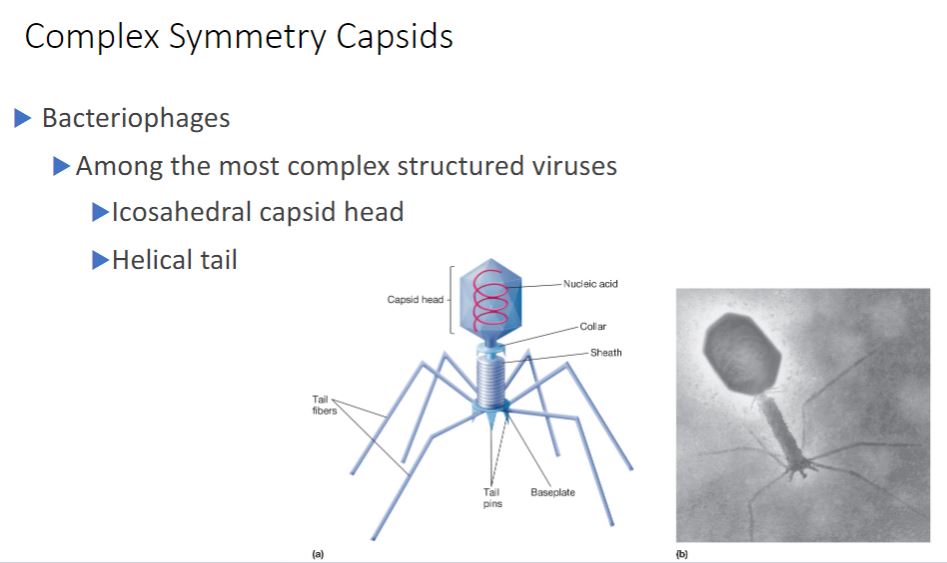
Some large bacteriophages have virions that are even more elaborate than those of poxviruses. The virions of T2, T4, and T6 phages (T-even phages) that infect Escherichia coli are said to have binal symmetry because they have a head that resembles an icosahedron and a tail that is helical. The icosahedral head is elongated by one or two rows of hexamers in the middle and contains the DNA genome (figure 18.6). The tail is composed of a collar joining it to the head, a central hollow tube, a sheath surrounding the tube, and a complex baseplate. In T-even phages, the baseplate is hexagonal and has a pin and a jointed long tail fiber and a short tail fiber at each corner.
29
New cards
Visual of bacteriophages
a
30
New cards
Viral descriptions
a
31
New cards
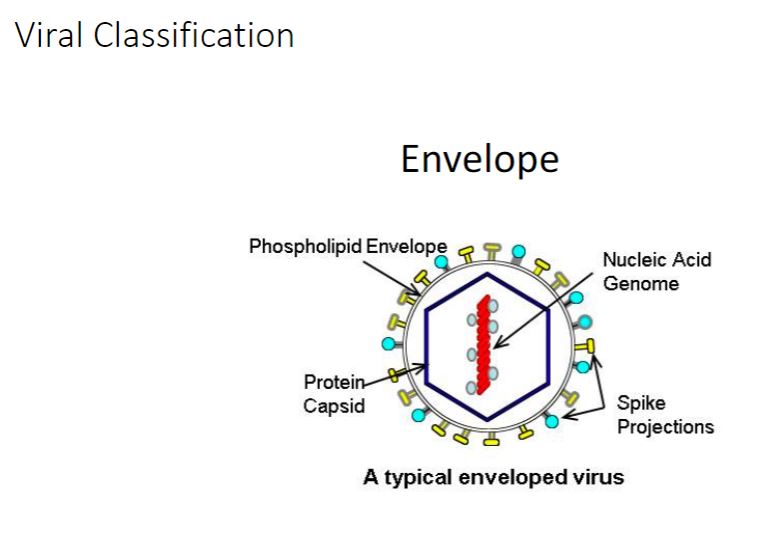
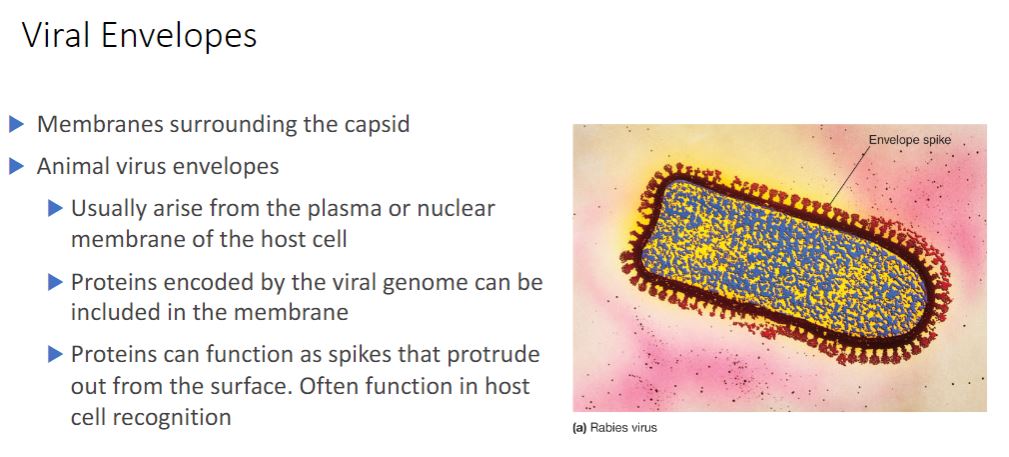
Viral envelopes
The nucleocapsids of many animal viruses, some plant viruses, and at least one bacterial
virus are surrounded by an outer membranous layer called an envelope (figure 18.26). Animal virus envelopes usually arise from the plasma or nuclear membranes of the host
cell. Envelope lipids and carbohydrates are therefore acquired from the host.
virus are surrounded by an outer membranous layer called an envelope (figure 18.26). Animal virus envelopes usually arise from the plasma or nuclear membranes of the host
cell. Envelope lipids and carbohydrates are therefore acquired from the host.
32
New cards
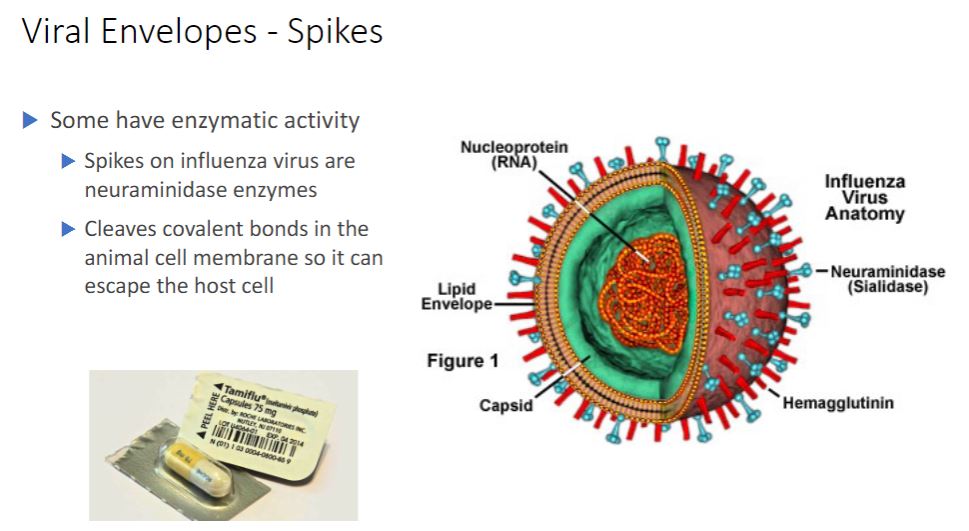
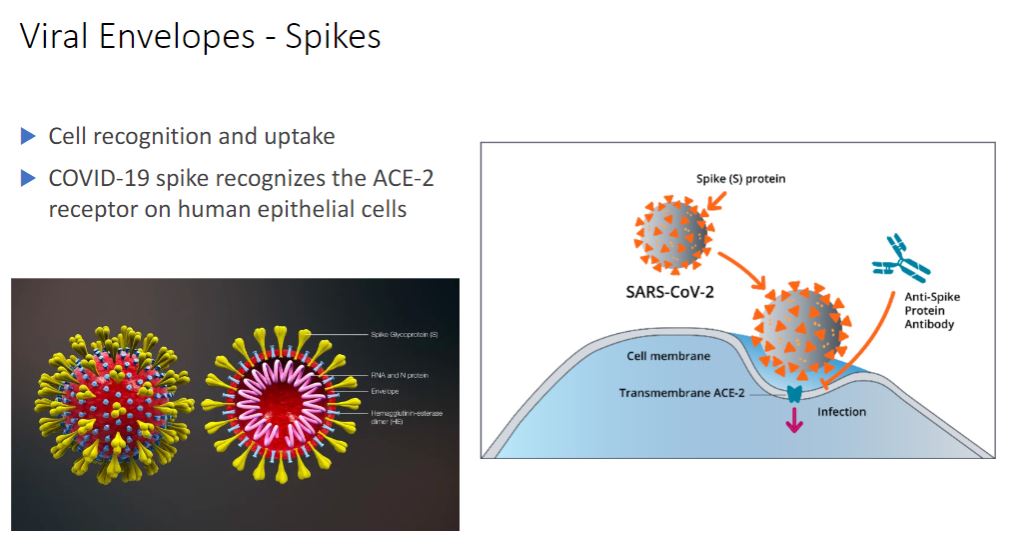
Spikes
-In contrast envelope proteins are encoded by viral genes and may even project from the envelope surface as spikes, which are also called peplomers. In many cases, spikes are involved in virion attachment to the host cell surface. Some surface proteins also have enzymatic activity needed for entry into or exit from the host cell. Because spikes differ among viruses, they also can be used to identify some viruses.
In addition to enzymes associated with the envelope or capsid, some viruses have enzymes within their capsids. Such enzymes are usually involved in nucleic acid replication. For example, influenza virus virions have an RNA genome and carry an enzyme that synthesizes RNA using an RNA template. Thus although viruses lack true metabolism and cannot reproduce independently of living cells, their virions may carry one or more enzymes essential to the completion of their life cycles.
- Just know that the spikes are what is interacting with the cells to either invade it inside or could spread to other cells. Now that the image below called Tamiflu is used to inhibit Neuraminidase, to prevent the further spreading to other cells but it's a small window as you only have 48 hrs but pregnant people can take it after the 48-hour window.
In addition to enzymes associated with the envelope or capsid, some viruses have enzymes within their capsids. Such enzymes are usually involved in nucleic acid replication. For example, influenza virus virions have an RNA genome and carry an enzyme that synthesizes RNA using an RNA template. Thus although viruses lack true metabolism and cannot reproduce independently of living cells, their virions may carry one or more enzymes essential to the completion of their life cycles.
- Just know that the spikes are what is interacting with the cells to either invade it inside or could spread to other cells. Now that the image below called Tamiflu is used to inhibit Neuraminidase, to prevent the further spreading to other cells but it's a small window as you only have 48 hrs but pregnant people can take it after the 48-hour window.
33
New cards
Spikes on COVID-19
a
34
New cards
Viral genomes
-One clear distinction between cellular organisms and viruses is the nature of their genomes. Cellular genomes are always double-stranded (ds) DNA. Viruses, on the other hand, employ all four possible nucleic acid types: dsDNA, single-stranded (ss) DNA, ssRNA, and dsRNA. All four types are used by animal viruses. Most plant viruses have ssRNA genomes, and most bacterial and archaeal viruses have dsDNA. The size of viral genomes also varies greatly. Very small genomes are around 4,000 nucleotides-just large enough to code for three or four proteins. Some viruses save additional space by using overlapping genes. At the other extreme are the genomes of pandoraviruses, which infect protists. They are about 2.5 x 10° nucleotides long, exceeding some bacteria and archaea in coding capacity.
Some RNA viruses have segmented genomes-genomes that consist of multiple pieces (segments) of RNA. In many cases, each segment codes for one protein and there may be as many as 10 to 12 segments. Usually all segments are enclosed in the same capsid.
Some RNA viruses have segmented genomes-genomes that consist of multiple pieces (segments) of RNA. In many cases, each segment codes for one protein and there may be as many as 10 to 12 segments. Usually all segments are enclosed in the same capsid.

35
New cards
Viral Genomes but specific
Know this!!!

36
New cards
Viral taxonomy
This is just telling that viruses are just under studied and that its very difficult to classify.

37
New cards
dsDNA Viruses
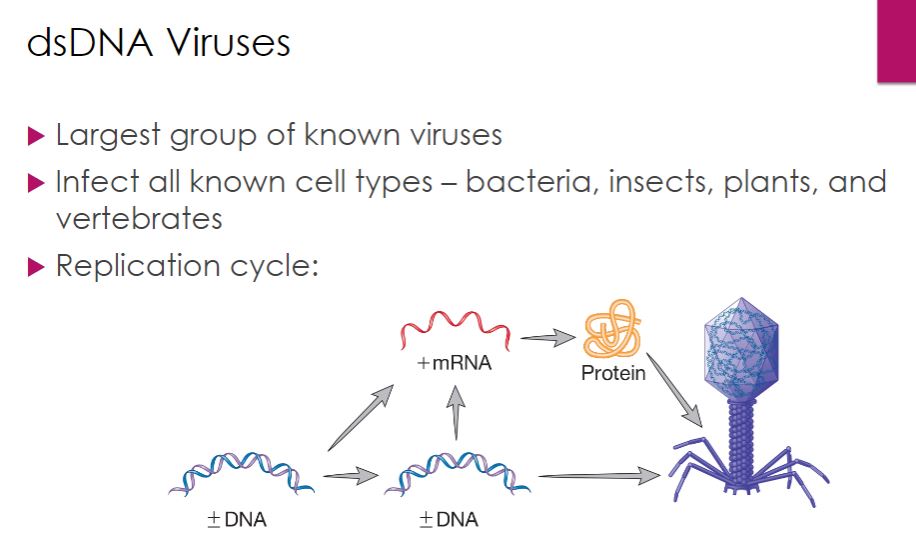
-Perhaps the largest group of known viruses is the double-stranded (ds) DNA viruses; most bacteriophages and archaeal viruses have dsDNA genomes, as do several insect viruses and a number of important vertebrate viruses, including herpesviruses and poxviruses. The pattern of multiplication for dsDNA viruses is shown in figure 18.19. The synthesis of DNA and RNA is similar to what occurs in cellular organisms; therefore many dsDNA viruses can rely entirely on their host's DNA and RNA polymerases.
38
New cards
Bacteriophage T4 ( dsDNA )
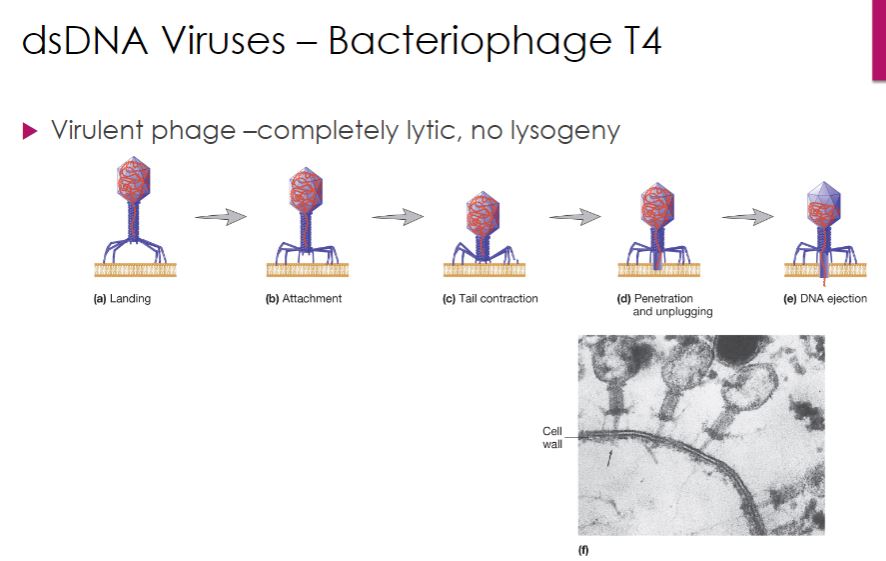
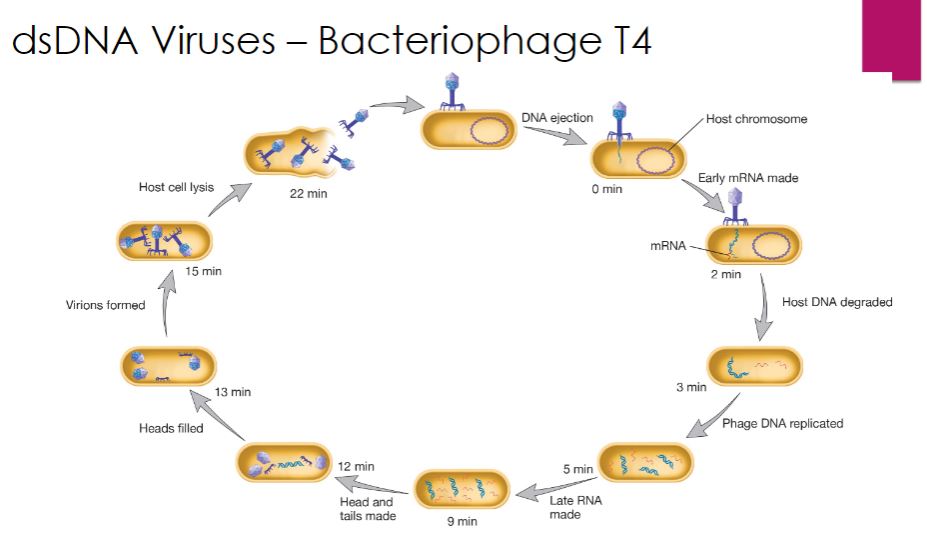
The life cycle of T4 bacteriophage (family Myoviridae, species Enterobacteria phage T4) serves as our example of a virulent (lytic) dsDNA phage. Cell lysis is the outcome of an infection with a lytic bacteriophage. T4 attachment begins when a long tail fiber contacts either the lipopolysaccharide or certain proteins in the outer membrane of its Escherichia coli host. This process is outlined in figure 18.20. Within 2 minutes after entry of T4 DNA into an E. coli cell, the E. coli RNA polymerase starts synthesizing T4 early mRNA. One of the first T4 genes to be expressed encodes a protein that binds to the host enzyme RNaseE and directs it to degrade host mRNA. This frees host ribonucleotides and ribosomes for transcription and translation of T4 genes. Within 5 minutes, viral DNA synthesis commences, catalyzed by a virus-encoded DNA-dependent DNA polymerase. DNA replication is initiated from several origins of replication and proceeds bidirectionally from each. Viral DNA replication is followed by the synthesis of late mRNAs, which are important in later stages of the infection.
-The linear dsDNA genome of T4 is generated in an interesting manner involving the formation of long DNA molecules called concatemers, which are composed of several genome units linked together in the same orientation (figure 18.22). This is made possible because each progeny viral DNA molecule has single-stranded 3' ends. These ends participate in homologous recombination with double-stranded regions of other progeny DNA molecules, generating concatemers. During assembly, concatemers are cleaved such that the genome packaged in the capsid is slightly longer than the T4 gene set. Thus each progeny virus has a genome unit that begins with a different gene and ends with the same set of genes. If each viral genome were circularized, the sequence of genes in each virion would be the same. Therefore the T4 genome is said to be terminally redundant and circularly permuted, and the genetic map of T4 is drawn as a circular molecule.
-The formation of new T4 phage particles is an exceptionally complex self-assembly process that involves viral proteins and some host cell factors (figure 18.11). A critical step in T4 virion construction is filling the head portion of the virion with the T4 genome. This is no simple matter-the dsDNA genome is somewhat rigid and has many negatively charged moieties. Therefore the dsDNA must be crammed into the capsid. This is accomplished by a complex of proteins sometimes called the "packasome." The T4 packasome has more power than an automobile engine.
The packasome includes a protein called terminase, which has two functions: to cut the concatemers formed during T4 genome replication and to coordinate the insertion of DNA into the T4 head. Terminase threads the end of the T4 genome through a portal, where it enters the phage head using energy supplied by ATP hydrolysis. The packasome has been proposed to move the DNA by causing a transition from B-form DNA to A- form DNA, and then reversing back to B-form DNA. This compresses the helix like a spring. When the phage head is filled with a DNA molecule roughly 3% longer than the length of one set of T4 genes, terminase then makes a second cut, and the packaging process for that head is complete. Terminase then leaves the head, and several other viral proteins bind at the portal through which the DNA entered. This seals the head and prepares it for addition of the tail and tail fibers.
Finally, virions are released so that they can infect new cells and begin the cycle anew. T4 lyses E. coli when about 150 virus particles have accumulated in the host cell. T4 encodes two proteins to accomplish this. The first, called holin, creates holes in the E. coli plasma membrane. The second, an endolysin called T4 lysozyme (distinct from the gp5 lysozyme used for bacteriophage entry), degrades peptidoglycan in the host's cell wall. Thus the activity of holin enables T4 lysozyme to move from the cytoplasm to the peptidoglycan so that both the plasma membrane and the cell wall are destroyed.
-The linear dsDNA genome of T4 is generated in an interesting manner involving the formation of long DNA molecules called concatemers, which are composed of several genome units linked together in the same orientation (figure 18.22). This is made possible because each progeny viral DNA molecule has single-stranded 3' ends. These ends participate in homologous recombination with double-stranded regions of other progeny DNA molecules, generating concatemers. During assembly, concatemers are cleaved such that the genome packaged in the capsid is slightly longer than the T4 gene set. Thus each progeny virus has a genome unit that begins with a different gene and ends with the same set of genes. If each viral genome were circularized, the sequence of genes in each virion would be the same. Therefore the T4 genome is said to be terminally redundant and circularly permuted, and the genetic map of T4 is drawn as a circular molecule.
-The formation of new T4 phage particles is an exceptionally complex self-assembly process that involves viral proteins and some host cell factors (figure 18.11). A critical step in T4 virion construction is filling the head portion of the virion with the T4 genome. This is no simple matter-the dsDNA genome is somewhat rigid and has many negatively charged moieties. Therefore the dsDNA must be crammed into the capsid. This is accomplished by a complex of proteins sometimes called the "packasome." The T4 packasome has more power than an automobile engine.
The packasome includes a protein called terminase, which has two functions: to cut the concatemers formed during T4 genome replication and to coordinate the insertion of DNA into the T4 head. Terminase threads the end of the T4 genome through a portal, where it enters the phage head using energy supplied by ATP hydrolysis. The packasome has been proposed to move the DNA by causing a transition from B-form DNA to A- form DNA, and then reversing back to B-form DNA. This compresses the helix like a spring. When the phage head is filled with a DNA molecule roughly 3% longer than the length of one set of T4 genes, terminase then makes a second cut, and the packaging process for that head is complete. Terminase then leaves the head, and several other viral proteins bind at the portal through which the DNA entered. This seals the head and prepares it for addition of the tail and tail fibers.
Finally, virions are released so that they can infect new cells and begin the cycle anew. T4 lyses E. coli when about 150 virus particles have accumulated in the host cell. T4 encodes two proteins to accomplish this. The first, called holin, creates holes in the E. coli plasma membrane. The second, an endolysin called T4 lysozyme (distinct from the gp5 lysozyme used for bacteriophage entry), degrades peptidoglycan in the host's cell wall. Thus the activity of holin enables T4 lysozyme to move from the cytoplasm to the peptidoglycan so that both the plasma membrane and the cell wall are destroyed.
39
New cards
The life cycle of Bacteriophage T4
a
40
New cards
Bacteria fights back
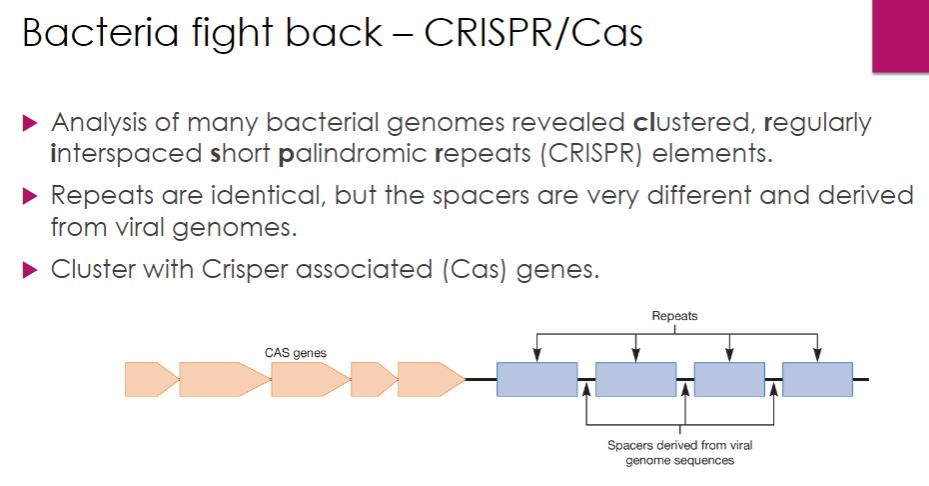
So the bacteria adaptive immunity. the space between the blue boxes contains DNA of teh bacteriophage and its information that is used to protect itself from the same/similar bacteriophage.
41
New cards
Bacteria fights back in details
a

42
New cards
Herpesviruses (dsDNA)
We now turn our attention to some important dsDNA viruses of eukaryotes, beginning
with herpesviruses, which include some familiar human pathogens (table 18.2). Herpesvirus virions are 150 to 200 nm in diameter, somewhat pleomorphic, and enveloped with spikes regularly dispersed over the surface. The envelope surrounds a layer of proteins called the tegument (Latin tegumentum, to cover), which in turn surrounds the nucleocapsid. Herpesvirus genomes are linear, about 125 to 295 kilobase pairs (kb) long, and encode 70 to over 200 proteins. When herpesviruses target cells of vertebrate hosts, some bind to epithelial cells, others to neurons. Host cell selection is mediated by the binding of envelope spikes to specific host cell surface receptors.
-Herpesviruses cause both productive infections and latent infections. In a productive infection, the virus multiplies explosively; between 50,000 and 200,000 new virions are produced from each infected cell. As the virus multiplies, the host cell's metabolism is inhibited and the host's DNA is degraded. As a result, the cell dies. The first exposure to a herpesvirus usually causes this type of infection. Some of the cells infected in the initial infection develop a latent infection. During the latent infection, virions cannot be detected. However, the virus can be reactivated in the host cells, leading to another productive infection. The viral genome remains in the host cell after reactivation; thus once infected, the host may experience repeated productive infections.
with herpesviruses, which include some familiar human pathogens (table 18.2). Herpesvirus virions are 150 to 200 nm in diameter, somewhat pleomorphic, and enveloped with spikes regularly dispersed over the surface. The envelope surrounds a layer of proteins called the tegument (Latin tegumentum, to cover), which in turn surrounds the nucleocapsid. Herpesvirus genomes are linear, about 125 to 295 kilobase pairs (kb) long, and encode 70 to over 200 proteins. When herpesviruses target cells of vertebrate hosts, some bind to epithelial cells, others to neurons. Host cell selection is mediated by the binding of envelope spikes to specific host cell surface receptors.
-Herpesviruses cause both productive infections and latent infections. In a productive infection, the virus multiplies explosively; between 50,000 and 200,000 new virions are produced from each infected cell. As the virus multiplies, the host cell's metabolism is inhibited and the host's DNA is degraded. As a result, the cell dies. The first exposure to a herpesvirus usually causes this type of infection. Some of the cells infected in the initial infection develop a latent infection. During the latent infection, virions cannot be detected. However, the virus can be reactivated in the host cells, leading to another productive infection. The viral genome remains in the host cell after reactivation; thus once infected, the host may experience repeated productive infections.
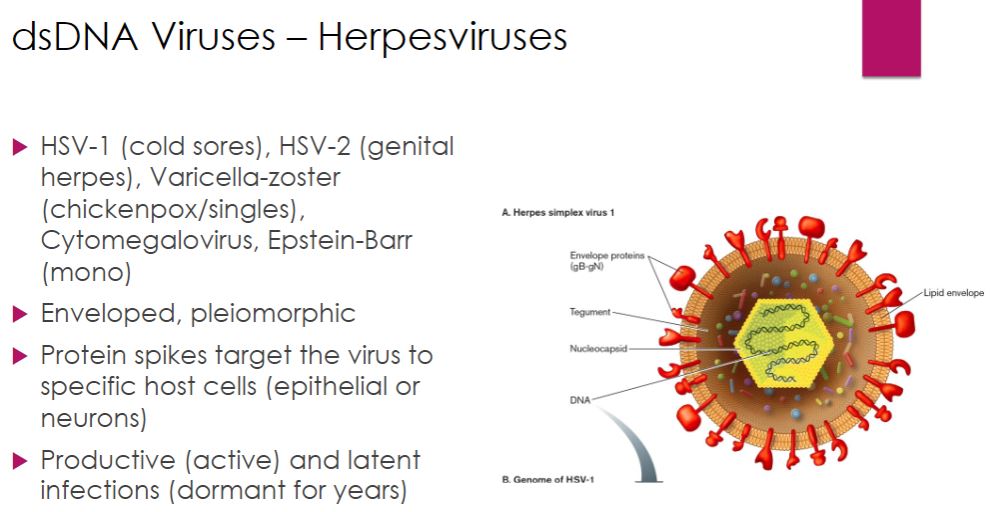
43
New cards
ssDNA Viruses
-Most DNA viruses are double-stranded, but several important viruses with single- stranded (ss) DNA genomes have been described. The life cycles of ssDNA viruses are similar to those of dsDNA viruses with one major exception. An additional step must occur in the synthesis stage because the ssDNA genome needs to be converted to a dsDNA molecule. A few ssDNA viruses are discussed next.
-Parvoviruses-
Parvoviruses (family Parvoviridae) infect numerous animal hosts, including crustaceans, dogs, cats, mice, and humans. Parvovirus virions are uniform, icosahedral, nonenveloped particles approximately 26 nm in diameter. Their genomes are composed of one ssDNA molecule of about 5,600 bases. Most of the genomes are negative-strand DNA molecules. That is, their sequence of nucleotides is complementary to that of the viral mRNA ( figure 18.18). Parvoviruses are among the simplest of the DNA viruses. The genome is so small that it directs the synthesis of only three proteins and some smaller polypeptides. None has enzymatic activity.
A productive parvovirus infection requires attachment to a particular cell type. Virions enter host cells by receptor-mediated endocytosis. Each virion escapes the endosome and is transported to the nucleus by the host cell's microtubules, directed by a nuclear localization signal on its capsid. The nucleocapsid enters the nucleus, followed by release of viral DNA from the capsid. Because the parvovirus genome does not code for any enzymes, the virus must use host cell enzymes for all biosynthetic processes. Viral infection causes arrest of the cell cycle in S phase, when the host cell can provide all of the components needed for viral replication. Because the genome is negative-strand DNA, it serves as the template for mRNA synthesis. Some of the RNA products encode polypeptides required for the interesting way the virus's genome is replicated. The ends of the parvovirus genome are palindromic sequences that can fold back on themselves. Formation of a complex composed of a viral protein and a hairpin at the 3' end of the genome provides the primer needed for replication (figure 18.36). This is recognized as a substrate by the host DNA polymerase, and DNA replication ensues by a process that is somewhat similar to rolling-circle replication. The parvovirus version of this replication method is often called rolling-hairpin replication. The process involves a dsDNA intermediate, much like $X174 (this is just symbols that I'm not bothering).
-Parvoviruses-
Parvoviruses (family Parvoviridae) infect numerous animal hosts, including crustaceans, dogs, cats, mice, and humans. Parvovirus virions are uniform, icosahedral, nonenveloped particles approximately 26 nm in diameter. Their genomes are composed of one ssDNA molecule of about 5,600 bases. Most of the genomes are negative-strand DNA molecules. That is, their sequence of nucleotides is complementary to that of the viral mRNA ( figure 18.18). Parvoviruses are among the simplest of the DNA viruses. The genome is so small that it directs the synthesis of only three proteins and some smaller polypeptides. None has enzymatic activity.
A productive parvovirus infection requires attachment to a particular cell type. Virions enter host cells by receptor-mediated endocytosis. Each virion escapes the endosome and is transported to the nucleus by the host cell's microtubules, directed by a nuclear localization signal on its capsid. The nucleocapsid enters the nucleus, followed by release of viral DNA from the capsid. Because the parvovirus genome does not code for any enzymes, the virus must use host cell enzymes for all biosynthetic processes. Viral infection causes arrest of the cell cycle in S phase, when the host cell can provide all of the components needed for viral replication. Because the genome is negative-strand DNA, it serves as the template for mRNA synthesis. Some of the RNA products encode polypeptides required for the interesting way the virus's genome is replicated. The ends of the parvovirus genome are palindromic sequences that can fold back on themselves. Formation of a complex composed of a viral protein and a hairpin at the 3' end of the genome provides the primer needed for replication (figure 18.36). This is recognized as a substrate by the host DNA polymerase, and DNA replication ensues by a process that is somewhat similar to rolling-circle replication. The parvovirus version of this replication method is often called rolling-hairpin replication. The process involves a dsDNA intermediate, much like $X174 (this is just symbols that I'm not bothering).
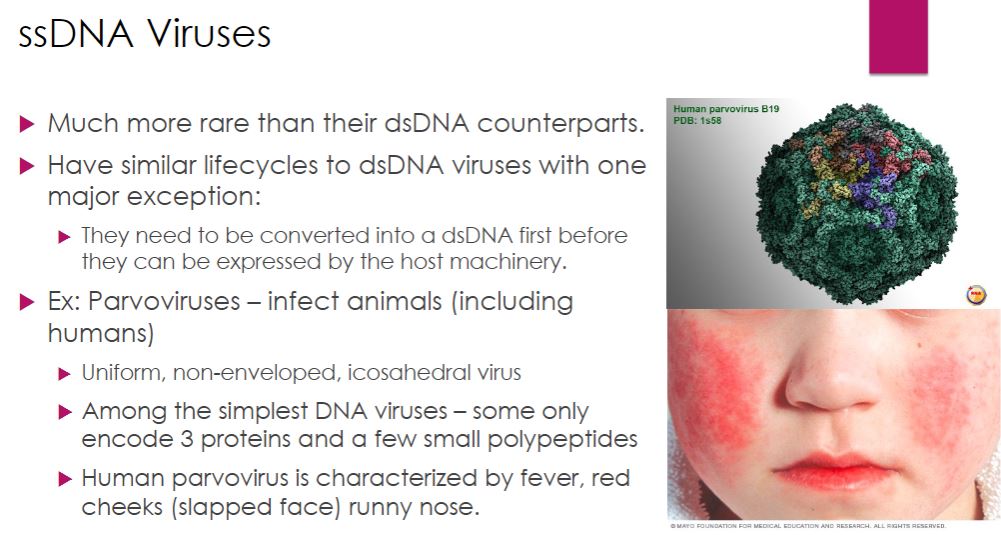
44
New cards
dsRNA question
a

45
New cards
dsRNA
-The Baltimore system divides viruses with RNA genomes into four groups (table 18.1). However, they all share the same dilemma: Their host cells have dsDNA genomes and thus lack a polymerase that can make RNA from an RNA template. Therefore RNA viruses classified in the Baltimore System as double-stranded must produce an enzyme called RNA-dependent RNA polymerase (RdRp). When the RdRp is used to replicate the viral RNA genome, it is often referred to as a replicase. When it is used to synthesize mRNA, the RdRp is often said to have transcriptase activity. In most cases, the same enzyme carries out both functions.
Among viruses with RNA genomes, those with dsRNA are uncommon. These viruses share a common multiplication strategy (figure 18.38). Here we discuss two representative dsRNA viruses: a bacteriophage and a vertebrate virus.
Among viruses with RNA genomes, those with dsRNA are uncommon. These viruses share a common multiplication strategy (figure 18.38). Here we discuss two representative dsRNA viruses: a bacteriophage and a vertebrate virus.
46
New cards
Rotaviruses (dsRNA)
-Human rotaviruses (family Reoviridae) are responsible for the vaccine preventable deaths of over 200,000 children worldwide each year. They cause severe diarrhea, which results in dehydration and death if appropriate therapy is not provided promptly. Because of their impact on humans, rotaviruses have been studied intensely to better understand their life cycles and pathogenesis.
Viewed by electron microscopy, rotavirus virions have a characteristic wheel-like appearance (Latin rota, wheel). Virions are nonenveloped and are composed of 11 segments of dsRNA surrounded by three concentric layers of proteins. The RNA segments code for six structural and six nonstructural proteins (NSPs).
When a rotavirus virion enters a host cell, it loses the outermost protein layer and is then referred to as a double-layered particle (DLP: figure 18.39). The genome is transcribed by viral transcriptase while still inside the DLP. The mRNA passes through channels in the DLP and is released into the cytosol of the host cell and translated by the host cell's ribosomes. The newly formed proteins cluster together, forming an inclusion called a viroplasm. It is within the viroplasm that new DLPs are formed. Initially the DLPs contain positive-strand RNA, but this is soon used as a template for synthesis of the negative strand. Replicase activity of the RdRp requires that the enzyme be in complex with the core shell protein. Thus the dsRNA molecules are synthesized within the developing DLP.
Viewed by electron microscopy, rotavirus virions have a characteristic wheel-like appearance (Latin rota, wheel). Virions are nonenveloped and are composed of 11 segments of dsRNA surrounded by three concentric layers of proteins. The RNA segments code for six structural and six nonstructural proteins (NSPs).
When a rotavirus virion enters a host cell, it loses the outermost protein layer and is then referred to as a double-layered particle (DLP: figure 18.39). The genome is transcribed by viral transcriptase while still inside the DLP. The mRNA passes through channels in the DLP and is released into the cytosol of the host cell and translated by the host cell's ribosomes. The newly formed proteins cluster together, forming an inclusion called a viroplasm. It is within the viroplasm that new DLPs are formed. Initially the DLPs contain positive-strand RNA, but this is soon used as a template for synthesis of the negative strand. Replicase activity of the RdRp requires that the enzyme be in complex with the core shell protein. Thus the dsRNA molecules are synthesized within the developing DLP.
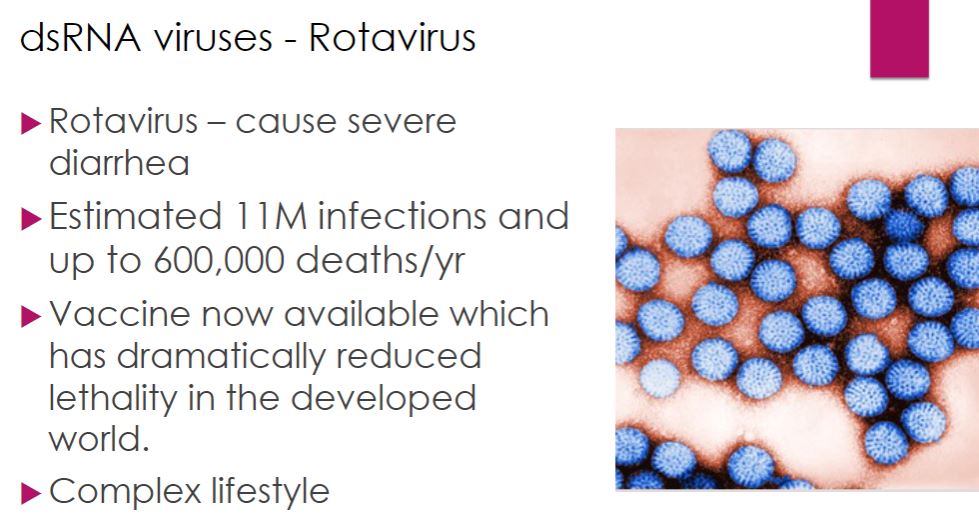
47
New cards
Types of ssRNA Viruses
a

48
New cards
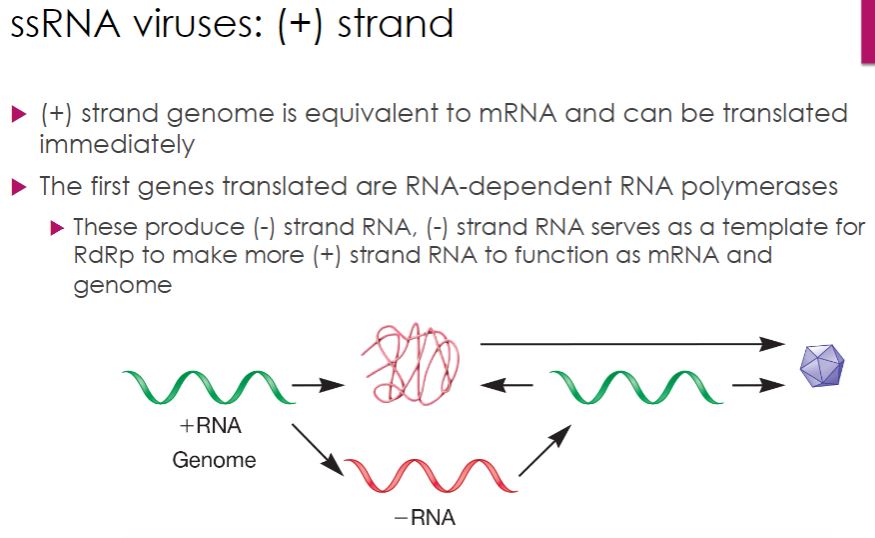
ssRNA: + Strand
-Positive (or plus) strand RNA viruses have genomes that act as mRNA and are therefore translated upon entry into the host cell. One of the first products is the RNA-dependent RNA polymerase, which catalyzes synthesis of negative (or minus) strand RNAs; these are then used to make more positive-strand RNAs (figure 18.40). In some cases, this occurs by way of a double-stranded RF, as seen with ssDNA viruses (section 18.8). In plant and animal viruses, viral genome replication and assembly of progeny virions occur in a structure formed within the cytoplasm called a replication complex (figure 18.10). Most plant viruses have positive-strand RNA genomes and there are a number of medically important positive-strand animal viruses, as we now discuss.
49
New cards
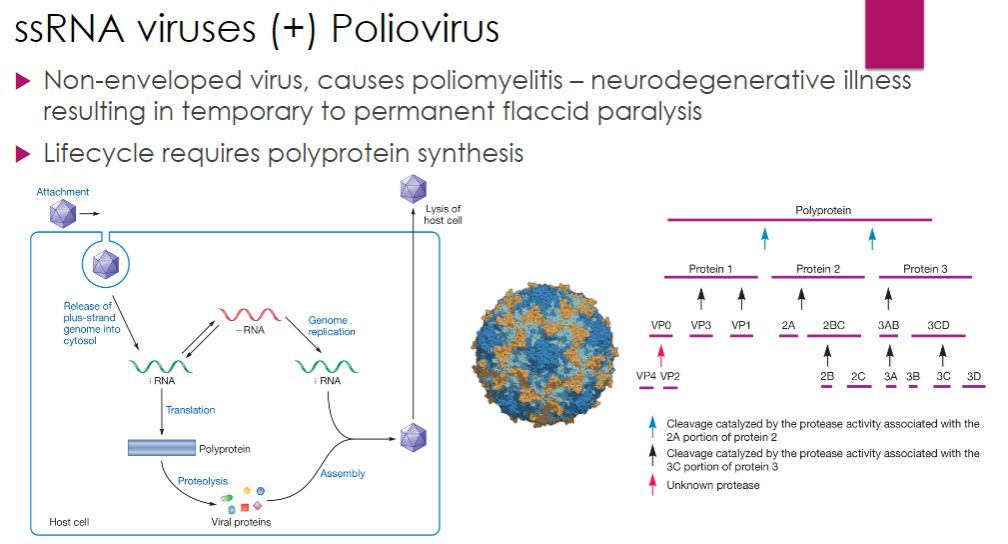
ssRNA: Poliovirus (+ strand)
-Picornaviruses include over 100 different enteroviruses; however, human Enterovirus C (commonly called poliovirus), the causative agent of poliomyelitis, is the most notable having caused disease in humans for centuries. It primarily targets children, with paralysis being the tragic result in some cases. The life cycle of poliovirus illustrates another strategy used by RNA viruses to ensure synthesis of needed proteins: synthesis of a polyprotein that is cleaved into individual proteins by a viral protease after translation.
Poliovirus enters the human host by ingestion. The virus multiplies in the mucosa of the throat or small intestine. From these sites, the virus invades the tonsils and lymph nodes of the neck and terminal portion of the small intestine. Generally there are either no symptoms or a brief illness characterized by fever, headache, sore throat, vomiting, and loss of appetite. The virus sometimes enters the bloodstream, causing viremia. In more than 99% of cases, the viremia is transient and clinical disease does not result. However, in a minority of cases, the viremia persists and the virus enters the central nervous system (CNS) and causes paralytic polio. It gains access to the CNS by attaching to a cell surface molecule called CD155, found on certain white blood cells and neurons.
Regardless of host cell type, the poliovirus nucleocapsid enters the host cell, and the positive-strand RNA genome is released into the cytosol while the virion is at the cell periphery and held within an endocytic vesicle. The genome acts as mRNA and is translated by host cell ribosomes. Like other viruses, polio virus RNA lacks the 5' cap found on eukaryotic mRNAs, which is important for ribosome binding. Poliovirus is among a group of viruses that "trick" the host into translating its capless RNA using a 5' region on the RNA called the internal ribosome entry site (IRES). In this region the ssRNA folds back on itself and forms extensive secondary structures (regions of dsRNA and numerous ssRNA loops), which are important for recognition of the RNA by ribosomes (figure 18.42). Other picornaviruses include rhinovirus, which has many strains that cause the common cold, and coxsackievirus, which causes hand, foot, and mouth disease in children.
Poliovirus enters the human host by ingestion. The virus multiplies in the mucosa of the throat or small intestine. From these sites, the virus invades the tonsils and lymph nodes of the neck and terminal portion of the small intestine. Generally there are either no symptoms or a brief illness characterized by fever, headache, sore throat, vomiting, and loss of appetite. The virus sometimes enters the bloodstream, causing viremia. In more than 99% of cases, the viremia is transient and clinical disease does not result. However, in a minority of cases, the viremia persists and the virus enters the central nervous system (CNS) and causes paralytic polio. It gains access to the CNS by attaching to a cell surface molecule called CD155, found on certain white blood cells and neurons.
Regardless of host cell type, the poliovirus nucleocapsid enters the host cell, and the positive-strand RNA genome is released into the cytosol while the virion is at the cell periphery and held within an endocytic vesicle. The genome acts as mRNA and is translated by host cell ribosomes. Like other viruses, polio virus RNA lacks the 5' cap found on eukaryotic mRNAs, which is important for ribosome binding. Poliovirus is among a group of viruses that "trick" the host into translating its capless RNA using a 5' region on the RNA called the internal ribosome entry site (IRES). In this region the ssRNA folds back on itself and forms extensive secondary structures (regions of dsRNA and numerous ssRNA loops), which are important for recognition of the RNA by ribosomes (figure 18.42). Other picornaviruses include rhinovirus, which has many strains that cause the common cold, and coxsackievirus, which causes hand, foot, and mouth disease in children.
50
New cards
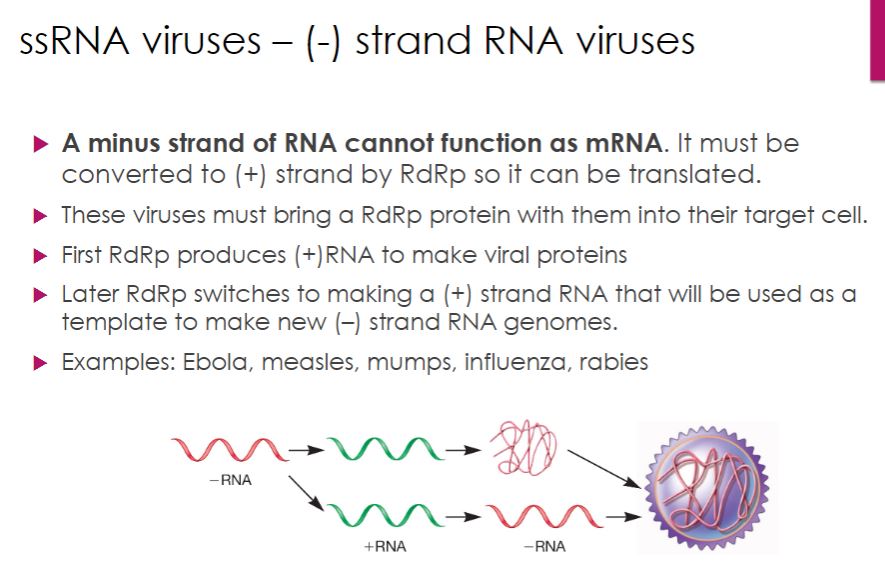
ssRNA: - strand
-Most negative (minus)-strand RNA viruses are enveloped viruses that vary in morphology from spherical, to filamentous, rod-shaped, bullet-shaped, and pleomorphic. Members of eight families have nonsegmented linear genomes and are grouped into the order Mononegavirales. The remaining negative-strand RNA viruses have segmented genomes that range from two to eight segments, each encoding usually one protein.
The genomes of negative-strand RNA viruses cannot function as mRNA. Therefore these viruses must bring at least one RNA-dependent RNA polymerase (RdRp) into the host cell during entry. Initially the viral genome serves as the template for mRNA synthesis (figure 18.44). Later the virus switches from mRNA synthesis to genome replication, as the RdRp synthesizes a distinct positive-strand RNA for replication. During this phase of the life cycle, the positive-strand RNA molecules synthesized from the negative-strand genome serve as templates for the manufacture of new negative-strand RNA genomes.
The genomes of negative-strand RNA viruses cannot function as mRNA. Therefore these viruses must bring at least one RNA-dependent RNA polymerase (RdRp) into the host cell during entry. Initially the viral genome serves as the template for mRNA synthesis (figure 18.44). Later the virus switches from mRNA synthesis to genome replication, as the RdRp synthesizes a distinct positive-strand RNA for replication. During this phase of the life cycle, the positive-strand RNA molecules synthesized from the negative-strand genome serve as templates for the manufacture of new negative-strand RNA genomes.
51
New cards
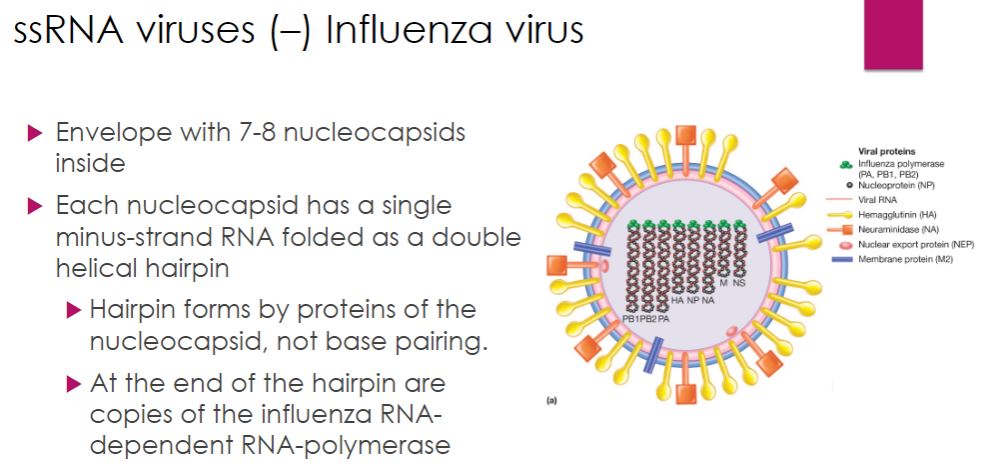
ssRNA: Influenza ( - Strand )
-Among the Orthomyxoviridae family, Influenzavirus A and B cause the human disease influenza (Italian un influenza di freddo, to be influenced by the cold). These influenza virus virions are composed of an envelope enclosing eight nucleocapsids. Each nucleocapsid consists of a single, negative-strand RNA that exists as a double helical hairpin (figure 18.45a). Formation and maintenance of the double helix do not involve base pairing. Rather, this is accomplished by numerous copies of the nucleocapsid protein (NP), which coat the RNA. The three polypeptides of the influenza RdRp are attached to the open end of the hairpin. The life cycle of the influenza virus is outlined in figure 18.45c.
52
New cards
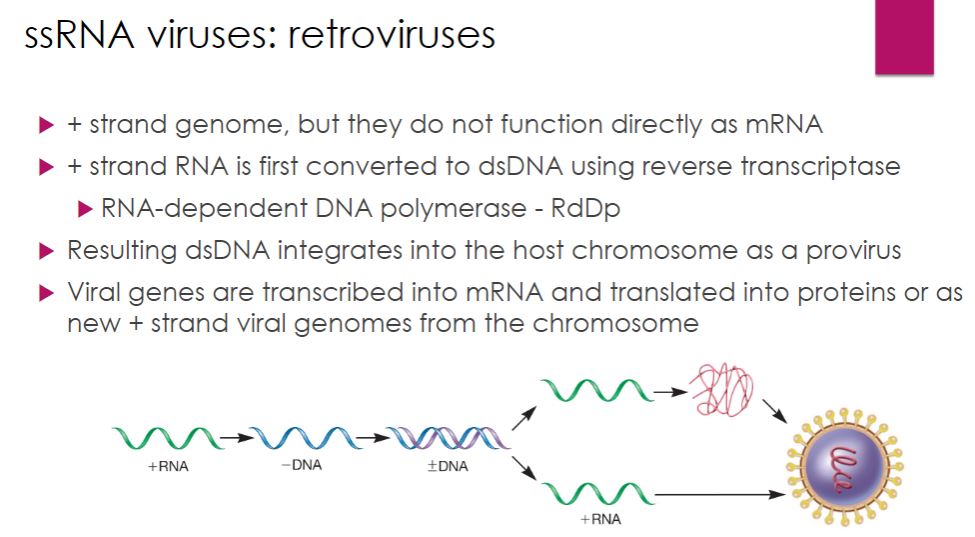
ssRNA: retroviruses
-Retroviruses have positive-strand RNA genomes. However, their genomes do not function as mRNA (figure 18.50). Instead, retroviruses first convert their ssRNA genomes into dsDNA using a multifunctional enzyme called reverse transcriptase (RT). The dsDNA is then integrated into the host's DNA, where it can serve as a template for mRNA synthesis and synthesis of the positive-strand RNA genome. The host cell's DNA- dependent RNA polymerase catalyzes both of these processes.
-Image below
Multiplication Strategy of Retroviruses. Retroviruses have a positive-strand RNA genome that is first converted into dsDNA by the enzyme reverse transcriptase. The viral dsDNA integrates into the host chromosome, where it serves as the template for synthesis of viral mRNA and viral genomes. Both are synthesized using the host cell's DNA-dependent RNA polymerase.
-Image below
Multiplication Strategy of Retroviruses. Retroviruses have a positive-strand RNA genome that is first converted into dsDNA by the enzyme reverse transcriptase. The viral dsDNA integrates into the host chromosome, where it serves as the template for synthesis of viral mRNA and viral genomes. Both are synthesized using the host cell's DNA-dependent RNA polymerase.
53
New cards
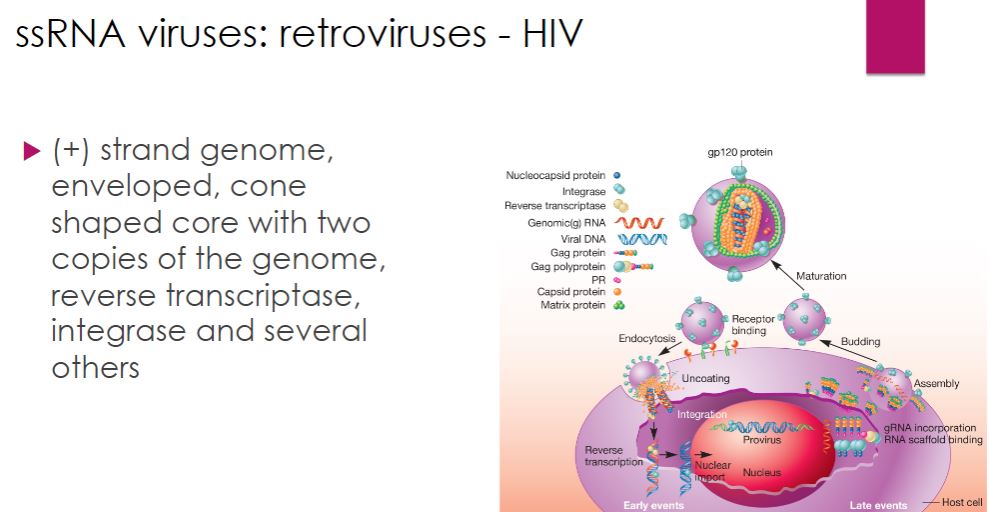
Retroviruses: HIV
-Many retroviruses have been identified and studied. However, human immunodeficiency virus (HIV), the cause of AIDS (acquired immune deficiency syndrome), is of particular interest. Because of its global importance, we focus exclusively on HIV in this section.
HIV is a member of the genus Lentivirus within the family Retroviridae. In the United States, HIV-1 is the predominant strain, while in Africa and Asia HIV-2 is prevalent. HIV is an enveloped virus. The envelope surrounds an outer shell, which encloses a cone- shaped core (figure 18.51). The core contains two copies of the 10,000-base HIV RNA genome and several enzymes, including reverse transcriptase and integrase. The genome possesses 9 genes that encode 15 virus-specific proteins.
HIV is a member of the genus Lentivirus within the family Retroviridae. In the United States, HIV-1 is the predominant strain, while in Africa and Asia HIV-2 is prevalent. HIV is an enveloped virus. The envelope surrounds an outer shell, which encloses a cone- shaped core (figure 18.51). The core contains two copies of the 10,000-base HIV RNA genome and several enzymes, including reverse transcriptase and integrase. The genome possesses 9 genes that encode 15 virus-specific proteins.
54
New cards
Retrovirus reverse transcriptase
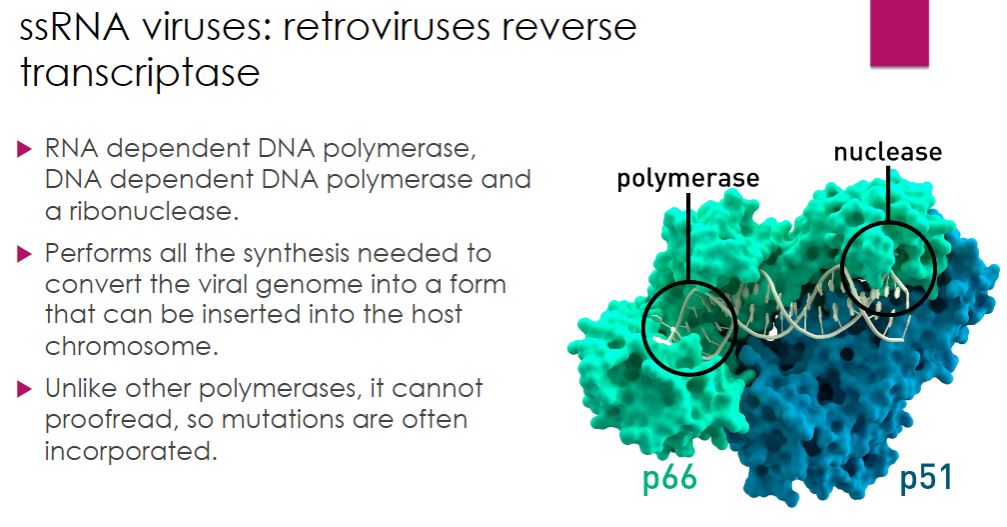
Example with HIV:
-Reverse transcription is a critical step in the life cycle of HIV, and RT is a remarkable enzyme with multiple activities. It is an RNA-dependent DNA polymerase, a DNA- dependent DNA polymerase, and a ribonuclease. Despite its versatility, RT lacks an important function observed in other DNA polymerases: proofreading. Thus it makes errors as it synthesizes DNA. This accounts for the rapid mutation rate seen in HIV.
-Reverse transcription is a critical step in the life cycle of HIV, and RT is a remarkable enzyme with multiple activities. It is an RNA-dependent DNA polymerase, a DNA- dependent DNA polymerase, and a ribonuclease. Despite its versatility, RT lacks an important function observed in other DNA polymerases: proofreading. Thus it makes errors as it synthesizes DNA. This accounts for the rapid mutation rate seen in HIV.
55
New cards
Summary of the genome of viruses
Know this

56
New cards
Microbial interaction
For most microbial ecologists, the nonobligatory aspect between host and symbiont differentiates cooperation from mutualism (Latin mutuus, borrowed or reciprocal) (figure 19.1). Unfortunately, it is often difficult to distinguish obligatory from nonobligatory because that which is obligatory in one habitat may not be in another (e.g., the laboratory). Nonetheless, the most useful distinction between cooperation and mutualism is the observation that cooperating organisms can grow independently, although they may not function as well.
Mutualism defines the relationship in which some reciprocal benefit accrues to both partners. This is an obligatory relationship in which the mutualist and the host are dependent on each other. In many cases, the individual organisms will not survive when separated. Several examples of mutualism are presented next.
-For many years bacterial symbioses were viewed as the relationship between one bacterial species and one host species. However, the wide application of metagenomics, single-cell sequencing, next-generation sequencing, and new approaches to culturing microbes have revealed that microbial symbioses can range from a single species of microorganism and its host to hosts with thousands of different kinds of microbes. In fact, there have been so many discoveries of new, and never imagined, symbiotic relationships that the vocabulary of microbial interactions is changing. For example, while the smaller organism in a symbiotic relationship has always been called the symbiont and the larger organism the host, these organisms are now sometimes regarded as partners. What was once considered a microbe-host relationship is now sometimes called a metaorganism, holobiont, or superorganism to reflect the critical nature of the interaction. Metagenomics provides access to uncultured microbes (section 32.3); Microbial biology relies on cultures (section 33.1)
All symbionts must undergo the process of colonization, reproduction, and persistence within the host, as well as transmission to a new host. Because microbial symbioses are the result of thousands, if not millions, of years of coevolution, these processes have been optimized. However, their specifics differ and make each symbiotic relationship seem like a clever feat of natural history. For example, host specificity can range from very narrow to very broad. Symbionts may be transmitted horizontally from host to host or vertically during reproduction, or each host may have to acquire the symbiont from the environment. Symbioses can range from those that are stable to interactions that are temporary, depending on the life cycle of the host or other variables. Symbionts can live within the host as an endosymbiont, or on the surface of the host as an ectosymbiont. Finally, symbionts alter not only the health of their hosts, but their behavior, evolution, and reproductive success as well
Mutualism defines the relationship in which some reciprocal benefit accrues to both partners. This is an obligatory relationship in which the mutualist and the host are dependent on each other. In many cases, the individual organisms will not survive when separated. Several examples of mutualism are presented next.
-For many years bacterial symbioses were viewed as the relationship between one bacterial species and one host species. However, the wide application of metagenomics, single-cell sequencing, next-generation sequencing, and new approaches to culturing microbes have revealed that microbial symbioses can range from a single species of microorganism and its host to hosts with thousands of different kinds of microbes. In fact, there have been so many discoveries of new, and never imagined, symbiotic relationships that the vocabulary of microbial interactions is changing. For example, while the smaller organism in a symbiotic relationship has always been called the symbiont and the larger organism the host, these organisms are now sometimes regarded as partners. What was once considered a microbe-host relationship is now sometimes called a metaorganism, holobiont, or superorganism to reflect the critical nature of the interaction. Metagenomics provides access to uncultured microbes (section 32.3); Microbial biology relies on cultures (section 33.1)
All symbionts must undergo the process of colonization, reproduction, and persistence within the host, as well as transmission to a new host. Because microbial symbioses are the result of thousands, if not millions, of years of coevolution, these processes have been optimized. However, their specifics differ and make each symbiotic relationship seem like a clever feat of natural history. For example, host specificity can range from very narrow to very broad. Symbionts may be transmitted horizontally from host to host or vertically during reproduction, or each host may have to acquire the symbiont from the environment. Symbioses can range from those that are stable to interactions that are temporary, depending on the life cycle of the host or other variables. Symbionts can live within the host as an endosymbiont, or on the surface of the host as an ectosymbiont. Finally, symbionts alter not only the health of their hosts, but their behavior, evolution, and reproductive success as well
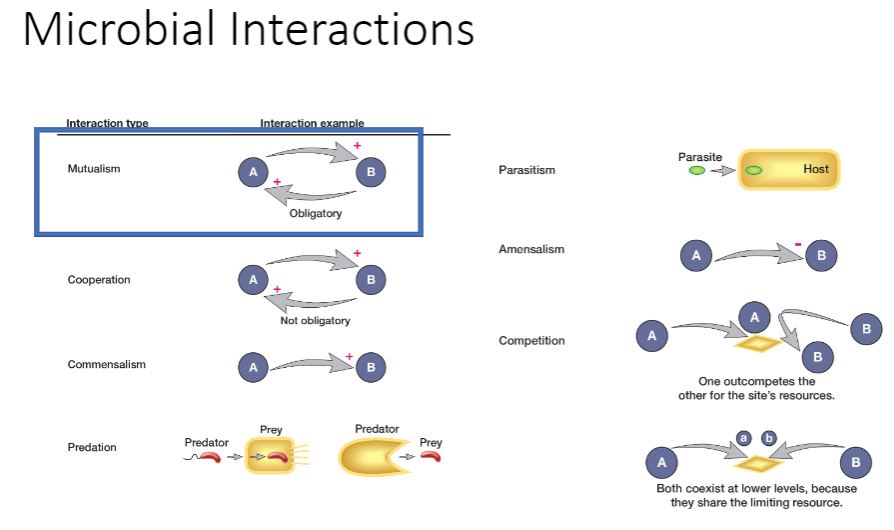
57
New cards
Insect Mutualism
-Mutualistic associations are common between microbes and insects partly because insects often consume plant sap or animal fluids lacking essential vitamins and amino acids. These are provided by bacterial symbionts in exchange for a secure habitat and ample nutrients. Aphids and their y-proteobacterium partner Buchnera aphidicola are a model system for the study of mutualistic symbioses (figure 19.2a). Strains of B. aphidicola provide their hosts with 10 essential amino acids absent in sap. If the insect is treated with antibiotics, it dies. Likewise, B. aphidicola is an obligate mutualistic symbiont. The inability of either partner to grow without the other indicates that the two organisms have evolved together. In fact, the pea aphid Acyrthosiphon pisum and its B. aphidicola symbionts share the pathway for the biosynthesis of several amino acids such that certain steps occur only in one organism.
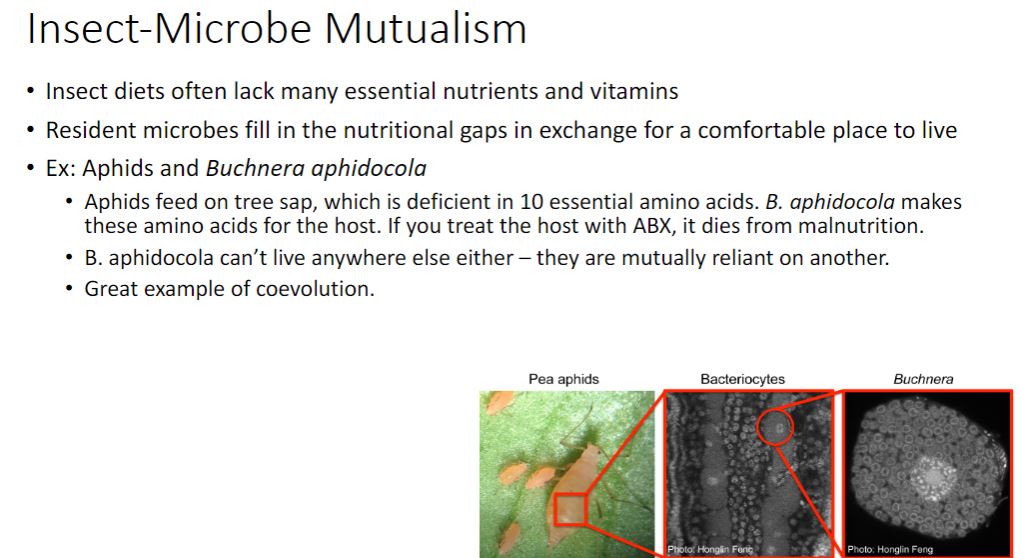
58
New cards
Insect mutualism with termites
- Termites are another good model system for the study of endosymbiosis. Some termites harbor protist, bacterial, and archaeal endosymbionts and eat only wood. The main structural polysaccharides of wood are cellulose and hemicellulose (glucose polymers), which combine with lignin (see figure 21.15) to form lignocellulose. A diet of wood poses two problems for the termite: how to degrade polysaccharides that may possess as many as 15,000 glucose monomers and where to get organic nitrogen needed for nucleotide and protein synthesis. Although termites produce a cellulolytic enzyme, only their mutualistic protists can complete lignocellulose degradation. Nitrogen-fixing bacteria that live in the termite gut solve the problem of obtaining organic nitrogen.◄ Nitrogen fixation section 8.5
Most termite-associated protists are members of the Excavata supergroup and as such are quite ancient, possessing hydrogenosomes rather than mitochondria. These protists ferment cellulose to acetate, CO₂, and H2. Acetate is the termite's preferred carbon source, and they rely on bacterial symbionts to convert the CO2 and H2 released by the protists to acetate by the reductive acetyl-CoA pathway (see figure 16.2). In addition, termites harbor methanogens that use these substrates to form CH4, which is excreted. Mitochondria related organelles and chloroplasts are involved in energy conservation section 4.6); Supergroup Excavata (section 17.2)
Some termite protists also harbor endosymbionts (i.e., endosymbionts of endosymbionts). For instance, Trichonympha spp. rely on a bacterial endosymbiont in the genus Elusimicrobium to convert glutamine to other amino acids and nitrogenous compounds (figure 19.26,c). In exchange, the protist supplies Elusimicrobium with glucose 6-phosphate, which can directly enter glycolysis. In addition, motility is often conferred by spirochetes that cover the protist surface. Motility is essential to prevent protist expulsion by the termite gut and to acquire food.
Most termite-associated protists are members of the Excavata supergroup and as such are quite ancient, possessing hydrogenosomes rather than mitochondria. These protists ferment cellulose to acetate, CO₂, and H2. Acetate is the termite's preferred carbon source, and they rely on bacterial symbionts to convert the CO2 and H2 released by the protists to acetate by the reductive acetyl-CoA pathway (see figure 16.2). In addition, termites harbor methanogens that use these substrates to form CH4, which is excreted. Mitochondria related organelles and chloroplasts are involved in energy conservation section 4.6); Supergroup Excavata (section 17.2)
Some termite protists also harbor endosymbionts (i.e., endosymbionts of endosymbionts). For instance, Trichonympha spp. rely on a bacterial endosymbiont in the genus Elusimicrobium to convert glutamine to other amino acids and nitrogenous compounds (figure 19.26,c). In exchange, the protist supplies Elusimicrobium with glucose 6-phosphate, which can directly enter glycolysis. In addition, motility is often conferred by spirochetes that cover the protist surface. Motility is essential to prevent protist expulsion by the termite gut and to acquire food.

59
New cards
Marine invertebrates
-Many marine invertebrates (sponges, jellyfish, sea anemones, corals, ciliated protists) harbor endosymbiotic dinoflagellates called zooxanthellae within their tissues. Because the degree of host dependency on the dinoflagellate is variable, only one well-known example is presented here. Alveolata (section 17.4)
The hermatypic (reef-building) corals satisfy most of their energy requirements using their zooxanthellae, which include members of the dinoflagellate genus Symbiodinium (figure 19.3a). These protists line the coral gastrodermal tissue at densities greater than 106 cells per square centimeter of coral animal. Symbiodinium cells give up to 95% of the carbon they fix in exchange for nitrogenous compounds, phosphates, CO₂, and protection from ultraviolet light from their coral hosts. In addition to dinoflagellates, corals host communities of bacteria, archaea, and protists in three habitats ( figure 19.36). The surface mucous layer supports the growth of nitrogen-fixing and chitin-degrading bacteria, while the animal tissue within the gastrodermal cavity and the coral skeleton feature other microbes. These microbial communities are responsible for extremely efficient nutrient cycling and tight coupling of trophic levels, which accounts for the stunning success of reef-building corals in developing vibrant ecosystems.
- During the past several decades, the number of coral bleaching events has increased dramatically. Coral bleaching is defined as a loss of either the photosynthetic pigments from the zooxanthellae or the complete expulsion of the dinoflagellates from the coral (figure 19.3c). Damage to photosystem II of the zooxanthellae generates reactive oxygen species (ROS); these ROS appear to directly damage many corals (recall that photosystem II uses water as the electron source, resulting in the evolution of oxygen). Coral bleaching appears to be caused by a variety of stressors, but one important factor is temperature. Temperature increases as small as 2°C above the average summer maxima can trigger coral bleaching. Sadly, evidence suggests that as many as one-third of corals and their zooxanthellae will be unable to evolve quickly enough to keep pace with predicted ocean warming if global climate change continues unchecked. Light reactions in oxygenic photosynthesis
The hermatypic (reef-building) corals satisfy most of their energy requirements using their zooxanthellae, which include members of the dinoflagellate genus Symbiodinium (figure 19.3a). These protists line the coral gastrodermal tissue at densities greater than 106 cells per square centimeter of coral animal. Symbiodinium cells give up to 95% of the carbon they fix in exchange for nitrogenous compounds, phosphates, CO₂, and protection from ultraviolet light from their coral hosts. In addition to dinoflagellates, corals host communities of bacteria, archaea, and protists in three habitats ( figure 19.36). The surface mucous layer supports the growth of nitrogen-fixing and chitin-degrading bacteria, while the animal tissue within the gastrodermal cavity and the coral skeleton feature other microbes. These microbial communities are responsible for extremely efficient nutrient cycling and tight coupling of trophic levels, which accounts for the stunning success of reef-building corals in developing vibrant ecosystems.
- During the past several decades, the number of coral bleaching events has increased dramatically. Coral bleaching is defined as a loss of either the photosynthetic pigments from the zooxanthellae or the complete expulsion of the dinoflagellates from the coral (figure 19.3c). Damage to photosystem II of the zooxanthellae generates reactive oxygen species (ROS); these ROS appear to directly damage many corals (recall that photosystem II uses water as the electron source, resulting in the evolution of oxygen). Coral bleaching appears to be caused by a variety of stressors, but one important factor is temperature. Temperature increases as small as 2°C above the average summer maxima can trigger coral bleaching. Sadly, evidence suggests that as many as one-third of corals and their zooxanthellae will be unable to evolve quickly enough to keep pace with predicted ocean warming if global climate change continues unchecked. Light reactions in oxygenic photosynthesis
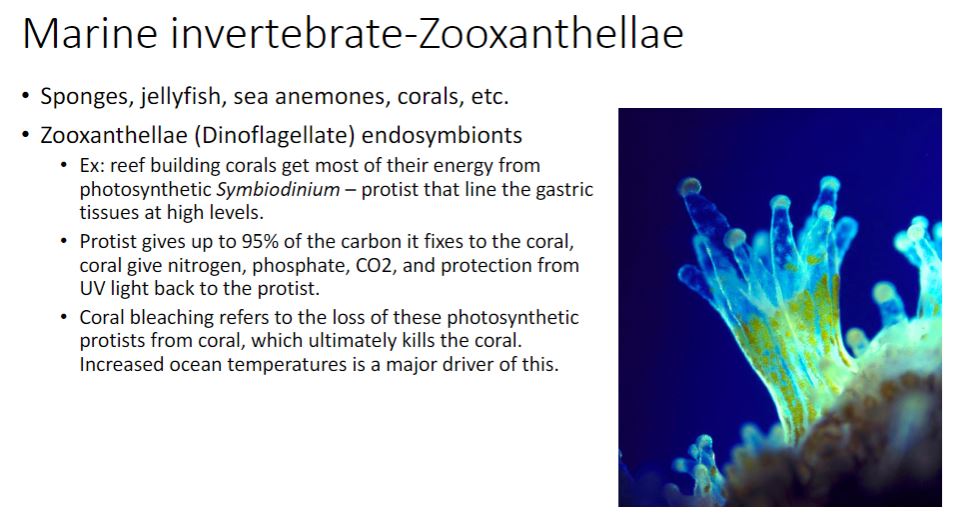
60
New cards
Sulfide - Mutualism
-Several thousand meters below the surface of the ocean, hydrothermal vents are found where the Earth's crustal plates are spreading apart (figure 19.4). Vent fluids are anoxic, contain high concentrations of hydrogen sulfide, and can reach a temperature of 350°C. The seawater surrounding these vents has sulfide concentrations around 250 μM and temperatures 10° to 20°C above the ambient seawater temperature of about 2°C. Although the high pressure at such depths prevents the water from boiling, one might think these conditions are inhospitable to life. But nothing could be further from the truth.
Giant red, gutless tube worms (Riftia spp., figure 19.5a) growing near hydrothermal vents exemplify a remarkably successful mutualism in which bacterial endosymbionts are maintained within specialized cells of the tube worm host (figure 19.5b, c, d). Riftia worms lack a mouth and a gut and rely on endosymbiotic bacteria to provide organic carbon. The bacteria are chemolithotrophic, using sulfide as an electron donor and oxygen as a terminal electron acceptor. In fact, Riftia worms have unique hemoglobin that provides both reduced sulfur and oxygen to the endosymbionts. Hydrogen sulfide (H₂S) and O₂ are removed from the seawater by the worm's
hemoglobin and delivered to an organ called the trophosome. The trophosome is packed with bacteriocytes, specialized cells that contain Candidatus Endoriftia persephone. These endosymbionts benefit from the stability provided by the worm in a turbulent
environment, and may reach densities of up to 1011 cells per gram of worm tissue. The endosymbionts use the Calvin-Benson cycle to fix CO₂ (see figure 8.4), which they reduce with electrons provided by H₂S. The CO₂ is carried to the endosymbionts in three ways: (1) freely in the bloodstream, (2) bound to hemoglobin, and (3) as organic acids such as malate and succinate. When these acids are decarboxylated, they release CO₂. This CO₂ is then fixed using the same pathway used by plants and cyanobacteria. This mutualism enables Riftia worms to grow to an astounding size (up to 2.5 m) in densely packed communities.
Giant red, gutless tube worms (Riftia spp., figure 19.5a) growing near hydrothermal vents exemplify a remarkably successful mutualism in which bacterial endosymbionts are maintained within specialized cells of the tube worm host (figure 19.5b, c, d). Riftia worms lack a mouth and a gut and rely on endosymbiotic bacteria to provide organic carbon. The bacteria are chemolithotrophic, using sulfide as an electron donor and oxygen as a terminal electron acceptor. In fact, Riftia worms have unique hemoglobin that provides both reduced sulfur and oxygen to the endosymbionts. Hydrogen sulfide (H₂S) and O₂ are removed from the seawater by the worm's
hemoglobin and delivered to an organ called the trophosome. The trophosome is packed with bacteriocytes, specialized cells that contain Candidatus Endoriftia persephone. These endosymbionts benefit from the stability provided by the worm in a turbulent
environment, and may reach densities of up to 1011 cells per gram of worm tissue. The endosymbionts use the Calvin-Benson cycle to fix CO₂ (see figure 8.4), which they reduce with electrons provided by H₂S. The CO₂ is carried to the endosymbionts in three ways: (1) freely in the bloodstream, (2) bound to hemoglobin, and (3) as organic acids such as malate and succinate. When these acids are decarboxylated, they release CO₂. This CO₂ is then fixed using the same pathway used by plants and cyanobacteria. This mutualism enables Riftia worms to grow to an astounding size (up to 2.5 m) in densely packed communities.
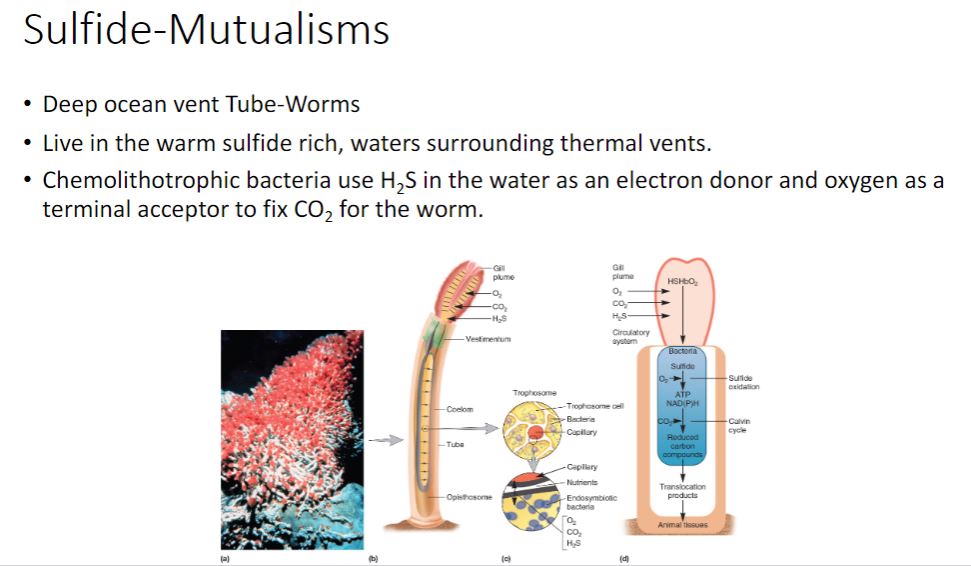
61
New cards
Cooperation
-So far our examples have featured symbionts that promote the growth of hosts in
exchange for a safe, nutrient-rich home. The example of the Gram-negative bacterium Xenorhabdus nematophila and its nematode (worm) host Steinernema carpocapsae is remarkable because the bacterium contributes directly to the reproductive success of its host. Juvenile nematodes harboring X nematophila within their guts live in the soil. To mature, the juvenile must find an insect to infect and consume; this is when things get really interesting (figure 19.7). As the hungry juvenile consumes insect blood (haemolymph), X. nematophila is excreted in its feces. X. nematophila is now free-living within the insect, and as the bacteria replicate, they use a type III secretion system to secrete enzymes to kill the insect. Once the insect is dead, the bacteria switch to the production of different compounds that protect the insect from degradation by other bacteria and from attack by ants, thereby protecting the home of their host nematode. Amazingly, as the process continues, X nematophila produces yet a different set of molecular signals that trigger S. carpocapsae development to adulthood. A single insect cadaver may host many adult nematodes that mate, yielding a cadaver that ultimately teems with S. carpocapsae eggs. As these eggs mature to the juvenile stage with X. nematophila symbionts, they leave to find new insect homes and start the cycle anew. Because the partners can be grown separately in the laboratory, it has been possible to dissect these events at the structural, molecular, and biochemical level, making this an
important model system.
exchange for a safe, nutrient-rich home. The example of the Gram-negative bacterium Xenorhabdus nematophila and its nematode (worm) host Steinernema carpocapsae is remarkable because the bacterium contributes directly to the reproductive success of its host. Juvenile nematodes harboring X nematophila within their guts live in the soil. To mature, the juvenile must find an insect to infect and consume; this is when things get really interesting (figure 19.7). As the hungry juvenile consumes insect blood (haemolymph), X. nematophila is excreted in its feces. X. nematophila is now free-living within the insect, and as the bacteria replicate, they use a type III secretion system to secrete enzymes to kill the insect. Once the insect is dead, the bacteria switch to the production of different compounds that protect the insect from degradation by other bacteria and from attack by ants, thereby protecting the home of their host nematode. Amazingly, as the process continues, X nematophila produces yet a different set of molecular signals that trigger S. carpocapsae development to adulthood. A single insect cadaver may host many adult nematodes that mate, yielding a cadaver that ultimately teems with S. carpocapsae eggs. As these eggs mature to the juvenile stage with X. nematophila symbionts, they leave to find new insect homes and start the cycle anew. Because the partners can be grown separately in the laboratory, it has been possible to dissect these events at the structural, molecular, and biochemical level, making this an
important model system.
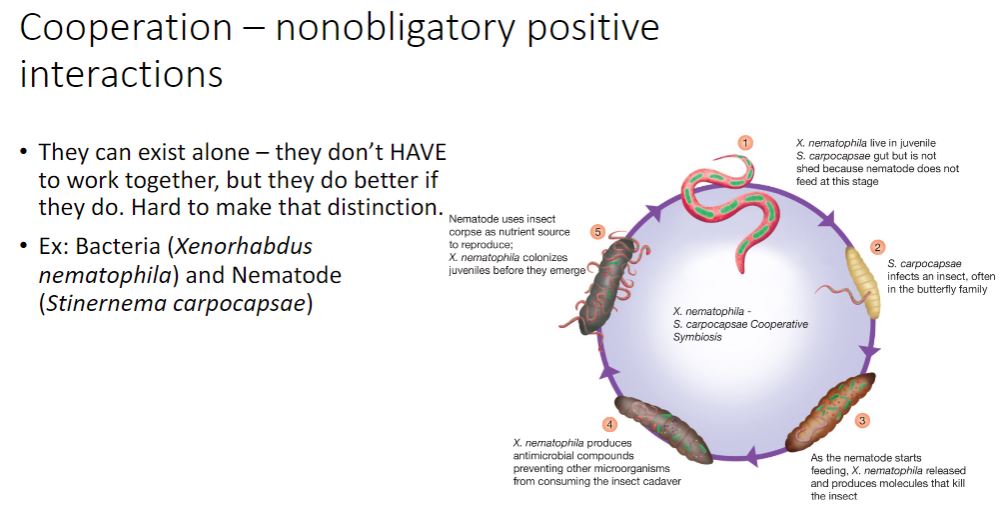
62
New cards
commensalism
Unidirectional symbiotic associations benefit only one partner. These may have either little effect on the other (commensalism) or a negative effect on the other (amensalism).
Commensalism (Latin com, together, mensa, table) is a relationship in which one symbiont, the commensal, benefits, while the host is neither harmed nor helped, as shown in figure 19.1. The spatial proximity of the two partners permits the commensal to feed on substances captured or ingested by the host, and the commensal often obtains shelter by living either on or in the host. The commensal is not directly dependent on the host's metabolism, so when it is separated from the host experimentally, it can survive without the addition of factors of host origin.
Commensalistic associations also occur when one microbial group modifies the environment to make it better suited for another organism. For instance, the synthesis of acidic waste products during fermentation stimulates the proliferation of more acid- tolerant microorganisms, which may be only a minor part of the microbial community at neutral pH. Biofilm formation is a good example of the unidirectional interaction that defines commensalism. The colonization of a newly exposed surface by one type of microorganism (an initial colonizer) makes it possible for other microorganisms to attach to the microbially modified surface. Biofilms are common in nature ( section 5.6)
The microorganisms that live in and on humans, known as our microbiome, have traditionally been called commensals. Although this term is still applied broadly to these microbes, it is important to keep in mind that the more we learn about the vast microbial ecology of our bodies, the more we come to understand that we depend on our microbes for a myriad of important functions that we cannot achieve without them. These include the synthesis of vitamins, maturation of our immune systems, maintaining a healthy body weight, and prevention of and recovery from disease. The term commensals does not apply to our microbiome, and it is being replaced by the more appropriate term, mutualists.
Commensalism (Latin com, together, mensa, table) is a relationship in which one symbiont, the commensal, benefits, while the host is neither harmed nor helped, as shown in figure 19.1. The spatial proximity of the two partners permits the commensal to feed on substances captured or ingested by the host, and the commensal often obtains shelter by living either on or in the host. The commensal is not directly dependent on the host's metabolism, so when it is separated from the host experimentally, it can survive without the addition of factors of host origin.
Commensalistic associations also occur when one microbial group modifies the environment to make it better suited for another organism. For instance, the synthesis of acidic waste products during fermentation stimulates the proliferation of more acid- tolerant microorganisms, which may be only a minor part of the microbial community at neutral pH. Biofilm formation is a good example of the unidirectional interaction that defines commensalism. The colonization of a newly exposed surface by one type of microorganism (an initial colonizer) makes it possible for other microorganisms to attach to the microbially modified surface. Biofilms are common in nature ( section 5.6)
The microorganisms that live in and on humans, known as our microbiome, have traditionally been called commensals. Although this term is still applied broadly to these microbes, it is important to keep in mind that the more we learn about the vast microbial ecology of our bodies, the more we come to understand that we depend on our microbes for a myriad of important functions that we cannot achieve without them. These include the synthesis of vitamins, maturation of our immune systems, maintaining a healthy body weight, and prevention of and recovery from disease. The term commensals does not apply to our microbiome, and it is being replaced by the more appropriate term, mutualists.

63
New cards
Predation
-Antagonistic interactions include killing (predation), exploitation (parasitism), and competition. Those that obtain biochemical precursors and energy after the victim is killed are predators; parasites benefit while the victim is alive. Competition is best described as an uneasy truce between the microbes involved. We present examples of each.
Microbial predators can be classified according to their predation strategy. Epibiotic predators attach to their prey organism's surface. Vampirococcus cells (figure 19.9a) attach to the outer membrane and secrete degradative enzymes that result in lysis and the release of the prey's cytoplasmic contents. Endobiotic predators invade the victim's cytoplasm or periplasm where they consume the contents to obtain the energy and precursors needed for cell division. Daptobacter (figure 19.9b) and Bdellovibrio (see figure 14.30) use this predation strategy. Order Bdellovibrionales includes predators that invade other Gram-negative bacteria.
-In contrast to the predatory bacteria shown in figure 19.9, some bacteria are facultative predators. Sometimes they consume organic matter released from dead organisms, whereas at other times, they actively prey on other microbes. Two examples are members of the y-proteobacterial genus Lysobacter and the &-proteobacterial genus Myxococcus. Both display predation whereby populations of cells use gliding motility to creep toward and cover their prey while releasing an arsenal of degradative enzymes. M. xanthus fruiting body formation can be triggered when its prey has been exhausted.
-Importantly, predation has many beneficial effects, especially when one considers predators and prey at the population and community levels. Ingestion and assimilation of a prey bacterium can lead to increased rates of nutrient cycling, critical for the functioning of the microbial loop (see figure 21.3). Although here we focus on bacterial predators, the impact of viral predation on communities, ecosystems, and even the global carbon budget is vast. As such, it is an active area of research. Marine viruses: mortality at sea
Microbial predators can be classified according to their predation strategy. Epibiotic predators attach to their prey organism's surface. Vampirococcus cells (figure 19.9a) attach to the outer membrane and secrete degradative enzymes that result in lysis and the release of the prey's cytoplasmic contents. Endobiotic predators invade the victim's cytoplasm or periplasm where they consume the contents to obtain the energy and precursors needed for cell division. Daptobacter (figure 19.9b) and Bdellovibrio (see figure 14.30) use this predation strategy. Order Bdellovibrionales includes predators that invade other Gram-negative bacteria.
-In contrast to the predatory bacteria shown in figure 19.9, some bacteria are facultative predators. Sometimes they consume organic matter released from dead organisms, whereas at other times, they actively prey on other microbes. Two examples are members of the y-proteobacterial genus Lysobacter and the &-proteobacterial genus Myxococcus. Both display predation whereby populations of cells use gliding motility to creep toward and cover their prey while releasing an arsenal of degradative enzymes. M. xanthus fruiting body formation can be triggered when its prey has been exhausted.
-Importantly, predation has many beneficial effects, especially when one considers predators and prey at the population and community levels. Ingestion and assimilation of a prey bacterium can lead to increased rates of nutrient cycling, critical for the functioning of the microbial loop (see figure 21.3). Although here we focus on bacterial predators, the impact of viral predation on communities, ecosystems, and even the global carbon budget is vast. As such, it is an active area of research. Marine viruses: mortality at sea
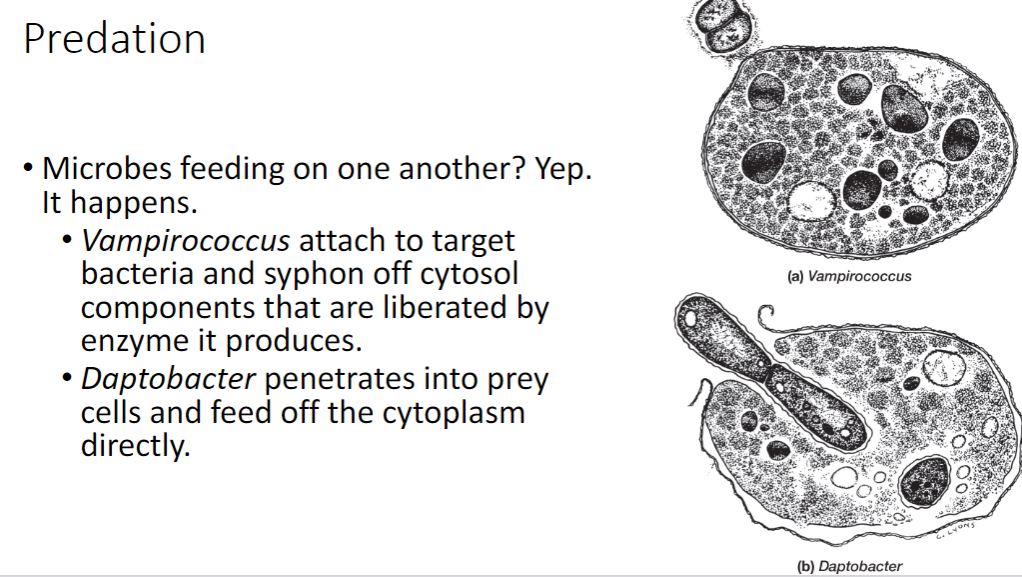
64
New cards
Parasitism
-Parasitism is one of the most complex microbial interactions; parasite and host must coexist at least temporarily, so the microbe has enough time to reproduce and colonize a new host. Coexistence may involve nutrient acquisition, physical maintenance in or on the host, or both. But what happens if the host-parasite equilibrium is upset? If the balance favors the host (perhaps by a strong immune defense or antimicrobial therapy), the parasite loses its habitat and may be unable to survive. On the other hand, if the equilibrium is shifted to favor the parasite, the host becomes ill and, depending on the specific host-parasite relationship, may die.
On the other hand, a controlled parasite-host relationship can be maintained for long periods of time and the parasitic nature of the relationship may not be obvious. For example, lichens (figure 19.11) are the association between specific ascomycetes (fungi) and either green algae or cyanobacteria. The fungal partner is termed the mycobiont and the algal or cyanobacterial partner, the phycobiont. The fungus obtains nutrients from its partner by projections of fungal hyphae called haustoria, which penetrate the phycobiont cell wall. It also uses 02 produced by the phycobiont for respiration. In turn, the fungus protects the phycobiont from high light intensities, provides water and minerals, and creates a firm substratum within which the phycobiont can grow protected from environmental stress. This is an ancient association, having developed prior to the evolution of vascular plants. Based on the earlier description, one could easily conclude that the relationship is mutualistic. However, some cyanobacteria and algae phycobionts grow more quickly when cultured without their mycobiont. These are considered to be in a relationship in which the mycobiont is the parasite. This is in contrast to cooperation, in which the symbiont often grows more slowly without its host. Phylum Cyanobacteria: oxygenic photosynthetic bacteria.
-An important aspect of many symbiotic relationships, including parasitism, is that over time, the symbiont will discard excess, unused genomic information, a process called genomic reduction. This occurs when the symbiont has become dependent on the host for specific functions, such as synthesis of key metabolites. It is observed in the aphid endosymbiont Buchnera aphidicola, it has also occurred in the human pathogens Mycobacterium leprae, Mycoplasma genitalia, and Encephalitozoon cuniculi, a microsporidium. These microorganisms now can only survive inside their host cells.
On the other hand, a controlled parasite-host relationship can be maintained for long periods of time and the parasitic nature of the relationship may not be obvious. For example, lichens (figure 19.11) are the association between specific ascomycetes (fungi) and either green algae or cyanobacteria. The fungal partner is termed the mycobiont and the algal or cyanobacterial partner, the phycobiont. The fungus obtains nutrients from its partner by projections of fungal hyphae called haustoria, which penetrate the phycobiont cell wall. It also uses 02 produced by the phycobiont for respiration. In turn, the fungus protects the phycobiont from high light intensities, provides water and minerals, and creates a firm substratum within which the phycobiont can grow protected from environmental stress. This is an ancient association, having developed prior to the evolution of vascular plants. Based on the earlier description, one could easily conclude that the relationship is mutualistic. However, some cyanobacteria and algae phycobionts grow more quickly when cultured without their mycobiont. These are considered to be in a relationship in which the mycobiont is the parasite. This is in contrast to cooperation, in which the symbiont often grows more slowly without its host. Phylum Cyanobacteria: oxygenic photosynthetic bacteria.
-An important aspect of many symbiotic relationships, including parasitism, is that over time, the symbiont will discard excess, unused genomic information, a process called genomic reduction. This occurs when the symbiont has become dependent on the host for specific functions, such as synthesis of key metabolites. It is observed in the aphid endosymbiont Buchnera aphidicola, it has also occurred in the human pathogens Mycobacterium leprae, Mycoplasma genitalia, and Encephalitozoon cuniculi, a microsporidium. These microorganisms now can only survive inside their host cells.

65
New cards
Amensalism
-Amensalism describes the adverse effect that one organism has on another organism (figure 19.1). This is a unidirectional process based on the release of a specific compound by one organism that has a negative effect on another. A classic example of amensalism is the production of antibiotics that can inhibit or kill other, susceptible microorganisms.
-Amensalisms can be quite complex. Attine ants (ants belonging to a New World tribe) are perhaps the most fascinating example. These ants cultivate for their own nourishment a garden of fungi belonging to the genus Leucocoprinus (figure 19.8). Their ability to accomplish this advanced task depends upon actinobacteria in the genus Pseudonocardia. These bacteria produce inhibitory compounds that prevent the growth of Escovopsis spp., a predatory fungus that would otherwise destroy the Leucocoprinus fungi crop.
-Amensalisms can be quite complex. Attine ants (ants belonging to a New World tribe) are perhaps the most fascinating example. These ants cultivate for their own nourishment a garden of fungi belonging to the genus Leucocoprinus (figure 19.8). Their ability to accomplish this advanced task depends upon actinobacteria in the genus Pseudonocardia. These bacteria produce inhibitory compounds that prevent the growth of Escovopsis spp., a predatory fungus that would otherwise destroy the Leucocoprinus fungi crop.

66
New cards
competition
-Competition arises when different organisms within a population or community try to acquire the same resource, whether this is a physical location or a particular limiting nutrient (figure 19.1). If one of the two competing organisms can dominate the environment, whether by occupying the physical habitat or by consuming a limiting nutrient, it will overtake the other. This phenomenon was studied by E. F. Gause, who in 1934 described it as the competitive exclusion principle. In chemostats, competition for a limiting nutrient may occur among microorganisms with transport systems of differing affinity. This can lead to the exclusion of the slower-growing population under a particular set of conditions. If the dilution rate in the chemostat is changed, the previously slower-growing population may become dominant. Often two microbial populations that appear to be similar nevertheless coexist. In this case, they share the limiting resource (space, a nutrient) and coexist while surviving at lower population levels. Competitive exclusion in part accounts for the phenomenon of colonization resistance the capacity of mutualistic gut microbes to prevent pathogen growth.

67
New cards
Is there a best of interaction? question
Maybe cooperation but doesn't mean its correct.

68
New cards
Microbiome and microbiota
-In 2001, renowned microbiologist and Nobel laureate Joshua Lederberg coined the term microbiome to describe all the microbes "that literally share our body space." Very few scientists at that time would have predicted the profound impact the discoveries made over the next two decades would have on our understanding of human physiology, pathophysiology, and what it means to be human. We have learned that humans cannot live a normal life without their microbial partners-that we are holobionts: hosts and microbes living together, and importantly, evolving together. After all, the average adult carries about three times more microbial cells (1014) than human somatic cells (average about 4 × 10¹³). With a combined microbial contribution of over 8 million protein-encoding genes (as compared to our roughly 22,000 genes), we have come to realize that many of these gene products serve essential functions our genome cannot provide.
Without technological advances associated with next-generation nucleotide sequencing and metagenomics, we would still know very little of our microbiomes, also known as normal microbiota. Through the application of these techniques, the many thousands of metagenomes derived from multiple body sites from hundreds of healthy volunteers have been sequenced. These data demonstrate that there is no single healthy microbiome. Rather, each person harbors a unique collection of microorganisms that assemble from a lifetime of interaction with their environment, diet, medications, and many other factors. We thus begin the chapter with the story of human colonization by their microbial partners.
Without technological advances associated with next-generation nucleotide sequencing and metagenomics, we would still know very little of our microbiomes, also known as normal microbiota. Through the application of these techniques, the many thousands of metagenomes derived from multiple body sites from hundreds of healthy volunteers have been sequenced. These data demonstrate that there is no single healthy microbiome. Rather, each person harbors a unique collection of microorganisms that assemble from a lifetime of interaction with their environment, diet, medications, and many other factors. We thus begin the chapter with the story of human colonization by their microbial partners.
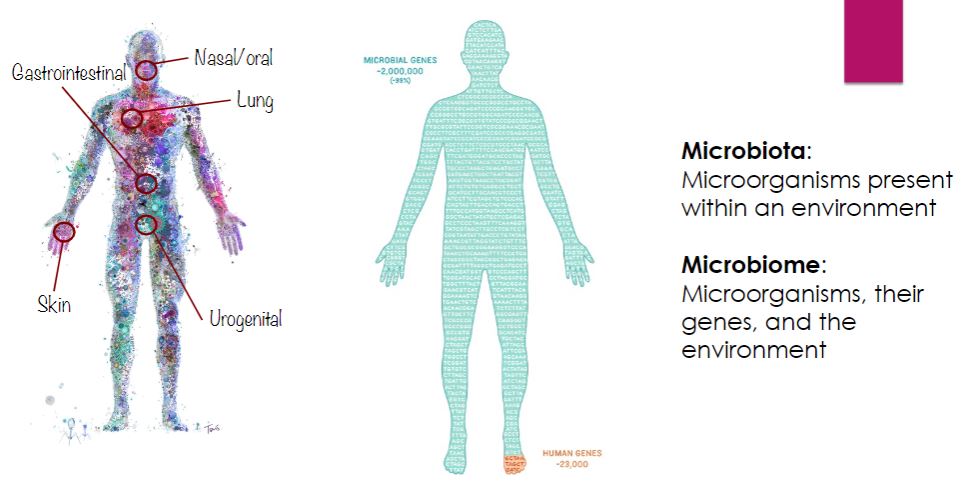
69
New cards
Microbiome variability
Body visual
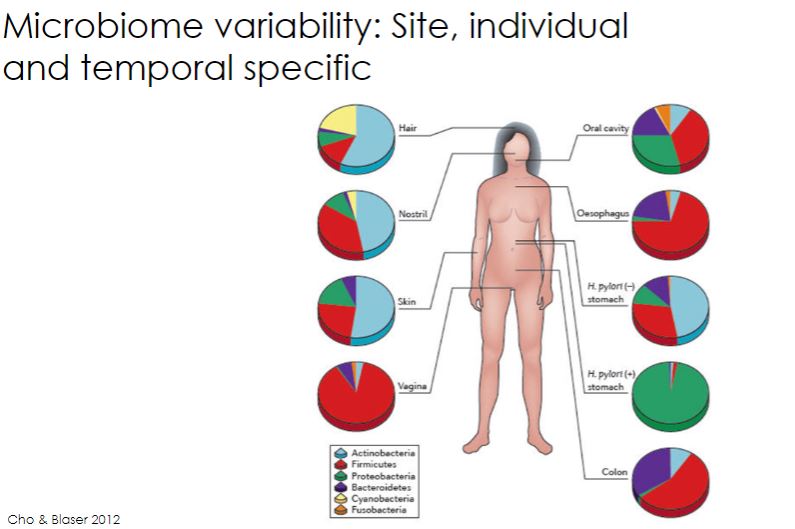
70
New cards
Microbiome variability
So what this image shows is clusters of microbial community. and other individual dots are different from one another, so their different microbial communities.
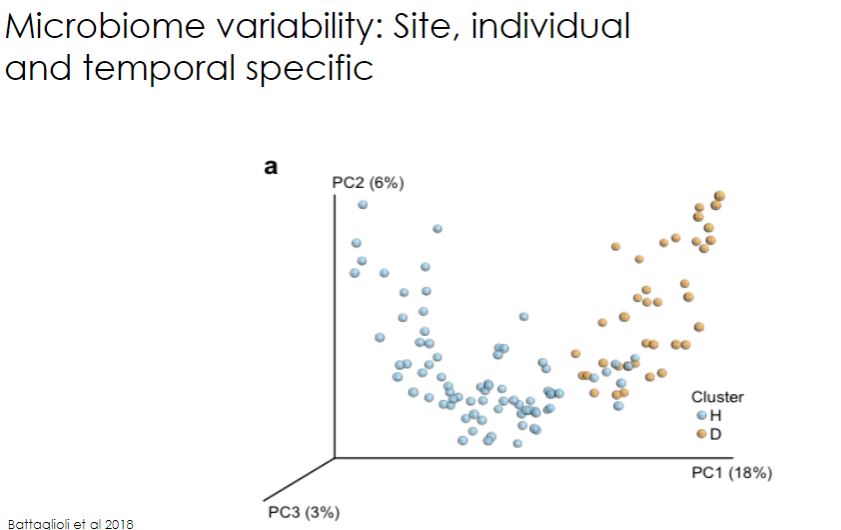
71
New cards
Development of a stable microbiome
So this data was created to observe an infant and how much its microbiome is develop and completed at a certain age. At 2.5 - 3 years old, its microbiome is solidified at these age, So in conclusion is that microbes are highly plastid ( if wrong wording then changing and increasing until 2-3 years old.)
-The normal community of mutualistic and commensal microbes is not static. It begins developing at birth and changes as we age. Infancy appears to be a "window of opportunity" for the development of the kinds of microbes that will be most beneficial for the maturation of our immune systems. By the time we are 3 years old, an adultlike community of microbes is established(figure 24.1). As we further explore, it is important to develop a microbiome rich in diversity.
-Newborns are initially colonized by microorganisms from the immediate environment. For example, babies born vaginally acquire most of their microorganisms from their mother's birth canal, whereas babies born by cesarean delivery acquire microorganisms from the skin of their initial caretakers. The importance of newborn colonization is so critical that some health-care providers swab the mouths of newborns with maternal vaginal secretions after cesarean delivery.
The early colonization of the infant intestinal tract by Escherichia coli and streptococci appears to establish a reducing environment for the growth of anaerobes such as bifidobacteria and bacteroidetes. In addition, human milk acts as a selective medium for nonpathogenic bacteria, as bottle-fed babies have a much smaller proportion of intestina bifidobacteria. Bifidobacteria transport some of the polymeric sugars found in human breast milk directly across their plasma membranes-a remarkable feat for a bacterium. This helps explain its dominance in a breastfed infant's gut. The fermentation of these sugars produces acetate and lactate, which provide calories for the growing baby, and lowers the gut pH, helping to limit the growth of pathogens. An interesting discovery is that a robust immune response to vaccination is linked to the presence of bifidobacteria in an infant's GI tract. Switching to cow's milk or solid food (mostly polysaccharide) appears to result in the loss of bifidobacteria predominance; proteobacteria, firmicutes, and bacteroidetes-specifically enterobacteria, enterococci, lactobacilli, clostridia, and Bacteroides spp.-increase and outcompete bifidobacteria.
-The normal community of mutualistic and commensal microbes is not static. It begins developing at birth and changes as we age. Infancy appears to be a "window of opportunity" for the development of the kinds of microbes that will be most beneficial for the maturation of our immune systems. By the time we are 3 years old, an adultlike community of microbes is established(figure 24.1). As we further explore, it is important to develop a microbiome rich in diversity.
-Newborns are initially colonized by microorganisms from the immediate environment. For example, babies born vaginally acquire most of their microorganisms from their mother's birth canal, whereas babies born by cesarean delivery acquire microorganisms from the skin of their initial caretakers. The importance of newborn colonization is so critical that some health-care providers swab the mouths of newborns with maternal vaginal secretions after cesarean delivery.
The early colonization of the infant intestinal tract by Escherichia coli and streptococci appears to establish a reducing environment for the growth of anaerobes such as bifidobacteria and bacteroidetes. In addition, human milk acts as a selective medium for nonpathogenic bacteria, as bottle-fed babies have a much smaller proportion of intestina bifidobacteria. Bifidobacteria transport some of the polymeric sugars found in human breast milk directly across their plasma membranes-a remarkable feat for a bacterium. This helps explain its dominance in a breastfed infant's gut. The fermentation of these sugars produces acetate and lactate, which provide calories for the growing baby, and lowers the gut pH, helping to limit the growth of pathogens. An interesting discovery is that a robust immune response to vaccination is linked to the presence of bifidobacteria in an infant's GI tract. Switching to cow's milk or solid food (mostly polysaccharide) appears to result in the loss of bifidobacteria predominance; proteobacteria, firmicutes, and bacteroidetes-specifically enterobacteria, enterococci, lactobacilli, clostridia, and Bacteroides spp.-increase and outcompete bifidobacteria.
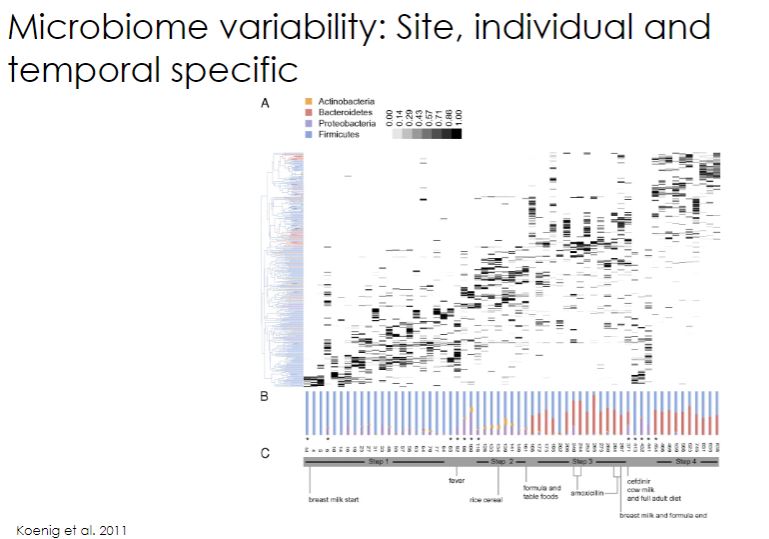
72
New cards
Assembling our microbiome
-Interestingly, once established, the adult microbiome is difficult to alter. Most changes are the result of substantial host physical or lifestyle changes (e.g., adopting a long-term vegetarian diet). Supporting this observation is the effect that infectious disease and its treatment have on the microbiome. Acute infection and brief antimicrobial chemotherapy result in short-term microbiome shifts but typically do not alter the long- term composition of the adult microbiota. However, long-term shifts in microbiota can occur with chronic infection and sustained use of antimicrobial agents.
-While the adult human microbiome is relatively stable over time, it is highly variable from person to person and at different sites within the same person. In other words, there are relatively few species of microorganisms common to all humans; each human has a relatively unique microbiome. Based on 16S rRNA and whole genome metagenomic data, bacteria common to human skin, the intestinal tract, and the other mucosal surfaces include five major phyla: Actinobacteria, Bacteroidetes, Firmicutes, Fusobacteria, and Proteobacteria (figure 24.2). Additionally, a number of archaea, fungi, and viruses are also present. Despite limited diversity at the phylum level, an enormous number of species are present. In fact the average adult gut is host to 500 to 1,000 different microbial species.
-This image below describes how you could build your microbiome in different events, such as giving birth or any interaction with objects and such.
-While the adult human microbiome is relatively stable over time, it is highly variable from person to person and at different sites within the same person. In other words, there are relatively few species of microorganisms common to all humans; each human has a relatively unique microbiome. Based on 16S rRNA and whole genome metagenomic data, bacteria common to human skin, the intestinal tract, and the other mucosal surfaces include five major phyla: Actinobacteria, Bacteroidetes, Firmicutes, Fusobacteria, and Proteobacteria (figure 24.2). Additionally, a number of archaea, fungi, and viruses are also present. Despite limited diversity at the phylum level, an enormous number of species are present. In fact the average adult gut is host to 500 to 1,000 different microbial species.
-This image below describes how you could build your microbiome in different events, such as giving birth or any interaction with objects and such.
73
New cards
Birth exposure
This image depicts on the birth, specifically on C-sections.
The C-section itself only cause the infant to be naturally exposed to the bacteria from the vaginal. so the solution would be to obtain bacteria from the vagina and perform the C-section and expose the bacteria from the vaginal to the infant to obtain a good microbial community or a good microbiome.
The C-section itself only cause the infant to be naturally exposed to the bacteria from the vaginal. so the solution would be to obtain bacteria from the vagina and perform the C-section and expose the bacteria from the vaginal to the infant to obtain a good microbial community or a good microbiome.
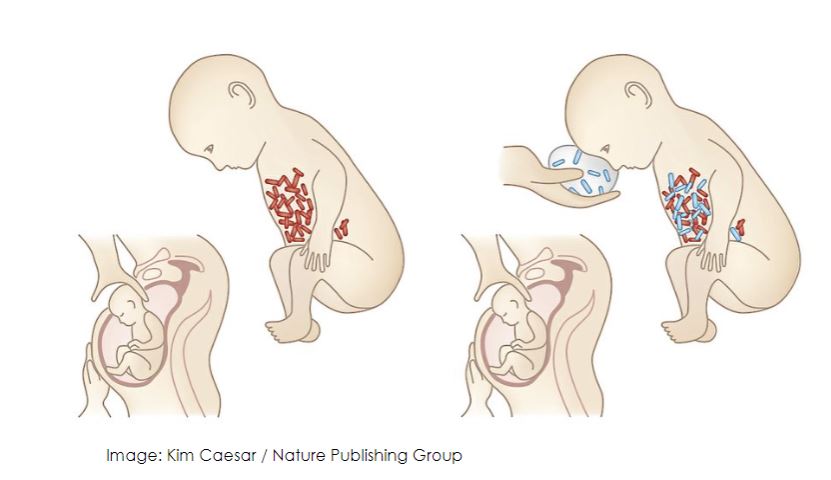
74
New cards
Guts, Glorius Guts
So this slide depicts other factors that alter the microbiota, like pets that are dogs because they lick a lot. so that results in having similarities to you pet because you share the microbe.
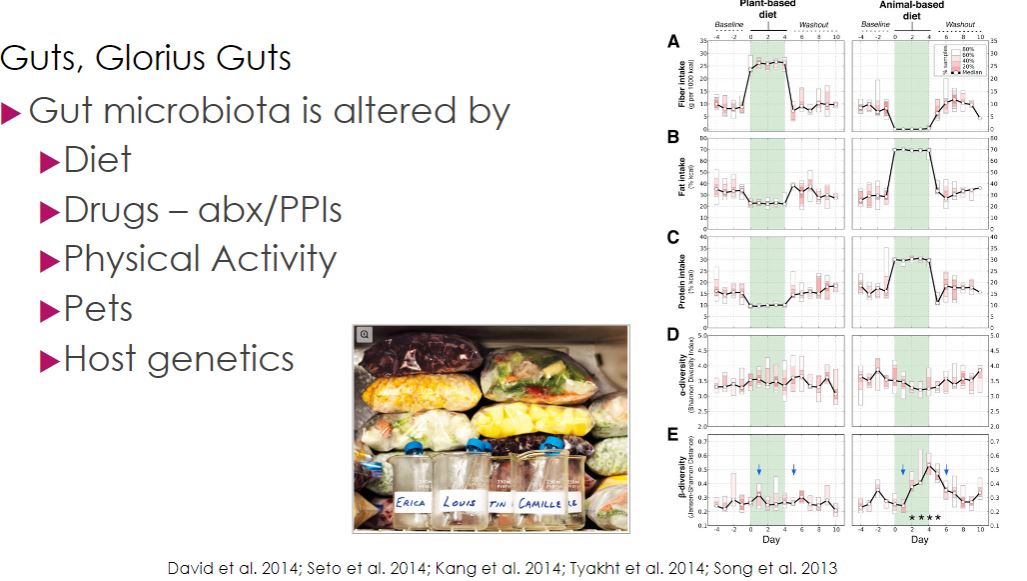
75
New cards
Early Antibiotic Use
-The first clue that the microbiome was critical to host immunity was gleaned from an observation made over 50 years ago. At that time it was noted that following antibiotic treatment, people had a much higher risk of GI tract infection by members of the Enterobacteraceae, such as E. coli or Salmonella enterica. It was rightly assumed that antibiotics disrupted the gut microbial community, and this phenomenon was explained by colonization resistance. Colonization resistance is based on the ecological principle of competitive exclusion. In this case, it was assumed that the commensal bacteria in the healthy colon outcompete pathogens for nutrients and space. Although this is true, like every other aspect of the microbiome, colonization resistance has turned out to be much more complex, involving not just direct microbiome-pathogen interactions, but indirect microbiome-host-pathogen interactions as well.
While the initial observations that led to an appreciation of colonization resistance were based on antibiotic therapy in people, sophisticated experiments using GF mice have been essential in advancing our understanding. Among the early results that signaled the importance of the microbiome were observations that the intestinal cellular architecture of GF mice differed from conventional mice. Specifically, the fingerlike projections (villi) that line the intestine were found to be longer, while the gulfs, or crypts, between villi were less developed in GF mice. To explore this change in morphology, scientists measured levels of antimicrobial peptides (AMPs) normally secreted by host Paneth cells located in the crypts. They discovered GF mice produced very low levels of AMPs, suggesting AMP production requires bacteria in the gut. This result was notable because it suggested that the host had to detect, rather than ignore, gut microbes.
While the initial observations that led to an appreciation of colonization resistance were based on antibiotic therapy in people, sophisticated experiments using GF mice have been essential in advancing our understanding. Among the early results that signaled the importance of the microbiome were observations that the intestinal cellular architecture of GF mice differed from conventional mice. Specifically, the fingerlike projections (villi) that line the intestine were found to be longer, while the gulfs, or crypts, between villi were less developed in GF mice. To explore this change in morphology, scientists measured levels of antimicrobial peptides (AMPs) normally secreted by host Paneth cells located in the crypts. They discovered GF mice produced very low levels of AMPs, suggesting AMP production requires bacteria in the gut. This result was notable because it suggested that the host had to detect, rather than ignore, gut microbes.
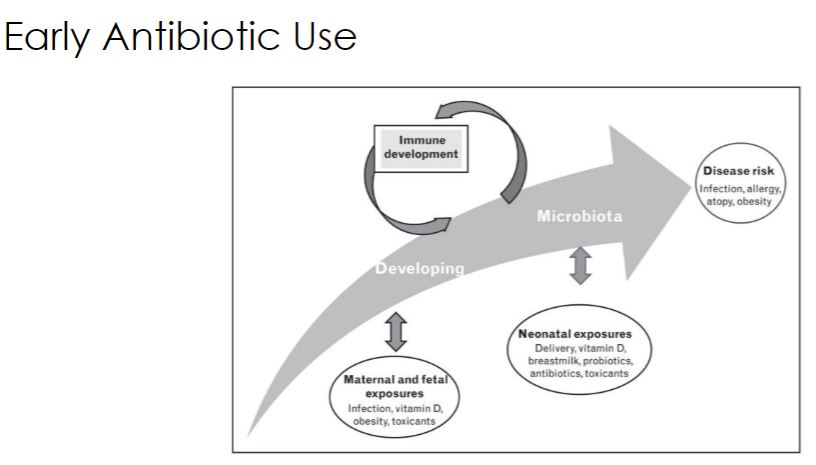
76
New cards
Antibiotics
This slide shows the damage antibiotics do to microbe because antibiotics kills every microbes even good ones.
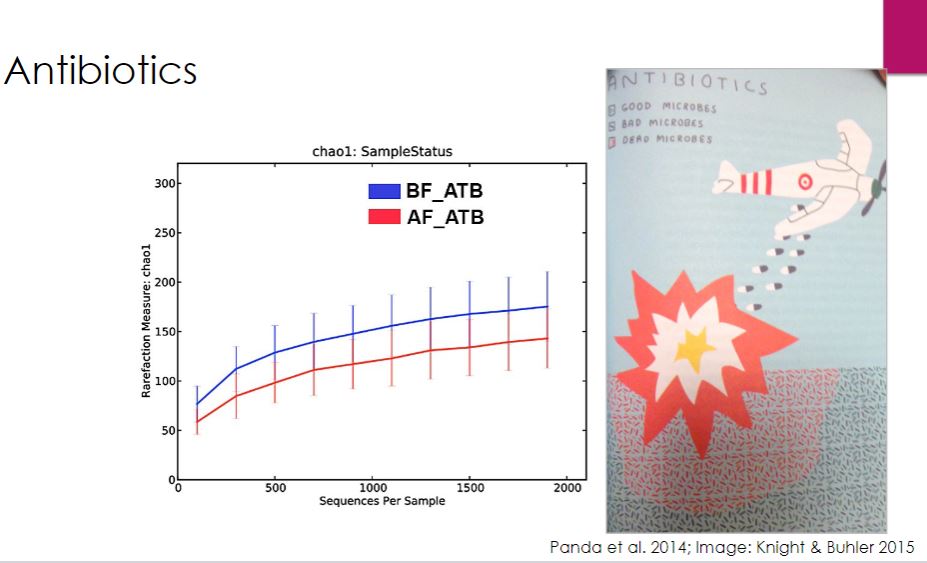
77
New cards
Dissecting host microbe
a

78
New cards
microbial Consortia
This slide is just how they measure the microbe communities and its very difficult to measure as it needs specialized tools.
So microbial consortia is referred as a group of diverse microorganisms that have the ability to act together in a community.
So microbial consortia is referred as a group of diverse microorganisms that have the ability to act together in a community.
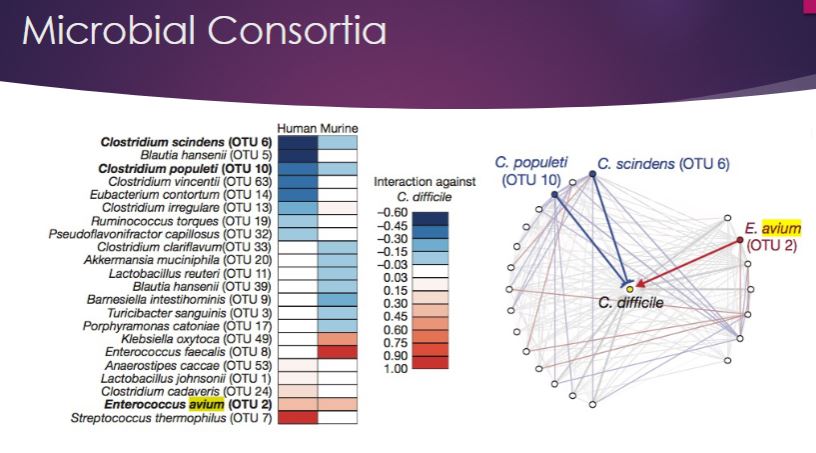
79
New cards
Tool: 16s sequencing
So 16s is a tool, I believe it uses the ribosomes and have a better visual of what's happening. Taxonomic resolution is very low, meaning very limited visuals. so you be able to distinguish the gene. like look through a tunnel and see straight to the other side, but you cannot see the side of teh tunnel.
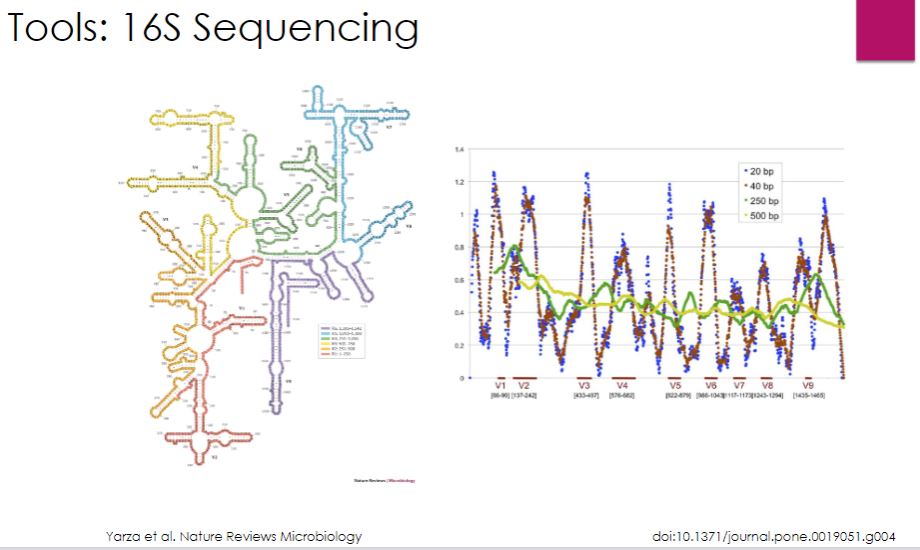
80
New cards
Microbial fingerprint
This slide describes how you can predict where people have been based on their microbe on their hands. So both your hands have very different microbial communities as one could be dominant and the other not, because you touch/interact with your dominant. So you can change the component of your left or right hand.
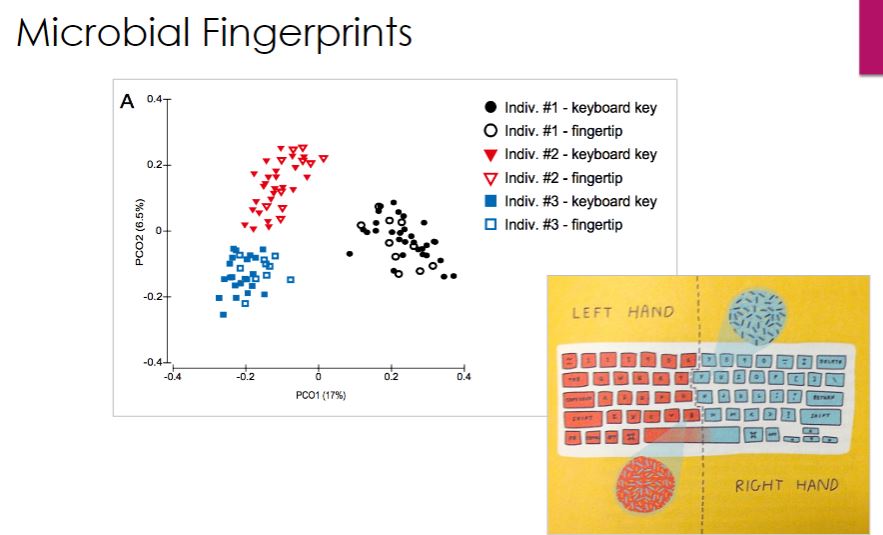
81
New cards
Germ-free animal model
This machine facilitates mice that have never been exposed to microbes. Other samples that can be used are fruit flies, zebra flies, hamsters, rats (tough), ferrets and calfs. So these blank slate of microbes are used to be able t manipulate the microbe community and be able to observe changes.

82
New cards
Gut Microbe and disease
-Body weight is most closely related not to the number of calories we eat, but the number of calories we absorb. Our cells are very good at taking up simple sugars and a few polysaccharides, but for the most part, we rely on our gut microbiota to convert our food into calories we can use. This was shown in the early 2000s in a series of experiments that used germ-free mice. Germ-free (GF) mice are born by cesarean delivery, reared in sterile enclosures, and fed sterile food and water. Although GF animals are nowhere near representative of normal life, comparing GF mice with those bearing a normal microbiome (in this context, known as conventional animals) allows scientists to test many of the complex relationships between microorganisms, hosts, and environmental factors in a controlled fashion.
These early microbiome studies showed that GF mice could eat more but gain less weight than conventional mice. For example, in one 8-week study, GF mice gained 50% less than conventional mice after both groups consumed a diet of 40% fat. While this may sound like an ad for a weight-loss plan ("Eat more! Weigh less!"), this study and others like it showed that gut microbiota convert much of the food eaten into degradation products that contribute to total caloric intake of the host. The causal nature of the microbiome population and weight gain has been demonstrated by fecal microbiome transplant (FMT) experiments (see chapter opener). FMT using conventional, obese mice as donors and GF mice as recipients causes the formerly GF recipient mice to become obese as well, despite no change to their diet or exercise regime. Additional experiments have shown that FMT from lean to obese mice can trigger weight loss in the recipients.
-It is frequently (but not always) observed that obese mice, like overweight and obese people, turn out to have higher relative concentrations of gut bacteria belonging to the phylum Firmicutes compared to bacteria belonging to the phylum Bacteroidetes, along with several other changes in gut microbiota population (table 24.2). Why is this important? Simply put, we cannot break down fiber very well but our bacteria can. Although the term fiber is not very specific, all fiber consists of a variety of linear and branch chained sugars. Because we can digest only a few complex polysaccharides, we feed most of them to our microbiome. Unlike our own cells, the typical gut microbiome produces thousands of enzymes that evolved to break down complex polysaccharides into their constituent monomers. Importantly many bacteria ferment these monomers into short-chain fatty acids (SCFAs). These SCFAs include butyrate, propionate, and acetate. Butyrate is the principle source of calories for intestinal epithelial cells (called colonocytes) that line the gut. In fact, butyrate increases colonocyte mitochondrial activity so that many carbon substrates are oxidized to CO₂-thus calories consumed are literally vaporized. Propionate travels to the liver where it inhibits cholesterol synthesis, and along with butyrate helps regulate weight by increasing the release of intestinal hormones that suppress hunger. On the other hand, acetate is readily absorbed by a variety of host cells and acts as a precursor for lipid synthesis in liver and fat cells. For this reason, acetate in this context is sometimes called "obesogenic." The shift in the population of gut microbiota observed in obese individuals results in more bacteria that produce acetate, which promotes weight gain and fewer bacteria that produce butyrate and propionate, which protect against obesity. The role of SCFAs in weight regulation and maintaining host homeostasis has refocused attention from the individual species that constitute the gut microbiome to the products these microorganisms secrete, called the metabolome As we now discuss, SCFAs and the microbes that produce them are key players in immune modulation as well.
These early microbiome studies showed that GF mice could eat more but gain less weight than conventional mice. For example, in one 8-week study, GF mice gained 50% less than conventional mice after both groups consumed a diet of 40% fat. While this may sound like an ad for a weight-loss plan ("Eat more! Weigh less!"), this study and others like it showed that gut microbiota convert much of the food eaten into degradation products that contribute to total caloric intake of the host. The causal nature of the microbiome population and weight gain has been demonstrated by fecal microbiome transplant (FMT) experiments (see chapter opener). FMT using conventional, obese mice as donors and GF mice as recipients causes the formerly GF recipient mice to become obese as well, despite no change to their diet or exercise regime. Additional experiments have shown that FMT from lean to obese mice can trigger weight loss in the recipients.
-It is frequently (but not always) observed that obese mice, like overweight and obese people, turn out to have higher relative concentrations of gut bacteria belonging to the phylum Firmicutes compared to bacteria belonging to the phylum Bacteroidetes, along with several other changes in gut microbiota population (table 24.2). Why is this important? Simply put, we cannot break down fiber very well but our bacteria can. Although the term fiber is not very specific, all fiber consists of a variety of linear and branch chained sugars. Because we can digest only a few complex polysaccharides, we feed most of them to our microbiome. Unlike our own cells, the typical gut microbiome produces thousands of enzymes that evolved to break down complex polysaccharides into their constituent monomers. Importantly many bacteria ferment these monomers into short-chain fatty acids (SCFAs). These SCFAs include butyrate, propionate, and acetate. Butyrate is the principle source of calories for intestinal epithelial cells (called colonocytes) that line the gut. In fact, butyrate increases colonocyte mitochondrial activity so that many carbon substrates are oxidized to CO₂-thus calories consumed are literally vaporized. Propionate travels to the liver where it inhibits cholesterol synthesis, and along with butyrate helps regulate weight by increasing the release of intestinal hormones that suppress hunger. On the other hand, acetate is readily absorbed by a variety of host cells and acts as a precursor for lipid synthesis in liver and fat cells. For this reason, acetate in this context is sometimes called "obesogenic." The shift in the population of gut microbiota observed in obese individuals results in more bacteria that produce acetate, which promotes weight gain and fewer bacteria that produce butyrate and propionate, which protect against obesity. The role of SCFAs in weight regulation and maintaining host homeostasis has refocused attention from the individual species that constitute the gut microbiome to the products these microorganisms secrete, called the metabolome As we now discuss, SCFAs and the microbes that produce them are key players in immune modulation as well.
83
New cards
The gut
Look for the audio that describes this. Just know that eating bad food or basically what microbes don't like to eat will eventually cause carcinoma,
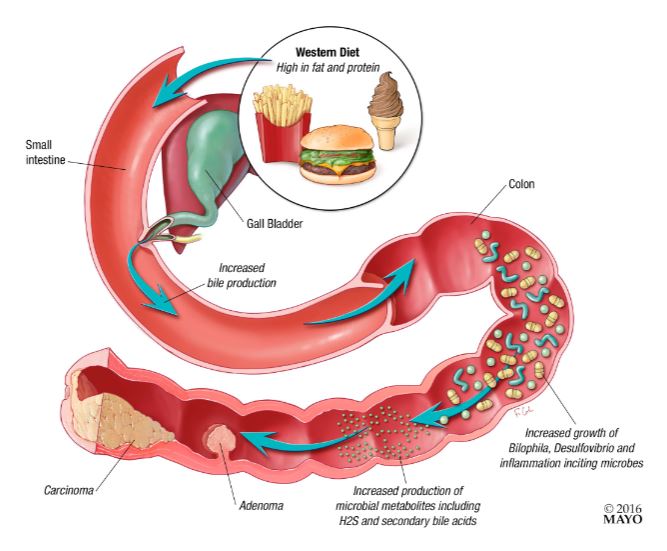
84
New cards
Immune development
So these are mice where the very left is with no microbes and the middle is with very little microbe exposure and the last one is normal and have a complete microbe community.

85
New cards
Xenobiotic Metabolism
Xenobiotic is what we excrete.
-modern medicine is to treat the avg. person, so not a useful curve because outside the avg curve,it wont be good for those outside and for their microbiome. (individualized medicine)
-modern medicine is to treat the avg. person, so not a useful curve because outside the avg curve,it wont be good for those outside and for their microbiome. (individualized medicine)
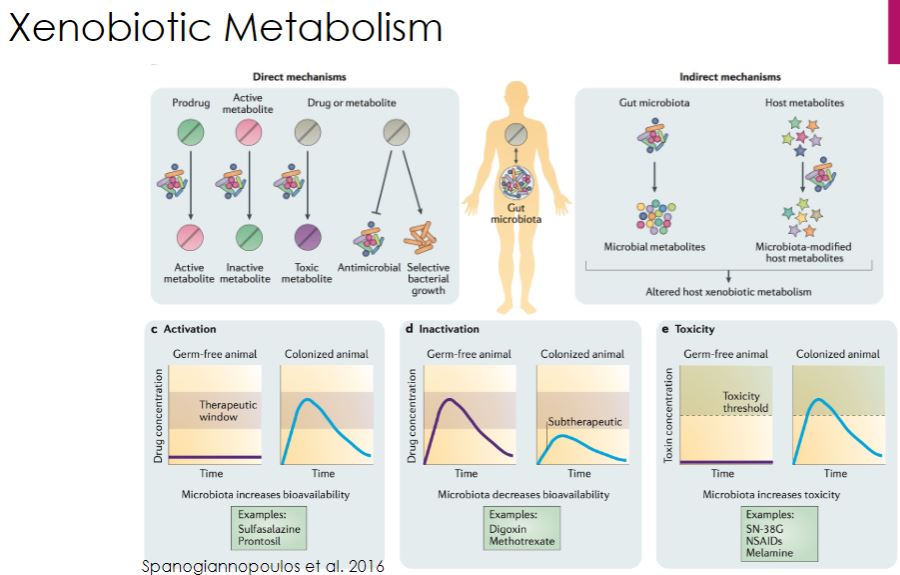
86
New cards
Table of metabolized by the microbes
-All of these are manipulated by microbes and others may experience these because of their own microbiome.
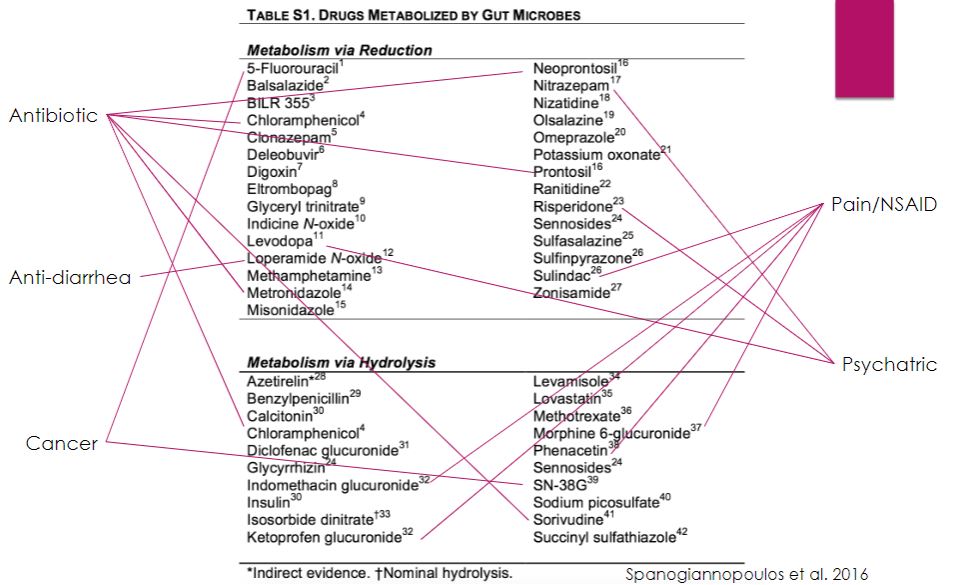
87
New cards
Microbiome effects on health and disease
-so mood is by your gut
-gut communicating to brain
-5-HT releases serotonin and made by colon
-gut communicating to brain
-5-HT releases serotonin and made by colon
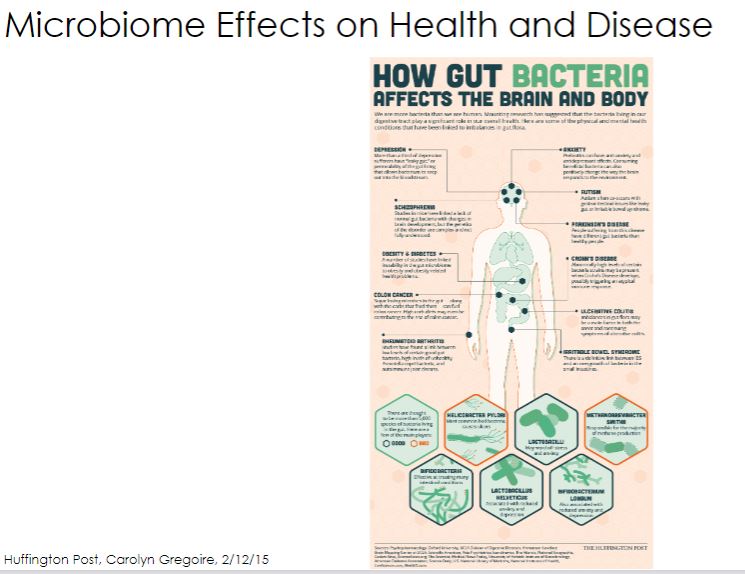
88
New cards
Gut-Brain Axis
One of the most intriguing areas of microbiome investigation concerns how our gut microbes affect our central nervous system (CNS). A variety of studies have shown that specific behavioral traits (inquisitiveness, sociability, anxiety, depression) differ when comparing GF mice and conventional mice. Amazingly, when GF mice are colonized by bacteria, these traits normalize. In addition, some behavioral traits are reproducibly transferred between mice via FMT. The influence of the microbiome is heritable as well, because the gut microbiome of pregnant mice influences the neurodevelopment of their offspring. Intriguingly, bacteria that stimulate maternal secretion of the pro-inflammatory cytokine IL-17 give rise to pups (baby mice) that display repetitive and antisocial behaviors.
How do we account for such curious observations? There are at least three ways the microbiome can influence the CNS (figure 24.6). First as we have seen, the microbiome has a profound impact on the immune system, which in turn modulates the CNS. One documented way in which this occurs is through microbial regulation of the balance between pro- and anti-inflammatory cytokines (figure 24.4). When pro- inflammatory cytokines reach the brain, they alter the function of neurons and CNS macrophages, called microglia. For example when lipopolysaccharide (LPS) crosses the gut, it triggers the production of pro-inflammatory cytokines, which in turn are responsible for the behaviors we associate with being sick: loss of appetite, decreased motor activity, loss of sociability, and reduced cognition.
-Another mechanism that helps mediate the gut-brain axis is a direct pathway from gut to brain. This is possible because the GI tract is lined with a network of nerves, called the enteric nervous system, that is connected to the CNS via the vagus nerve. Gut bacteria directly stimulate neurons within the enteric nervous system, which then communicate with the vagus nerve. The vagus nerve in turn transmits signals to the brain. The vagus nerve can differentiate between stimuli from normal microbiota and pathogens even in the absence of inflammation, although how this fascinating feat is possible remains under investigation.
Finally, as suggested by experiments in which butyrate and FMT had similar effects on the behavior of GF mice, soluble microbial products can impact the CNS. To understand this, we must first consider the CNS as a site that is set apart from the remainder of the body by the blood-brain barrier (BBB). The BBB is lined with cells that protect the CNS from normal blood components as well as toxins and infectious agents. This is largely due to impermeable connections, called tight junctions, between the cells that make up the BBB. Once again, a flurry of research was triggered by an observation made in GF mice: Their BBBs have the undesirable quality of being more permeable than conventional mice. And once again, it is SCFAs, particularly butyrate, that help maintain the BBB as an impenetrable barricade. Because SCFAs are most abundant when mice or humans eat a high-fiber diet, the broader implication is that one's diet can govern the entry of metabolites and other blood-borne compounds to the CNS.
The unexpected influence gut microbiota has on behavior, documented in animals and some human trials, has unleashed a torrent of studies into how the microbiome may mediate neurodegenerative diseases like Parkinson's and Alzheimer's diseases. Autism spectrum disorder (ASD) has also garnered a great deal of attention. This reflects evidence that the establishment of a functional microbiome has long-lasting and important effects on behavior and perhaps on neural development within the first 3 years of life, when most children are diagnosed with ASD. How our microbiomes impact both neurodevelopment and degeneration are intriguing areas of investigation that will no doubt continue to reveal unexpected findings.
How do we account for such curious observations? There are at least three ways the microbiome can influence the CNS (figure 24.6). First as we have seen, the microbiome has a profound impact on the immune system, which in turn modulates the CNS. One documented way in which this occurs is through microbial regulation of the balance between pro- and anti-inflammatory cytokines (figure 24.4). When pro- inflammatory cytokines reach the brain, they alter the function of neurons and CNS macrophages, called microglia. For example when lipopolysaccharide (LPS) crosses the gut, it triggers the production of pro-inflammatory cytokines, which in turn are responsible for the behaviors we associate with being sick: loss of appetite, decreased motor activity, loss of sociability, and reduced cognition.
-Another mechanism that helps mediate the gut-brain axis is a direct pathway from gut to brain. This is possible because the GI tract is lined with a network of nerves, called the enteric nervous system, that is connected to the CNS via the vagus nerve. Gut bacteria directly stimulate neurons within the enteric nervous system, which then communicate with the vagus nerve. The vagus nerve in turn transmits signals to the brain. The vagus nerve can differentiate between stimuli from normal microbiota and pathogens even in the absence of inflammation, although how this fascinating feat is possible remains under investigation.
Finally, as suggested by experiments in which butyrate and FMT had similar effects on the behavior of GF mice, soluble microbial products can impact the CNS. To understand this, we must first consider the CNS as a site that is set apart from the remainder of the body by the blood-brain barrier (BBB). The BBB is lined with cells that protect the CNS from normal blood components as well as toxins and infectious agents. This is largely due to impermeable connections, called tight junctions, between the cells that make up the BBB. Once again, a flurry of research was triggered by an observation made in GF mice: Their BBBs have the undesirable quality of being more permeable than conventional mice. And once again, it is SCFAs, particularly butyrate, that help maintain the BBB as an impenetrable barricade. Because SCFAs are most abundant when mice or humans eat a high-fiber diet, the broader implication is that one's diet can govern the entry of metabolites and other blood-borne compounds to the CNS.
The unexpected influence gut microbiota has on behavior, documented in animals and some human trials, has unleashed a torrent of studies into how the microbiome may mediate neurodegenerative diseases like Parkinson's and Alzheimer's diseases. Autism spectrum disorder (ASD) has also garnered a great deal of attention. This reflects evidence that the establishment of a functional microbiome has long-lasting and important effects on behavior and perhaps on neural development within the first 3 years of life, when most children are diagnosed with ASD. How our microbiomes impact both neurodevelopment and degeneration are intriguing areas of investigation that will no doubt continue to reveal unexpected findings.
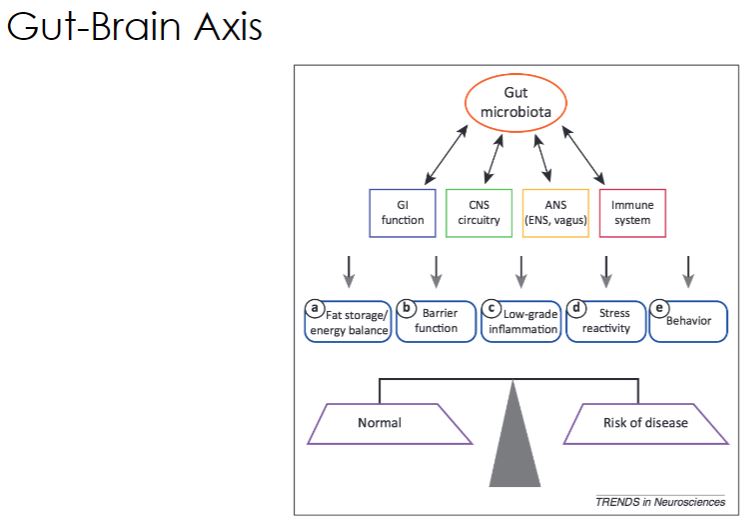
89
New cards
Behavior
Parkinson Disease
-Germ free mice are more laid back then normal mice.
-autism was also experiment like the obesity and resulted teh same.
-Germ free mice are more laid back then normal mice.
-autism was also experiment like the obesity and resulted teh same.
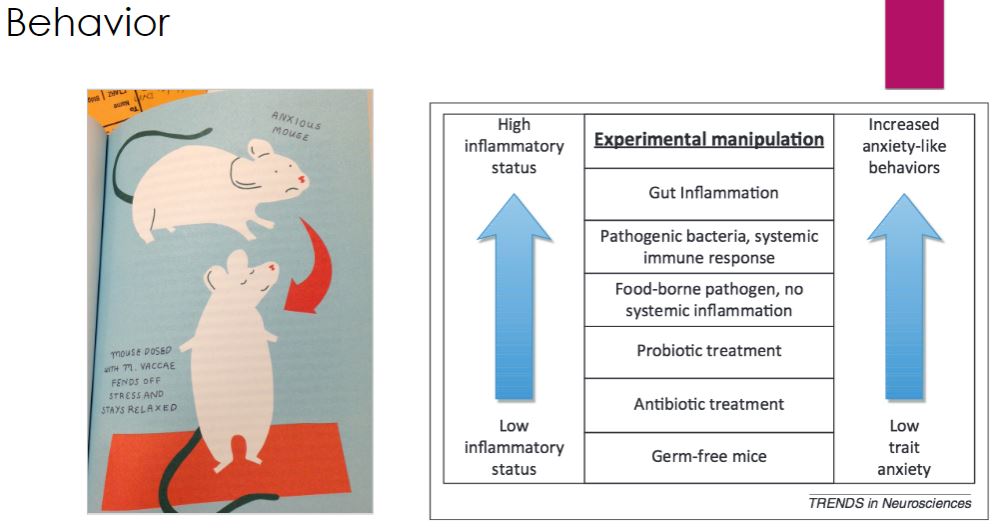
90
New cards
Mating preference
Microbes influence on mating preferences.
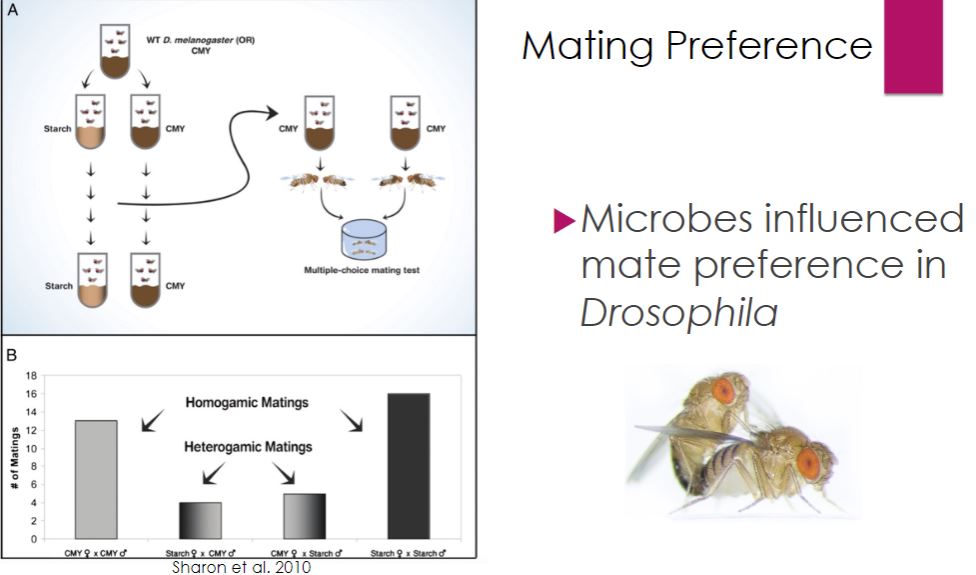
91
New cards
Clinical Applications of microbiome therapy
-Many years ago, a television commercial for yogurt featured a town full of centenarians (at least they were advertised as 100-year-old people) living in Eastern Europe who attributed their longevity to eating yogurt of a specific brand. It turns out that in 1908, microbiologist and Nobel laureate Élie Metchnikov also knew about these villagers and postulated that lactic acid-producing bacteria found in yogurt promoted their health. In the microbiome era, the probiotic industry has blossomed into a US$30 billion enterprise that continues to expand.
Despite this burgeoning market, many consumers are not really certain what the term "probiotic" means. In an effort to standardize the use of the term probiotic, the Food and Agricultural Organization of the United Nations-World Health Organization (FAO- WHO) has defined it as "live microorganisms, which, when administered in adequate amounts, confer a health benefit to the host" and provide specific requirements that should be met for foods and supplements to meet this standard.
Despite this burgeoning market, many consumers are not really certain what the term "probiotic" means. In an effort to standardize the use of the term probiotic, the Food and Agricultural Organization of the United Nations-World Health Organization (FAO- WHO) has defined it as "live microorganisms, which, when administered in adequate amounts, confer a health benefit to the host" and provide specific requirements that should be met for foods and supplements to meet this standard.
92
New cards
Fecal microbial transplant
-Despite this effort by the FAO-WHO, the U.S. Food and Drug Administration (FDA) does not regulate probiotic foods and supplements, so the health benefits the manufacturers of such products claim have not been rigorously tested. By contrast, if viable microbes are to be used as a medicine, the FDA must approve their use. Examples include the use of Mycobacterium bovis BCG for superficial bladder cancer and live, attenuated vaccines (e.g., measles and rubella vaccines). These products have a long track record of safe, therapeutic success and are regulated as drugs. The next phase of probiotic development will likely include products consisting of microorganisms used to treat a specific condition rather than to convey unspecified health benefits. These newer probiotics will require approval by the FDA (or similar agencies in other countries) after extensive safety and efficacy clinical trials. For example, bacterial mixtures marketed to prevent or treat C. difficile infection will be classified as a drug; this contrasts to fecal microbiome transfer, which is currently considered a procedure and is (at the time of this writing) permitted only for treatment of recurrent C. diff infection (see chapter opening story).

93
New cards
FMT
-Another confounding factor has been that until recently it has been difficult to evaluate whether probiotic bacteria actually colonize the gut or are retained long enough to have an effect. The discovery that many so-called probiotic microorganisms do not effectively colonize the intestines has led to the development of symbiotics. Synbiotics are foods or supplements that include both a prebiotic and a probiotic. The prebiotic is a compound(s) added to enhance the colonization and positive health benefits of probiotic microbes.
Although it is much harder to control the uniformity and effectiveness of a product that contains microbes than a drug with a unique chemical structure, the recognition that the microbiome is, in essence, a newly discovered organ needed for human homeostasis is propelling the unstoppable development of probiotics for preventative medicine. It is likely that hospitalized patients will routinely be given an oral dose of a synbiotic to protect against the development of infection with antibiotic-resistant pathogens. Similarly, outpatients requiring antibiotic therapy may also be prescribed a probiotic cocktail for microbiome reconstitution following treatment. Tourists may soon be packing probiotics to prevent travelers' diarrhea along with their sunscreen and flip-flops.
Until researchers, pharmaceutical companies, and regulatory agencies synchronize their approach to probiotics, it is instructive to consider the successful use of probiotics in animals. For example, Lactobacillus acidophilus is used in beef cattle feed. When these bacteria are sprayed on feed, cattle appear to have markedly lower (as much as 60%) carriage of the pathogenic E. coli strain O157:H7. This can make it easier to produce beef that meets current standards for microbiological quality at the time of slaughter. Probiotics are also used successfully with poultry. For instance, Salmonella enterica can be controlled by spraying a patented blend of 29 bacteria, isolated from the chicken cecum, on day-old chickens. As they preen themselves, the chicks ingest the bacterial mixture, establishing a functional microbiome that limits Salmonella colonization of the gut. In an age of increasing antimicrobial resistance, the use of probiotics may prove to be valuable for both humans and the animals they consume.
Although it is much harder to control the uniformity and effectiveness of a product that contains microbes than a drug with a unique chemical structure, the recognition that the microbiome is, in essence, a newly discovered organ needed for human homeostasis is propelling the unstoppable development of probiotics for preventative medicine. It is likely that hospitalized patients will routinely be given an oral dose of a synbiotic to protect against the development of infection with antibiotic-resistant pathogens. Similarly, outpatients requiring antibiotic therapy may also be prescribed a probiotic cocktail for microbiome reconstitution following treatment. Tourists may soon be packing probiotics to prevent travelers' diarrhea along with their sunscreen and flip-flops.
Until researchers, pharmaceutical companies, and regulatory agencies synchronize their approach to probiotics, it is instructive to consider the successful use of probiotics in animals. For example, Lactobacillus acidophilus is used in beef cattle feed. When these bacteria are sprayed on feed, cattle appear to have markedly lower (as much as 60%) carriage of the pathogenic E. coli strain O157:H7. This can make it easier to produce beef that meets current standards for microbiological quality at the time of slaughter. Probiotics are also used successfully with poultry. For instance, Salmonella enterica can be controlled by spraying a patented blend of 29 bacteria, isolated from the chicken cecum, on day-old chickens. As they preen themselves, the chicks ingest the bacterial mixture, establishing a functional microbiome that limits Salmonella colonization of the gut. In an age of increasing antimicrobial resistance, the use of probiotics may prove to be valuable for both humans and the animals they consume.
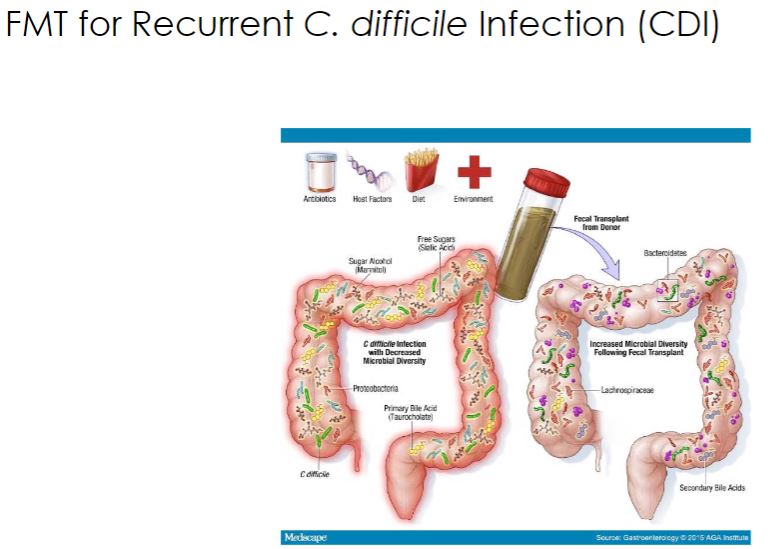
94
New cards
Pathogens
Statement
95
New cards
Pathogens definition
The virulence increases causes a bad disease but if its low, its not too bad. its more of tool.

96
New cards
What makes a pathogen? question
-When a microorganism is growing and multiplying on or within a larger organism, the latter, known as the host, is said to have an infection. The nature of an infection can vary widely with respect to severity, location, and type of organisms involved. An infection may or may not result in overt disease. Any organism that causes disease is known as a pathogen (Greek patho, disease; gennan, to produce), and its ability to cause disease is called pathogenicity. An organism's virulence is the degree of harm (pathogenicity) inflicted on its host. An infectious disease is any change from a state of health in which part or all of the host is incapable of carrying on its normal functions due to the presence of a pathogen or its products (e.g., toxins). Even resident microbiota, such as those associated with the gut or skin, can become opportunistic pathogens if they infect a host away from their typical niche, especially in a host with a weakened immune system (a compromised host). Host and tissue specificity offer microorganisms shelter and access to resources; however, relationships between organisms can be complex. Microorganisms actively seek host-specific sites that meet their unique growth and replication requirements.

97
New cards
Infection vs. Colonization
-To cause disease, microorganisms must not only contact the host but be able to survive in it. Bacteria, viruses, protists, and fungi need a suitable environment in which they can grow and reproduce. The range of factors required-temperature, pH, moisture, oxygen concentration, etc.-must be provided by the human host. These conditions can be met outside or inside host cells, depending on the pathogen. Infectious microorganisms are considered extracellular pathogens if they remain in tissues and fluids but never enter host cells during the course of disease. For instance, some bacteria and fungi can actively grow and multiply in the blood or tissue spaces. Yersinia pestis, which causes plague (also called Black Death; see section 14.3), is a good example of an extremely virulent extracellular pathogen. Microbes that grow and multiply within host cells are called intracellular pathogens; they can be further subdivided into two groups. Facultative intracellular pathogens are those organisms that reside within the cells of the host or in the environment but can also be grown in pure culture without host-cell support. Brucella abortus is a bacterium that grows and replicates within macrophages, neutrophils, and trophoblast cells (cells that surround the developing embryo; see chapter 14 opening story) and Histoplasma capsulatum is a fungus that grows within macrophages, yet both can be grown in vitro (see section 17.10). In contrast, obligate intracellular pathogens are incapable of growth and multiplication outside a host cell. By definition, all viruses are obligate intracellular pathogens in that they require a host cell for replication, often to the detriment of that cell. Some bacteria are also obligate intracellular bacterial pathogens, such as Chlamydia spp. and rickettsias (which cause diseases such as Rocky Mountain spotted fever and typhus; see sections 14.1 and 15.5). These microbes cannot be grown in the laboratory outside of their host cells. Malarial parasites (Plasmodium spp.) are examples of protozoa that require host cells for growth and differentiation
98
New cards
What is infection?
-infection
The invasion of a host by a microorganism with subsequent establishment and multiplication of the agent. An infection may or may not lead to overt disease.
The invasion of a host by a microorganism with subsequent establishment and multiplication of the agent. An infection may or may not lead to overt disease.

99
New cards
Infection dynamics
-Clinically, the course of an infectious disease usually has a characteristic pattern and can be divided into several phases (figure 25.1). The incubation period is the time between pathogen entry and development of signs and symptoms. The pathogen is reproducing but has not reached a sufficient level to cause clinical manifestations. This period's length varies with pathogen. Next, the prodromal stage occurs with an onset of signs and symptoms that are not yet specific enough to make a clear diagnosis. However, the patient often is contagious. This is followed by the illness period, when the disease is most severe and displays characteristic signs and symptoms. The host immune response is typically triggered at this stage. Finally, during the period of decline, the signs and symptoms begin to disappear. The recovery stage often is referred to as convalescence.
-Image below
The Course of an Infectious Disease. Most infectious diseases occur in four stages. The duration of each stage is a characteristic feature of each disease. The shaded area represents when many diseases are communicable. Should the host be unable to overcome illness, death results.
-Image below
The Course of an Infectious Disease. Most infectious diseases occur in four stages. The duration of each stage is a characteristic feature of each disease. The shaded area represents when many diseases are communicable. Should the host be unable to overcome illness, death results.
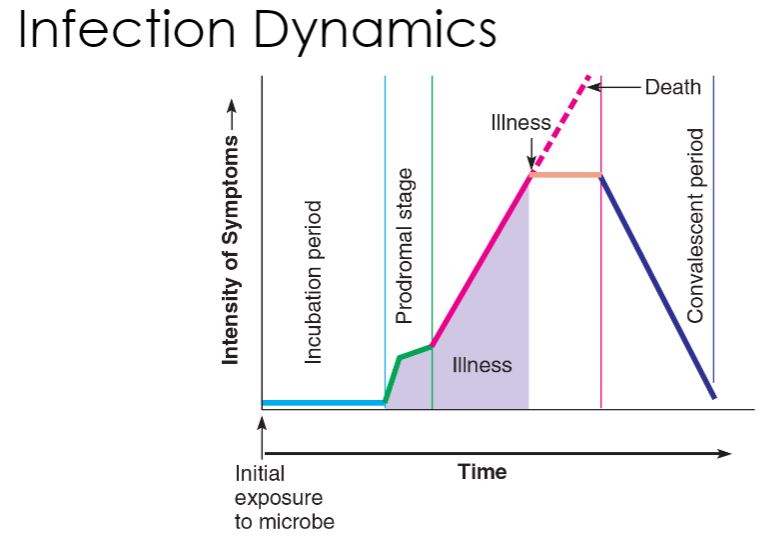
100
New cards
What determines if you get sick?
-Infectious diseases are typically not detected until distinct physical and/or physiological events alert the host of cellular battle. Infectious diseases often have characteristic signs and symptoms. Signs are objective changes in the body, such as a fever or rash, that can be directly observed. Symptoms are subjective changes, such as pain and loss of appetite, that are experienced by the patient. The term symptom is often used in a broader scope to include clinical signs. A disease syndrome is a set of signs and symptoms characteristic of a particular illness.
The fundamental process of infection is essentially a competition for resources. The host is a source of protection, nutrients, and energy for the pathogen to use. Infectious agents must therefore develop mechanisms to access and exploit their hosts. Furthermore, to continue surviving, pathogens must also devise methods to move on to better environments once the immediate one declines in value, at which point dissemination to another location or a new host must occur.
For a pathogen to infect a host and subsequently cause disease, specific events must take place. First, the organism must be transmitted from the previous host or reservoir and enter the new host. Certain factors will affect the success of this process: the virulence of the organism, the number of invading organisms, and the immediate response of the host. Second, the organism must be able to outcompete resident microbes for resources and survive the continued onslaught of host defense mechanisms. Third, disease ensues when the organism either produces molecules that directly damage host cells, stimulates the immune cells to destroy infected tissue, or alters the host-cell genome, which affects normal function. It is important to note that often the factors that help the pathogen avoid the host defenses are the same molecules that damage the host, hence there is a great deal of overlap when discussing these stages of the infection process.
The fundamental process of infection is essentially a competition for resources. The host is a source of protection, nutrients, and energy for the pathogen to use. Infectious agents must therefore develop mechanisms to access and exploit their hosts. Furthermore, to continue surviving, pathogens must also devise methods to move on to better environments once the immediate one declines in value, at which point dissemination to another location or a new host must occur.
For a pathogen to infect a host and subsequently cause disease, specific events must take place. First, the organism must be transmitted from the previous host or reservoir and enter the new host. Certain factors will affect the success of this process: the virulence of the organism, the number of invading organisms, and the immediate response of the host. Second, the organism must be able to outcompete resident microbes for resources and survive the continued onslaught of host defense mechanisms. Third, disease ensues when the organism either produces molecules that directly damage host cells, stimulates the immune cells to destroy infected tissue, or alters the host-cell genome, which affects normal function. It is important to note that often the factors that help the pathogen avoid the host defenses are the same molecules that damage the host, hence there is a great deal of overlap when discussing these stages of the infection process.
