BCH - Exam 3
1/108
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
109 Terms
Biological cell membrane
Thin
Semi-permeable
Control movement of substances into and out of cell
Selective for different substances
Phospholipid bilayer with proteins embedded and sugars associated
Define borders of cells, tissues, and organelles
Membrane composition
Phospholipids, proteins, cholesterol, and carbohydrates (sugars)
Different cells & organelles have different amounts of each type of constituent and this is related to function
Properties of lipids
Significant # of C-H bonds
Insoluble in water
Amphipathic: hydrocarbon (hydrophobic) tail + hydrophilic head
Integral membrane
Protein whose amino acids are inserted into membrane e.g., transmembrane protein
Peripheral membrane
Associate proteins do not span membrane
Proteins often in cytoplasm and associated with membrane upon PTM
Fluid Mosaic model
Membranes are fluid
Lipids form a bilayer embedded primarily with proteins
Components "float" around in 3D space
Evidence: Frye and Edidin experiment showed protein mixing when cells fuse
Fatty acid composition
Carboxylic acids (head) + long-chain hydrocarbon side groups
Even number of carbon atoms
Most common have 14-20 carbons
Rarely found "free" in nature
Biosynthesized by connection of C2 units
Occur in esterified form → modified to form “more complex” lipids
Saturated fatty acids
Only single C-C bonds
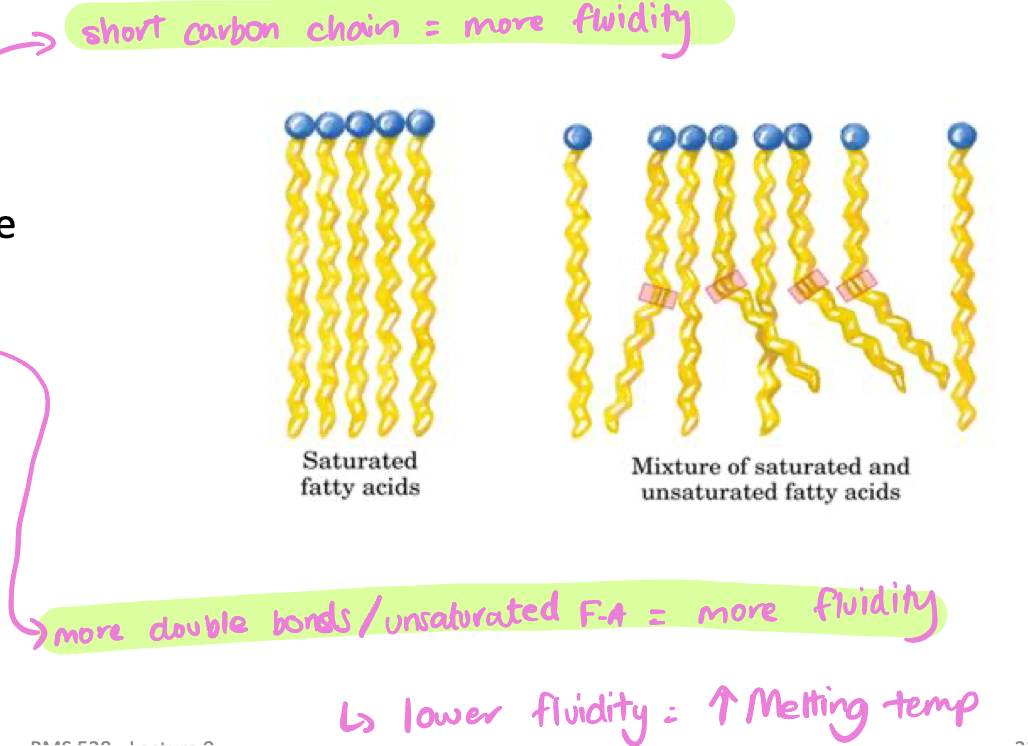
Less fluid → higher melting temperature
Longer chains = more solid
Unsaturated fatty acids
Contain one or more C=C (double) bonds
More fluid → lower melting point
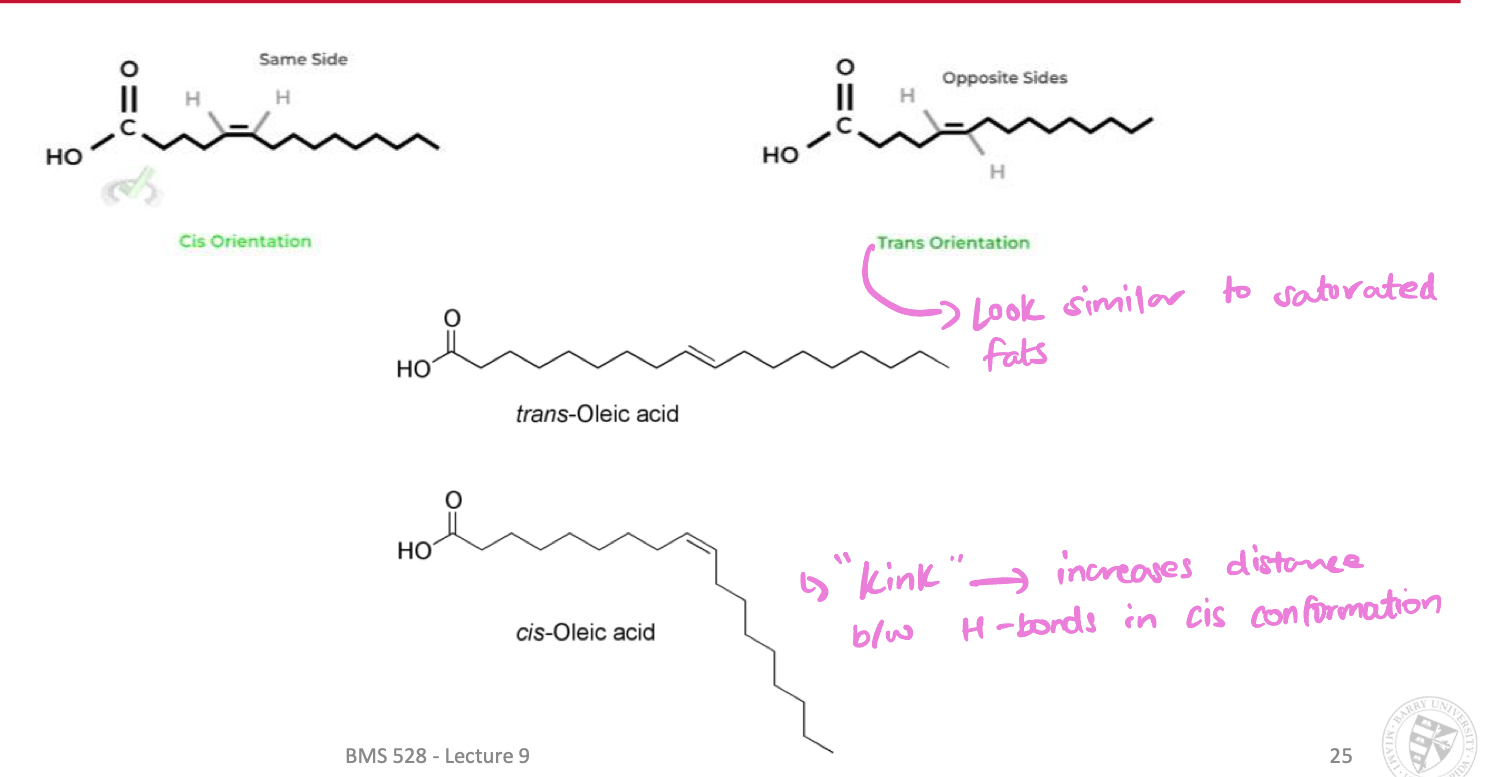
Cis vs. Trans configurations affect properties
“kink” in cis configurations → increases distance between H-bonds
Factors affecting fluidity of fatty acids
Carbon chain length
long chain = more solid/rigid = lower fluidity = higher melting temp
Number of cis double bonds
fewer cis bonds = less kinked = more solid/rigid = lower fluidity = higher melting temp
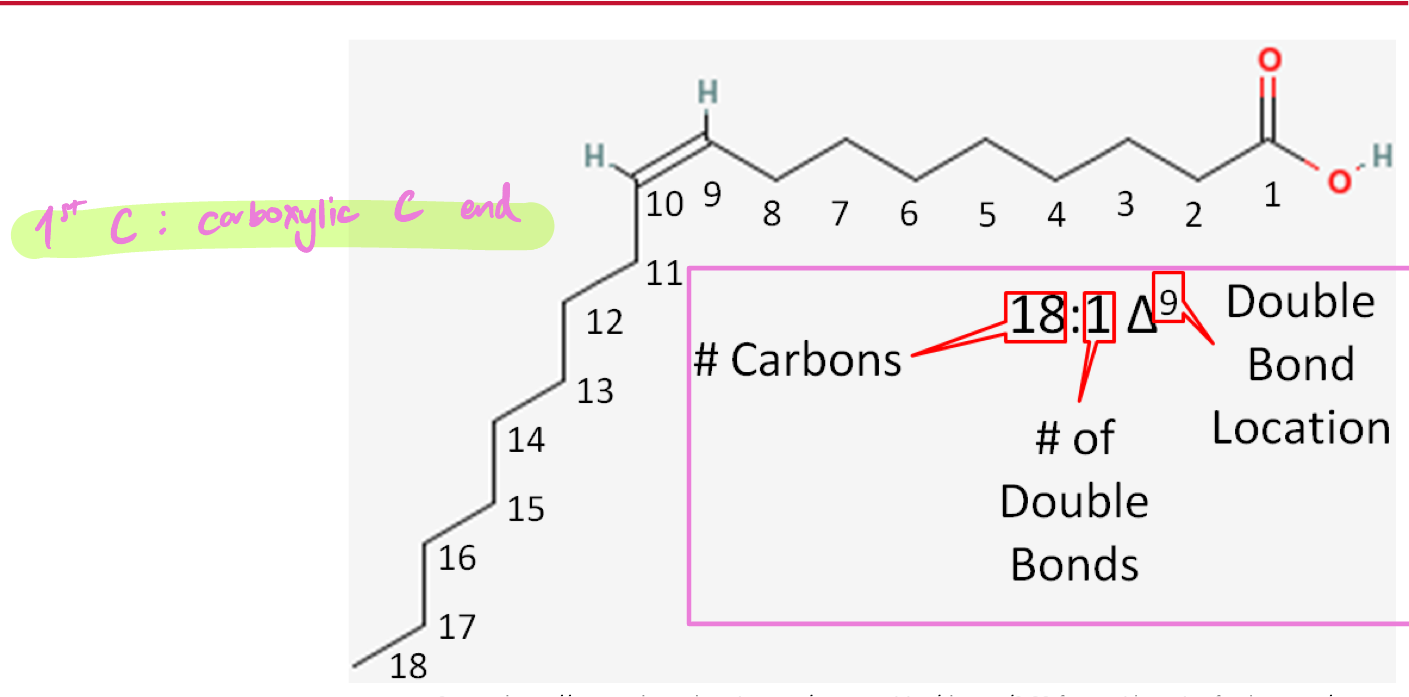
Δ nomenclature of FA
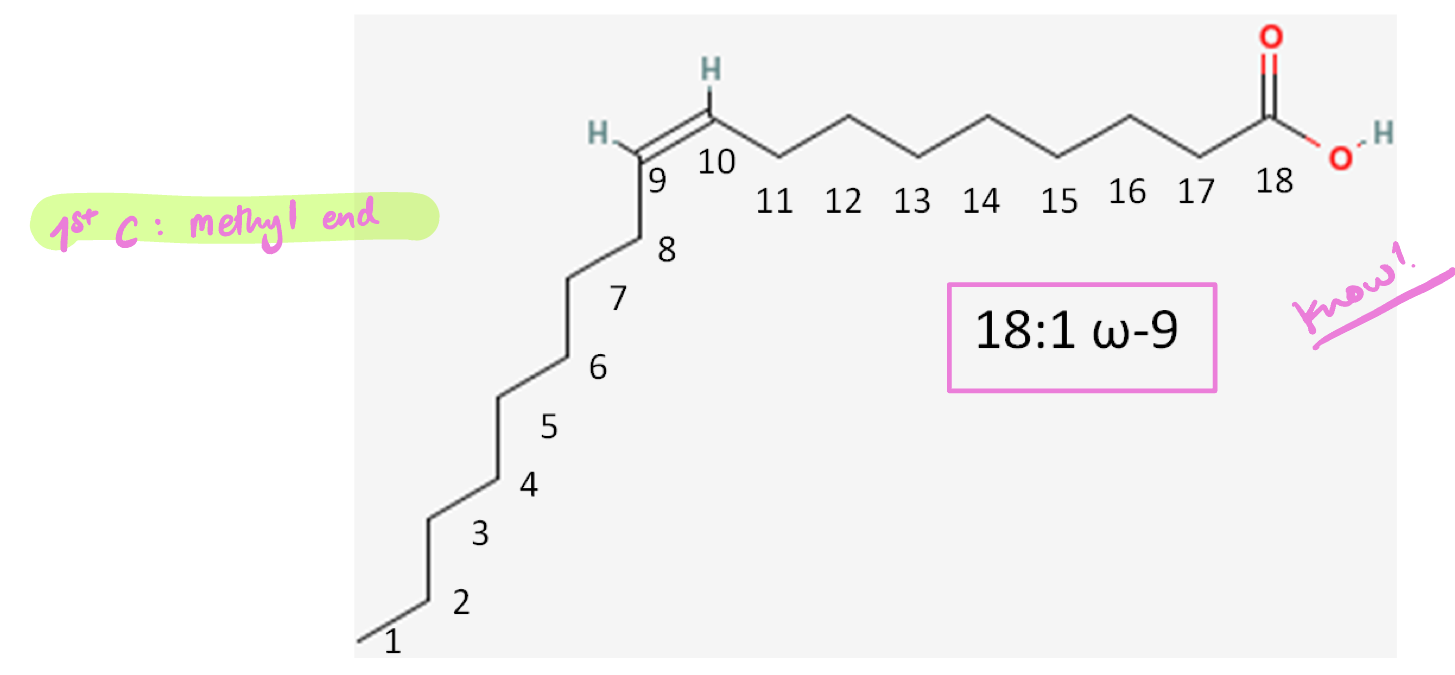
ω nomenclature of FA
Micelle
Unilayer assembly of lipids that have a hydrophobic core → transport of hydrophobic substances
Membrane lipids
Phospholipids
Sphingolipids
Cholesterol
Phospholipids
Amphipathic:
Polar hydrophilic head: phosphate & alcohol
2 hydrophobic fatty acid tails
Head & tail joined by phosphodiester linkage
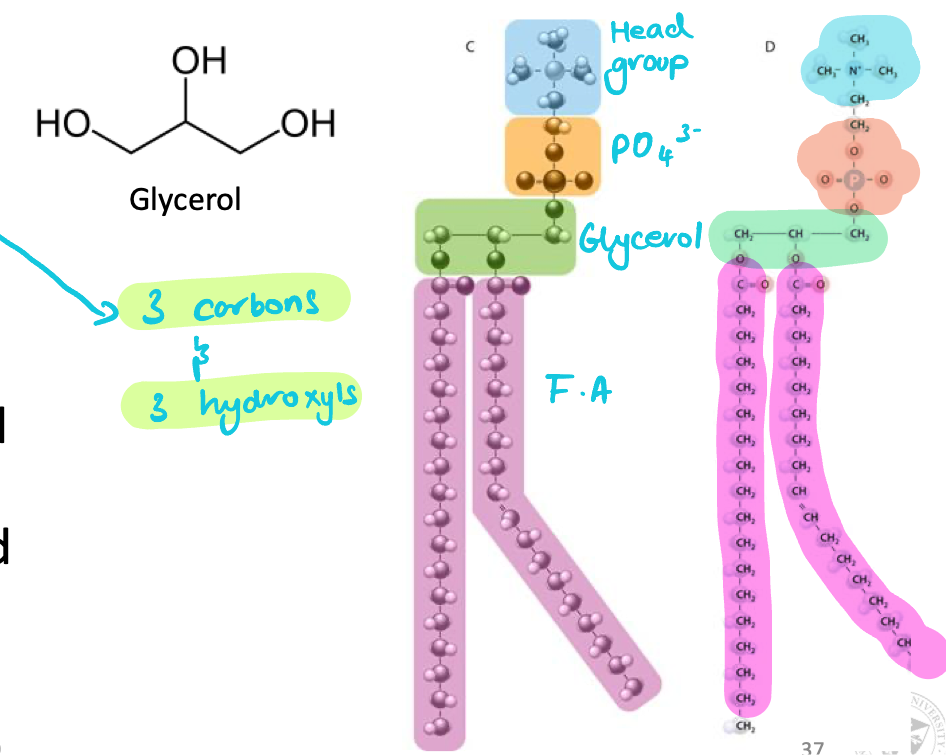
Glycerophospholipids = glycerol backbone
Sphingophospholipids = spingosine backbone
Major constituents of biological membranes
Glycerophospholipids
Major lipid component of biological membranes
Glycerol backbone: 3 carbons & 3 OH
2 carbons esterified to fatty acid tails
Bonded to a fatty acid by ester link & polar head group via a phosphate group
Characterized by variable head groups (different molecular size & charges)
Largest glycerophospholipid
Phosphatidylinositol 4,5-bisphosphate (PI)
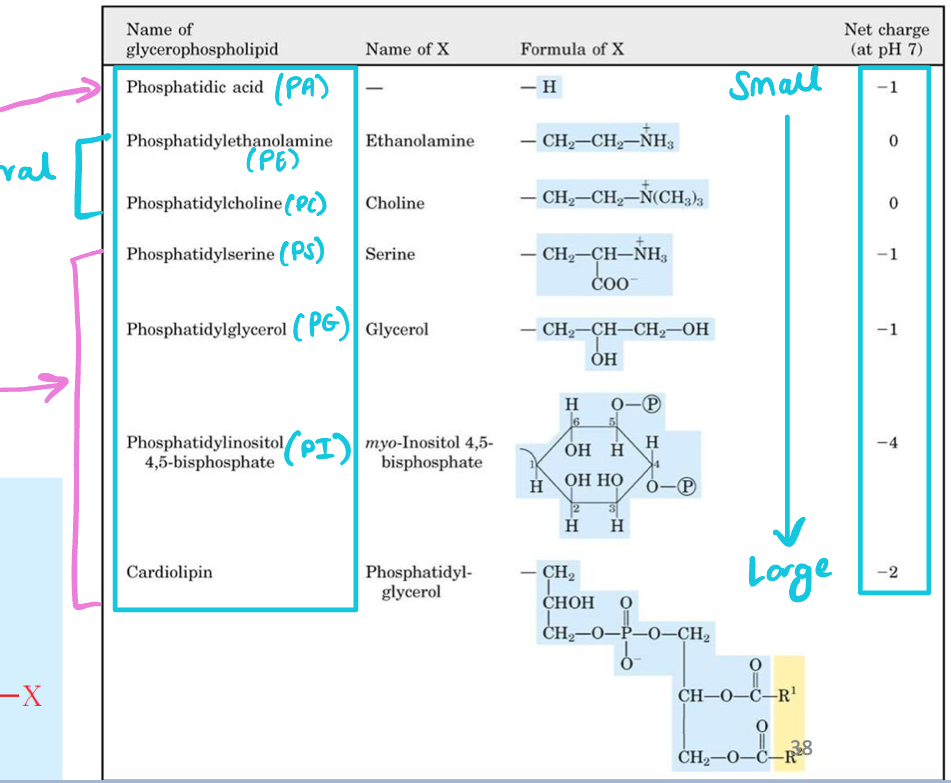
Smallest glycerophospholipids
Phosphatic acid (PA)
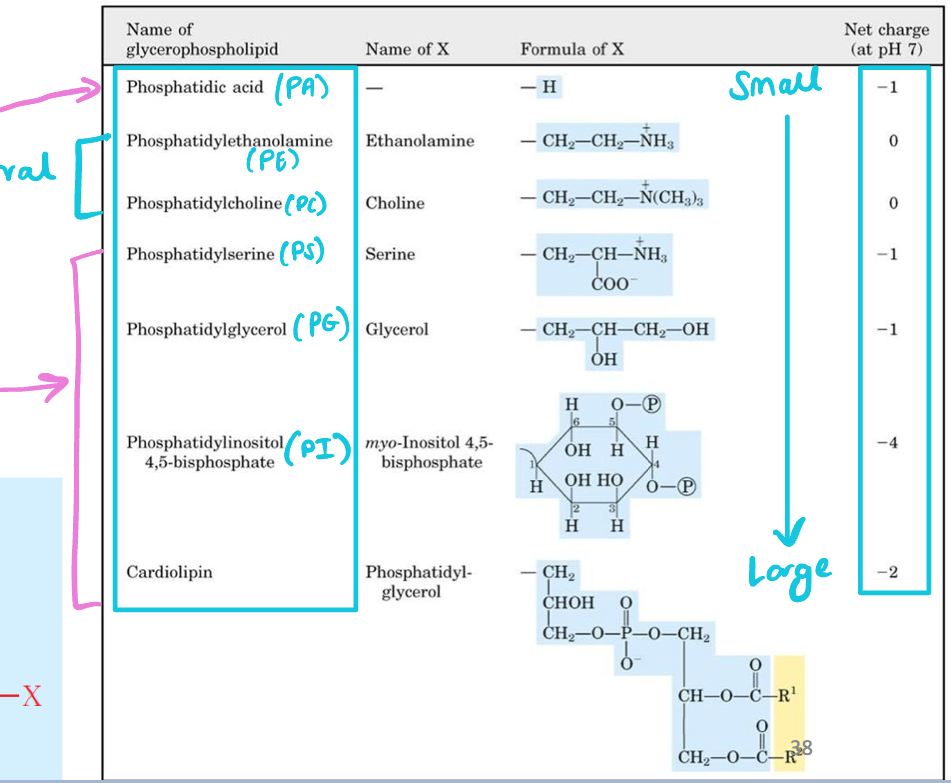
Neutral glycerophospholipids
Phosphatidylethanolamine (PE)
Phosphatidylcholine (PC)
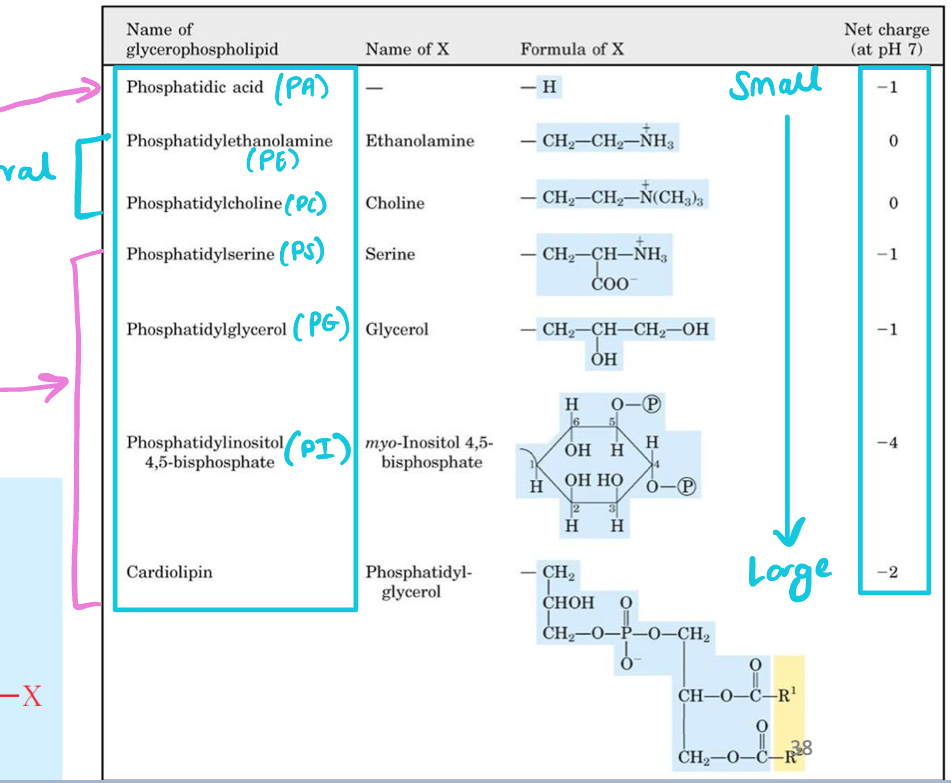
Negative-charged glycerophospholipids
Phosphatidylserine (PS)
Phosphatidylinositol 4,5-biphosphate (PI)
Phosphatidylglycerol (PG)
Phosphatidic acid (PA)
Cardiolipin
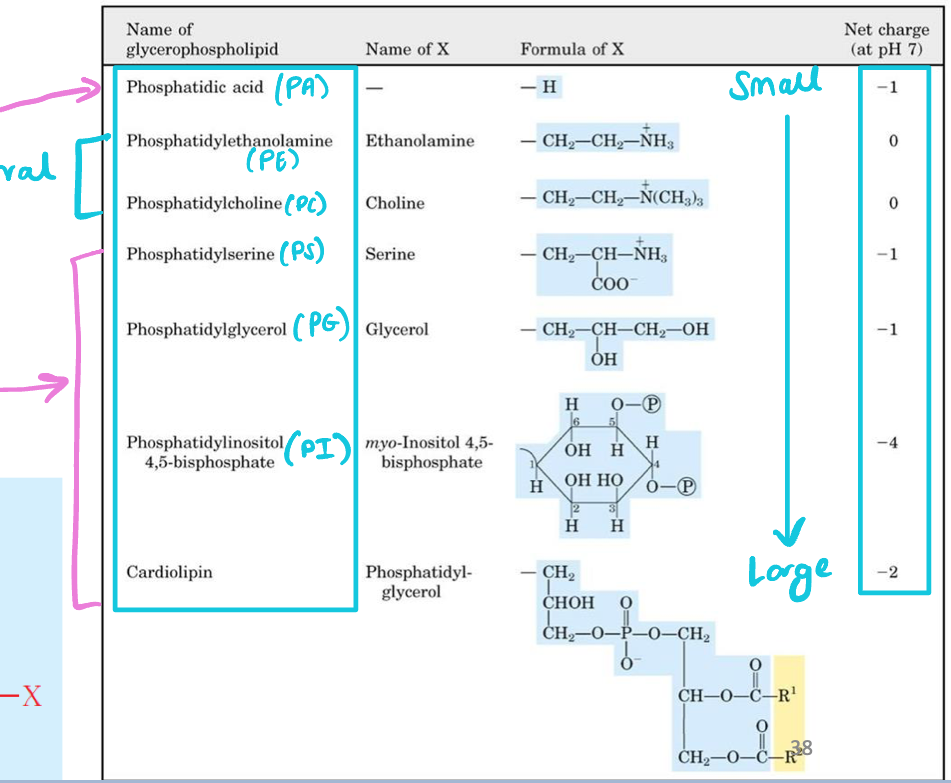
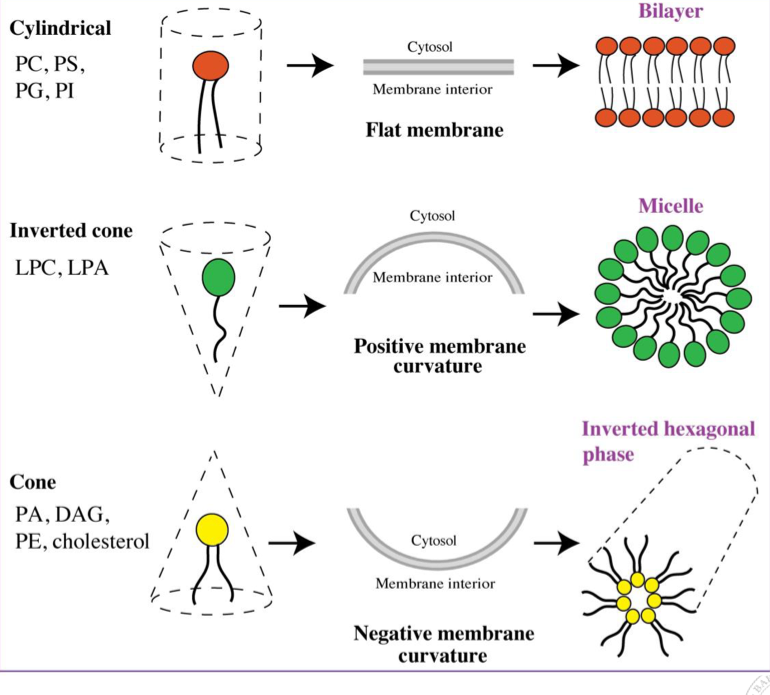
Shape of glycerophospholipids
Head group (size and charge) + fatty acid (saturated vs unsaturated) dictates “shape” of a lipid
Small head, unsaturated tails = cone, negative curvature
Cylindrical lipid (head = tail size) = flat membrane
Distribution of lipids between membrane leaflets
Inner leaflet of bilayer contains higher % of negatively charged glycerophospholipids (PS and PI) than outer leaflet (neutral lipids)
Phospholipid asymmetry → protein folding
Sphingolipids
Backbone: sphingosine
Can be phospholipids or not
Derivatives of 18 carbon unsaturated, amino alcohol sphingosine (relatively long)
Fatty acid linked to amino alcohol sphingosine via an amide bond to form a ceramide
Very stable, rigid & provides insulation → common in neurons
Ceramide as a common precursor
Cerebroside: ceramide + glucose head group
Ganglioside: ceramide + oligosaccharide head group
Cholesterol
Unique tetracyclic structure
Helps membrane flexibility → keeps membranes from solidifying when cold and breaking apart when hot
Stiffens & maintains fluidity of membrane lipids
Weakly amphipathic because of hydroxyl group
Bulky, rigid structure disrupts regular fatty acid chain packing in membranes
Enriched in lipid rafts
Membrane rafts
Membranes can have “domains”
Lipid rafts: type of membrane domain (enriched with cholesterol)
Contain tightly packed lipids
Enriched in saturated lipids
Function: cell signaling and sorting of proteins into organelles
Cardiolipin
Double glycerophosphate backbone + 4 fatty acid tails
Conical in shape
More IMM = more ETC proteins = more energy!
Found in inner mitochondrial membrane
Both outer & inner mitochondrial membranes are bilayers
GPCR
7 transmembrane domains
C-terminus intracellular (cytoplasmic) → binds G-proteins
N-terminus extracellular
“Inside positive” rule
Negative charges of phospholipids on inner leaflet of membrane interact via charge-charge interactions with +ve charged amino acids on cytoplasmic side of transmembrane proteins
Protein translocon (spans ER membrane) → passes proteins
Passive transport
No energy required (ΔG < 0)
Non-mediated transport (simple diffusion)
Mediated (facilitated diffusion): diffusion of solutes accelerated by membrane proteins (pores, carriers, permeases)
Non-mediated transport
Simple (passive) diffusion
Slow & down concentration gradient
Easier for hydrophobic (non-polar) than for hydrophilic molecules
Results in equal concentrations on both side of membrane
For non-polar and uncharged molecules e.g., gases (O2 & CO2)
Mediated (facilitated) transport
Passive transport
Accelerated diffusion for molecules that cannot do simple diffusion (fast enough)
Important for polar and charged molecules e.g., ions, glucose, water
Molecules that facilitate diffusion (mediated transport)
Transmembrane protein pores (aquaporin) → fast and selective (Na, K, Cl)
Carrier molecules (ionophores) → highly selective
Permeases (GLUT transporters) → generally slower, but more specific than protein pores
also transmembrane
Aquaporins
Water channel proteins
Structure = function → hydrophilic inside channel & hydrophobic outside channel
Crucial in cells like erythrocytes
Rapid transport of water → maintain osmotic balance & ion gradients
Glucose Transporters (GLUT)
Carrier protein (facilitated diffusion)
Facilitate glucose movement down concentration gradient (passive: high → low)
Transport can occur in both directions depending on concentration gradient of glucose
Active mediated transport
Energy/ATP required from ion gradients (ΔG > 0)
Crucial for maintenance of membrane potentials
Movement against concentration gradient
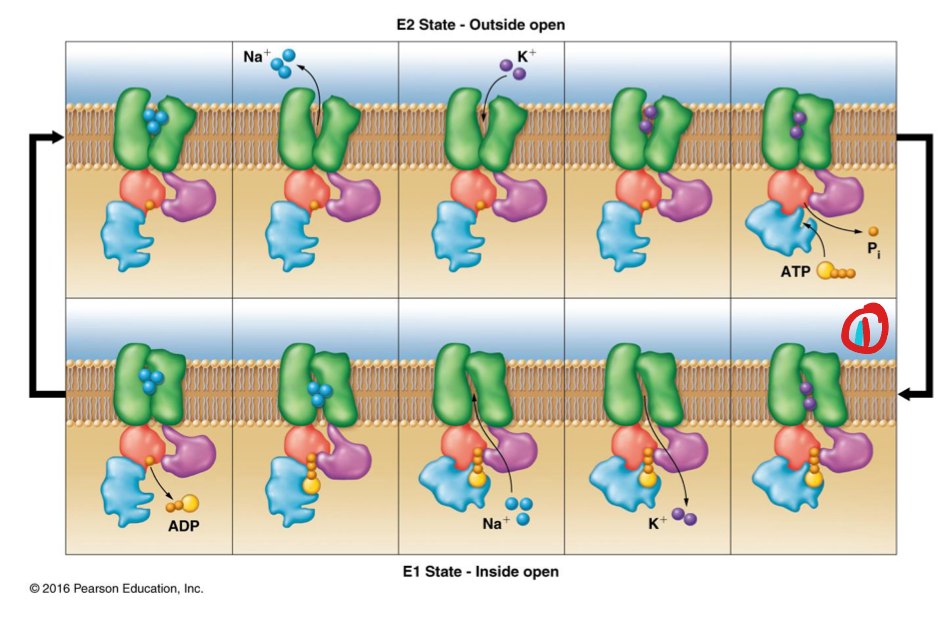
Na/K ATPase
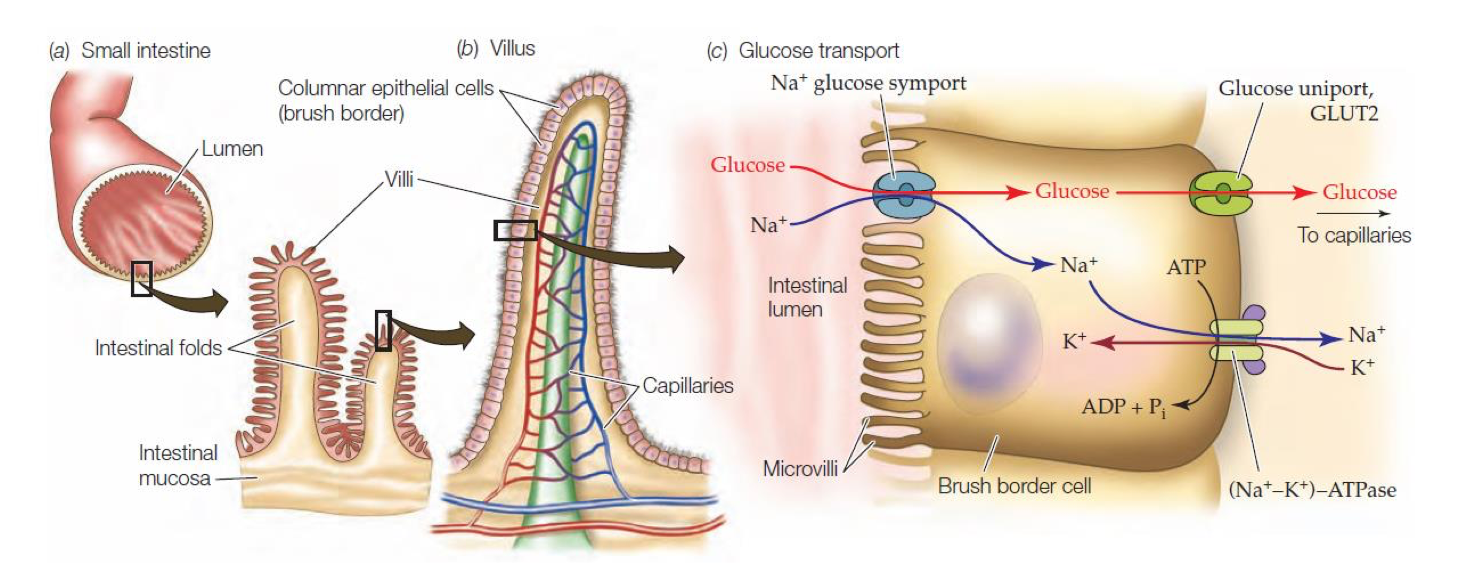
Na-Glucose Symport
Na+/K+ ATPase
Active transport of ions using ATP hydrolysis
Antiport: 3 Na+ ions out & 2 K+ ions in
E1 conformation: open on inside of cell, closed on outside, and has affinity for Na+
E2 conformation: open on outside of cell, closed on inside and has affinity for K+
Mechanism of Na/K ATPase
E1 state binds intracellular ATP via intracellular subunits and binds 3 Na+
Transporter is phosphorylated → conformation change (“high-energy”)
Protein adopts E2 conformation → 3 Na+ released out of cell (through membrane)
E2 binds 2 K+ from outside cell
Phosphate group on protein (transporter) is hydrolyzed → conformational change to E1 (binds ATP)
2 K+ released into cell
Na-Glucose symport
Secondary active transport (does not use ATP directly)
Generates potential energy in the form of ion gradients
Symport: Na+ & glucose transported in same direction
Cell signaling
Ability of a cell to receive, process, and transmit signals
Occurs between: cell & environment, neighboring cells and within the cell itself
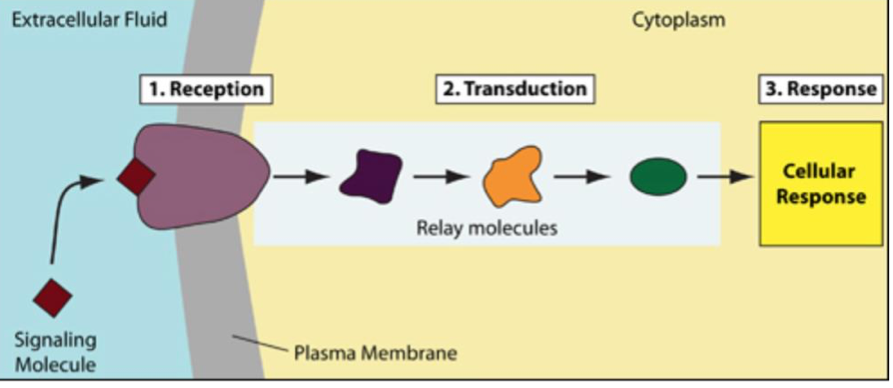
Signal transduction
Process by which a chemical or physical signal is transmitted through a cell as a series of molecular events
Stages of cell signaling
Reception: cell detects stimulus (molecule) by receptor
Intracellular: if ligand is trapped inside cell or is small/hydrophobic enough to pass through membrane
Extracellular: on cell surface if ligand is not permeable across membrane
Transduction: signaling molecule binding induces receptor protein change
Response (effect): receptor change triggers specific cellular change
Resetting/Recycling (optional): cell and receptor return to original state
Types of receptors
Cytokine receptors: IL-2R
Receptor tyrosine kinase: insulin receptor, epidermal growth factor receptor (EGFR),
GPCR: acetylcholine signaling, 𝛼1-adrenergic receptor
Channel proteins (ligand-gated ion channel): acetylcholine
Receptor Characteristics
Proteins made of amino acids
Amphipathic:
At least one hydrophobic transmembrane domain
Hydrophilic extracellular domain for ligand binding
Hydrophilic intracellular domain for effector binding
Receptor Tyrosine Kinases (RTKs)
Single transmembrane domain
Built-in cytoplasmic enzyme (kinase) domain → phosphorylate tyrosine amino acids to form recognition sites for scaffolding or effector proteins
Must dimerize to be functional
Kinase domains of each RTK monomer cross-phosphorylate cytoplasmic domain on other receptor unit
G-protein coupled receptors (GPCR)
7 transmembrane domains
N-terminus always extracellular
C-terminus always intracellular → binds G proteins for signal transduction
G proteins bind GDP or GTP
Channel Proteins
Form hydrophilic membrane pores
Narrow & highly selective → gated (unlike pores = open)
Ion channels:
Voltage-gated channels
Mechanically gated channels
Ligand-gated channels: respond to ligand-binding
Endocrine ligands
Long range
Signals travel from distant cells via bloodstream
Slow, long-lasting response
Paracrine ligands
Short range
Local signals between nearby cells
Quick, short-lived response
Autocrine ligands
Very short range
Cell produces and responds to its own signals
Ligand classification by effect
Hormones: endocrine glands; involved in homeostasis
Growth factors: endocrine, paracrine or autocrine; stimulate growth
Cytokines: paracrine or autocrine; immune response
Neurotransmitters: paracrine; stimulate neurons
Types of transducing (effector molecules)
When receptors bind their ligands, they “change conformations” and activate intracellular processes (protein activation)
Enzymes: kinases, phosphatases, hydrolyases
Second messenger molecules: cAMP, inositol triphosphate, diacylglycerol
Post-Translational Modifications
Phosphorylation & lipid addition (isoprenylation) to proteins
Can:
Switch proteins on/off
Change protein location
Target proteins for degradation
Kinase
Catalyzes transfer of phosphate group from ATP to a molecule
Can elicit an ON or OFF effect
Phosphatase
Removes phosphate group from its substrate without ATP
Phosphorylation
Covalent, dynamic modification
Very common in signal transduction pathways
Involves transfer of phosphate group from ATP to hydroxyl group on amino acid
Adds negative charge to protein
Does not involve water addition or loss → not a hydrolysis reaction (unlike GTPase)
Isoprenylation
Addition of hydrophobic molecules (fatty acid) to a protein
Farnesylation of Ras proteins (GTPase protein)
Ras proteins only functional in membrane (ineffective in cytoplasm)
C-terminal tail w/ lipid attached enters
GTPases (G-proteins)
Bind to and hydrolyze guanosine triphosphate (GTP) molecules
Also known as guanine nucleotide-binding proteins (G-proteins)
Hydrolase enzymes → bind to and hydrolyze GTP molecules to GDP
Ras proteins
Guanine Nucleotide Exchange Factors (GEF): GDP → GTP (turn GTPase ON)
GTPase Activating Protein (GAP): GTP → GDP (turn GTPase OFF)
Second messenger molecules
Small molecules that are enzymatically generated and transmit signals within a cell
Inositol phosphate
Diacylglycerol
cAMP: synthesized from ATP by adenylate cyclases
cAMP
Intracellular signaling molecule
Activates protein kinase A (PKA) by binding to regulatory subunits of PKA relieving inhibition
PKA phosphorylates (activates) transcription factors to promote gene expression
Adenylate cyclase → cAMP → PKA → CREB (phosphorylated in transactivation domain)
Inositol phosphates and diacylglycerol
Intracellular signaling molecules
IP3
Binds to intracellular calcium channels to release calcium
Formed by phospholipase: cleaves head group from PI → DAG in membrane & IP2 in cytoplasm
Derived from phosphatiyl inositol (glycerolipid)
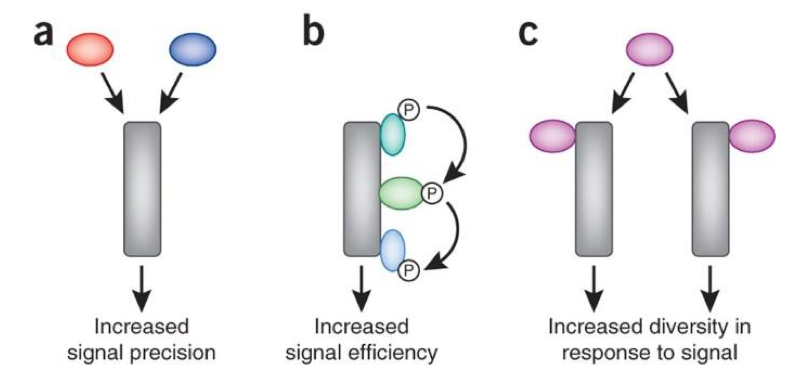
Signal integration
Processing multiple simultaneous signals to coordinate a unified cellular response
Increased signal precision & efficiency
Increased diversity in response to signal
Two or more pathways downstream of common receptor
Converging pathways: two phosphorylation events needed to activate an enzyme requiring two different pathways
Opposing pathway effects
Signal amplification
Increase in intensity of a signal through networks of intracellular reactions
Involves enzymes and production of second messenger molecules
SH2 domain effectors
Src homology 2 domains (SH2 domains) bind phosphorylated tyrosines on RTK
Serve as scaffolds for other effector proteins e.g. bring nucelotide exchange factor proteins (GEF) close to GTPases to facilitate nucleotide loading (GTPase activation)
SOS (GEF for Ras protein) bound to Grb2 (SH2 domain) → activates GTPase in membrane
Insulin signaling
Insulin receptor (receptor tyrosine kinase)
Phosphotidylinositol-3-kinase phosphorylates PIP2 → PIP3
PH domain binds PIP3 → upregulates expression of GLUT4 transporter on cell surface → increase glucose uptake by cells, lowering blood sugar levels
Epidermal Growth factor signaling
Paracrine
EGF binds Epidermal Growth factor receptor (EGFR) → signaling events that allow for cell cycle progression, cell growth
Effectors: PI3K, GTPase, Phospholipase C
G-proteins
Found in cytoplasm
Heterotrimers: Gα (binds GDP or GTP), Gβ, Gγ
In inactive state, Gα bound to GDP
GPCR binds ligand → receptor changes conformation → allosteric change in Gα → GDP replaced by GTP
GTP activates Gα causing it to dissociate from GβGγ (remain linked as dimer)
Activated Gα activates another effector
Gα subunit
Gα: inhibits adenylate cyclase → cAMP decreases → PKA decreases
Gαs: stimulates adenylate cyclase → cAMP increases → PKA increases
Gαq: stimulates phospholipase C (PLC) → converts phosphatidylinositol (PI2) into diacylglyercol (DAG) and inositolphosphates (IP3)
𝛼1- adrenergic receptor
GPCR: leads to muscle contraction by increasing calcium
𝛼1- AR binds to Gαq
Gαq activates PLC
PLC converts PIP2 → IP3 + DAG
Acetylcholine signaling
Neurotransmitter = paracrine signaling molecules
Signaling pathway:
Ligand gated ion channel: ion diffusion across membrane → rapid depolarization
GPCR
Interleukin-2 signaling
Autocrine: expressed by activated T cells & stimulate cell proliferation
IL-2 binds to portions of IL-2 receptor (IL-2R) → receptor forms trimer (oligomeric) → kinases associate with trimeric complex to stimulate signaling
IL-2R is not a RTK!
Signaling Termination
Reverse activating modifications e.g., kinase vs. phosphatase
Receptor internalization (endocytosis via β-arrestin) and degradation
Ligand-Receptor Pharmacology
Characterized by:
Binding rates (on/off rates)
Dissociation constant (Kd)
Receptor affinity
Ligands can switch signaling on/off & moderate signaling intensity
Kon rate
On-rate: how readily ligands bind to receptors
Koff rate
Off-rate: how readily ligands unbind from receptors
Dissociation constant (Kd)
Concentration of ligand required to occupy 50% of receptor binding sites
Kd is inversely related to binding affinity
High Kd = low receptor-binding affinity
Low Kd = high receptor-binding affinity
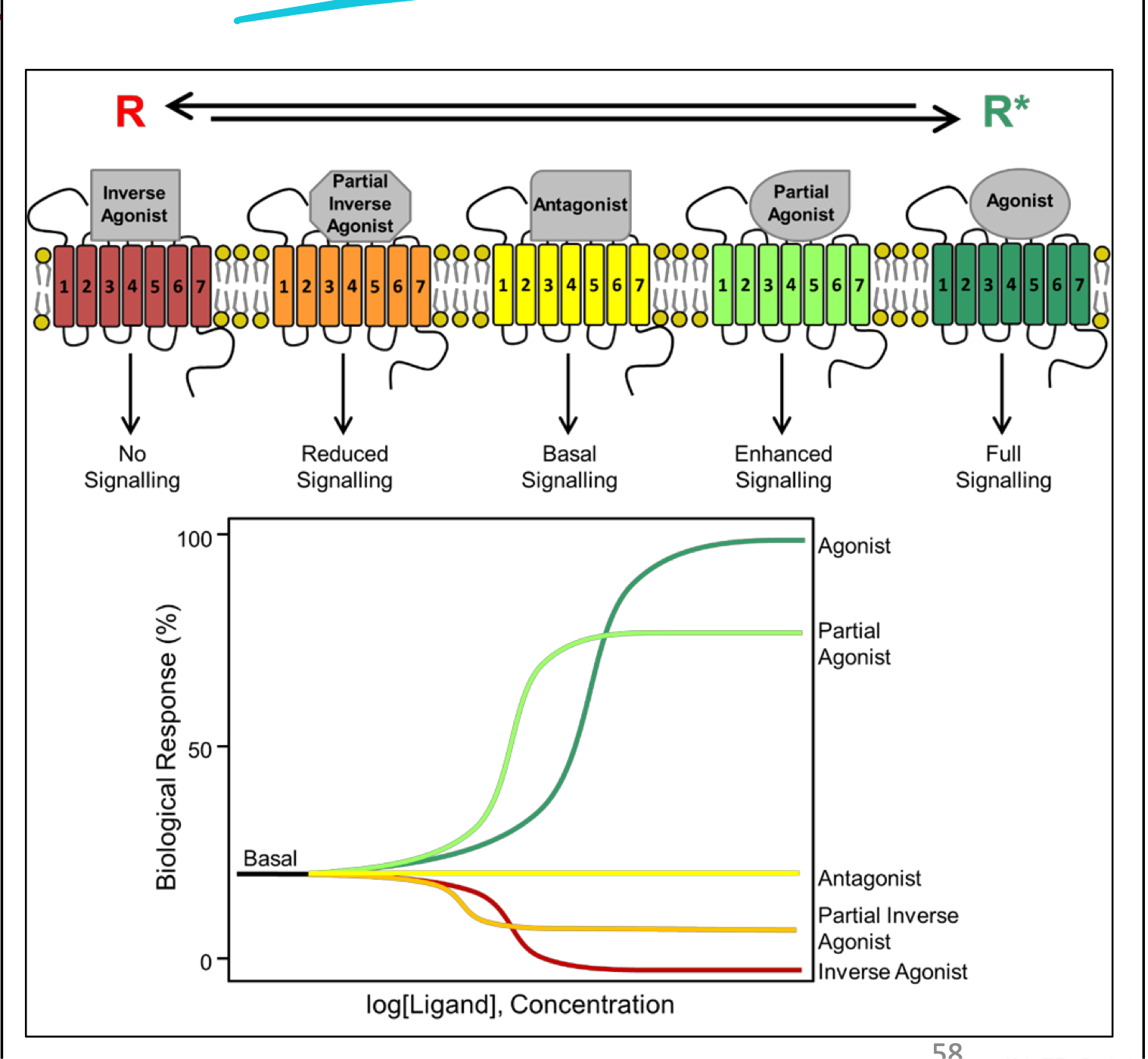
Ligand binding efficacy
Agonists: always elicit a biological response (ON or OFF)
Inverse agonists: elicit OFF response
Antagonists: elicit NO biological response
Can inhibit agonists (biological response elicited)
Restriction endonuclease enzyme
Cut DNA at specific target sequences
Create “blunt” or "sticky” ends (short, single-stranded overhangs) for DNA manipulation
If two molecules have these “sticky ends” complementary overhangs, they can base-pair and stick together → use DNA ligase to seal two molecules together and join gaps in DNA backbone
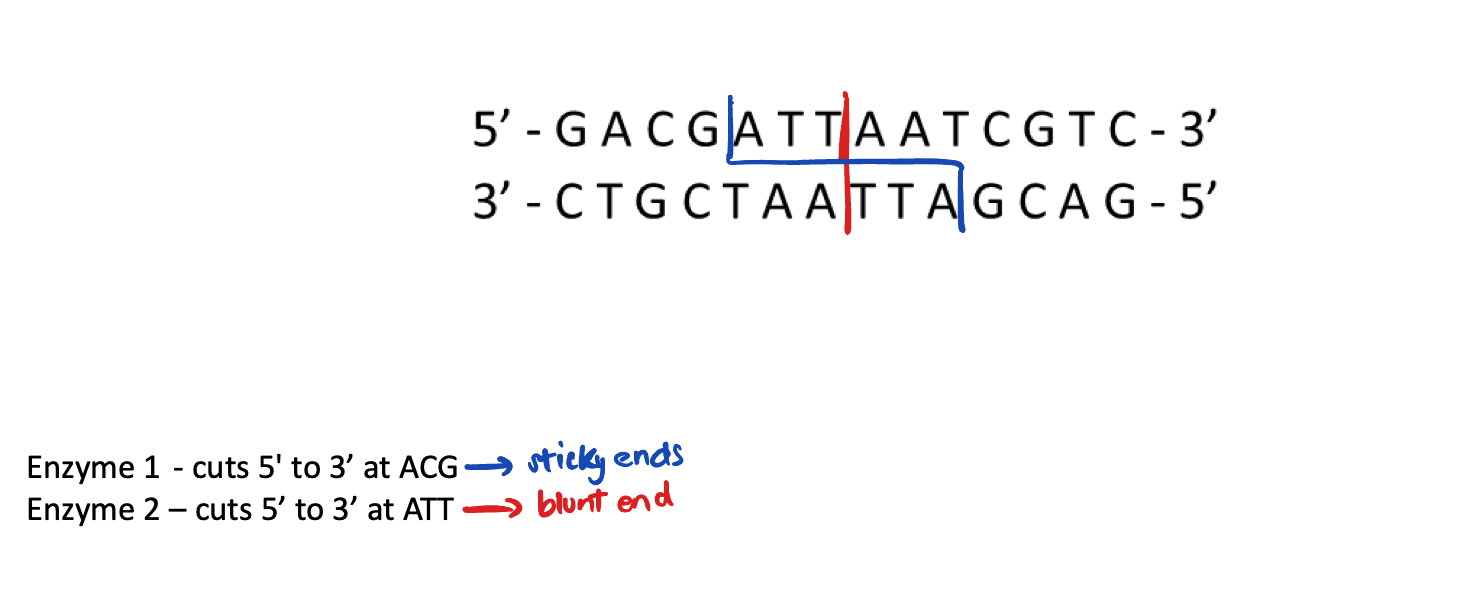
Challenges of gene isolation and cloning
Genes are not discrete entities
Difficult to purify specific genes from complex DNA mixtures
Plasmid
Small, circular, self-replicating extrachromosomal DNA molecules
Naturally occur in bacteria
Advantages/uses of inserting gene of interest into plasmid:
Protect genes from degradation
Replicate genes in bacteria
We can stick DNA into plasmids → put plasmid into bacteria to produce lots of DNA
Cannot be inserted into humans:
Don’t contain right transcription regions for humans
Would be rapidly degraded
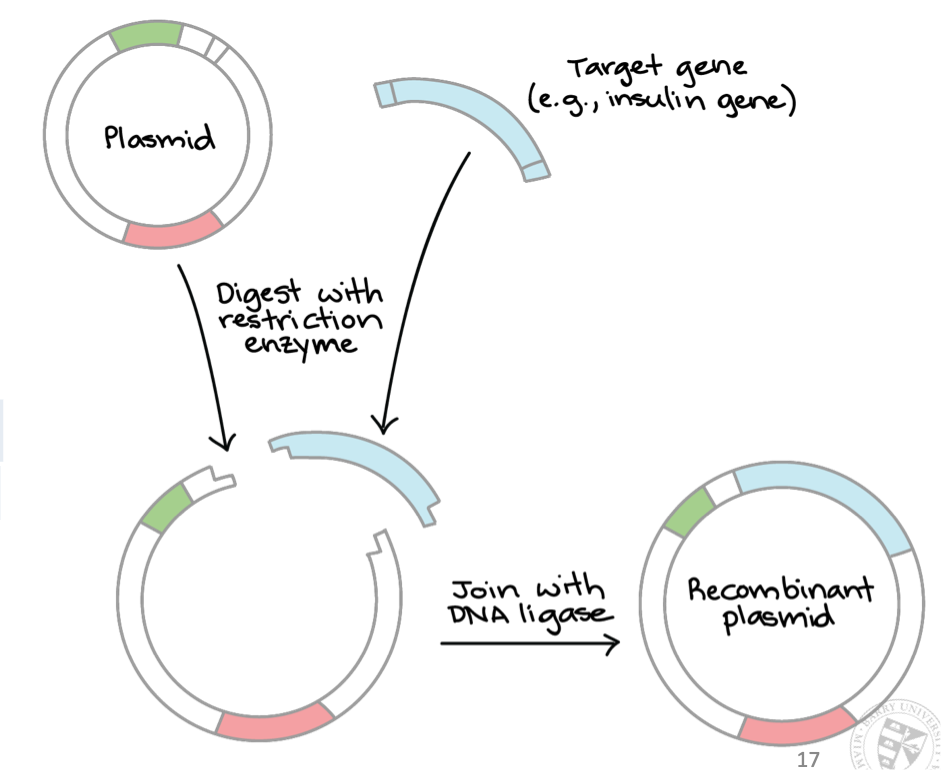
Recombinant DNA
DNA molecules formed by laboratory methods
Normally made by bringing together multiple pieces of DNA from different sources and incorporating them into one piece of DNA
Possible because while DNA differs between species in terms of sequence, structure of DNA is identical
“Recombinant” piece of DNA can then be inserted into cells for various uses
plasmid propagation
gene therapies
industrial scale production of proteins
Transforming bacteria with our plasmid containing gene
Once plasmid is inside bacteria → culture bacteria and generate billions of copies
How do we know the plasmid is in the bacteria?
Selection genes in plasmid
Antibiotic resistance
Features of a plasmid
ORI site (origin of replication)
Multiple cloning sites (restriction enzyme site)
Selectable markers (antibiotic resistance gene)
Promoter site
Genomic DNA libary
Contains entire genome of organism, including coding (exons) & non-coding (introns) regions
cDNA library
Collection of cDNAs made from mRNA, representing only expressed genes (exons)
Making a genetic library
Digest (cut up) DNA into fragments using restriction endonuclease enzymes
“Dilute” fragments into solution of digested plasmid → provides
conditions where one DNA fragment is ligated into one plasmid
Dilute plasmids and “transform” them → one plasmid into one organism
Genomic DNA library generated
Polymerase Chain Reaction
DNAP: duplicates DNA & needs primer
PCR used when you know which gene is needed from genome
Amplifies specific DNA segments
Uses gene-specific primers
Requires:
ssDNA template with sequence of interest/gene
Specific primers with complementary sequence to gene
DNA source
dNTPs, divalent cations (Mg²+), ATP, free 3’OH end
Heat-stable DNAP from bacteria
Denaturation: dsDNA separated using heat (H-bonds broken, NOT phosphodiester bonds/DNA backbone)
Annealing: heat reduced → primers specifically annealed to gene (target sequence)
Extension: heat-stable DNAP extends primers → synthesizes new DNA strands
Agarose gel electrophoresis
Used to separate/isolate DNA fragments by size (# of base pairs)
not applicable to proteins
Smaller fragments move faster through gel
-ve charge of DNA allows movement through gel towards positive terminus of gel
Reverse transcription of mRNA to make cDNA
mRNA (starting material)
Extract mRNA because it contains lots of A’s on its 3’ tail (poly A tail) → use primer rich in ‘T” to bind to tail
Primers:
Oligo-dT primers: bind to poly-A tail of mRNA; non-specific (good for whole transcriptome → make cDNA library of all mRNA)
Gene-specific primers (with complementary sequence): bind to a particular mRNA sequence (if targeting a specific gene)
cDNA
Complementary DNA synthesized from mRNA during reverse transcription
Contains only exons
How do we isolate a plasmid of interest from bacteria?
Alkaline lysis: isolate plasmid and not bacterial genomic DNA
Add alkaline-OH groups (makes solution alkaline) + detergent (SDS) to bacterial cell preparations
Detergent solubilizes (disrupts) cell membrane & alkaline-OH unzips DNA (disrupts hydrogen bonding between DNA bases converting dsDNA → ssDNA )
Neutralize the pH → easy for small circular plasmid DNA to re-nature (plasmid isolated)
Impossible for larger genomic DNA to properly anneal → precipitates out of solution
Filter out large precipitates (genomic DNA) using a filter and collect/concentrate plasmid DNA
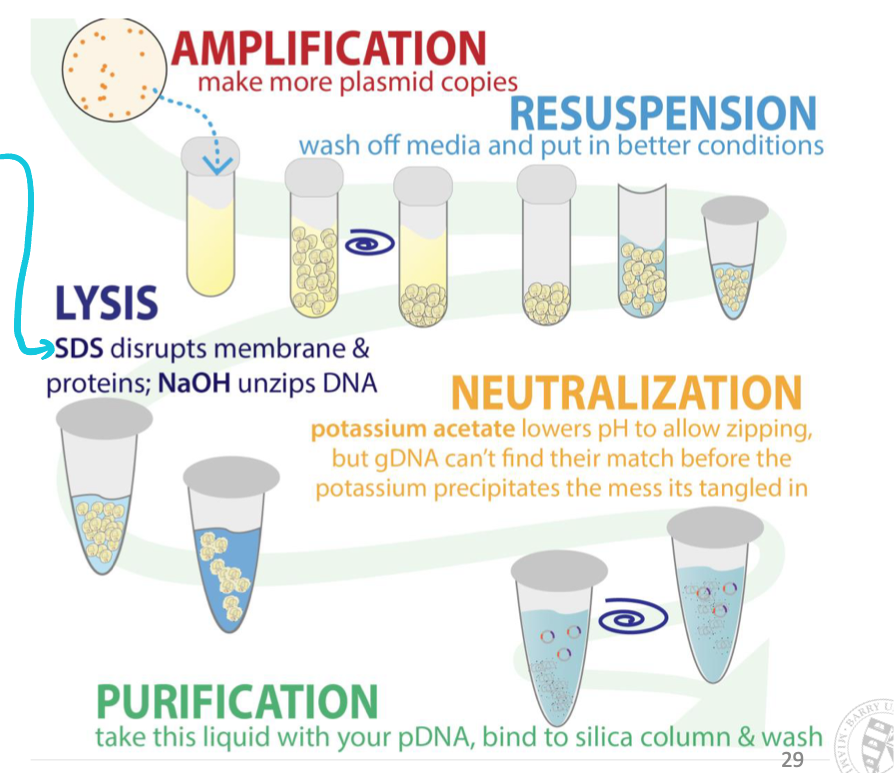
How do we know what the inserted DNA sequence in the plasmid is?
Sanger sequencing
Determines DNA sequence inserted into plasmids
Uses tagged ddNTP (dideoxynucleoside triphosphates) → terminates DNA strand elongation → length of fragments indicates sequence
Add template DNA; primer; DNAP; dNTPs; ddNTPs
DNAP extends primer by adding dNTPs
Occasionally, ddNTP is incorporated instead of dNTP
Once ddNTP is added, no more nucleotides can be added → synthesis terminated
Creates fragments of different lengths, each ending in a fluorescently labeled ddNTP
Gel electrophoresis: fragments separated by size (smallest move fastest)
Order of fluorescent colors indicates sequence of bases
Sources of recombinant genes
Genomic DNA
cDNA synthesized from mRNA
Isolating a gene
From mRNA: reverse transcribe to cDNA → PCR with gene-specific primers → gel electrophoresis → purify band
From genomic DNA: PCR with specific primers → gel electrophoresis → extract and purify band
From a library: screen cDNA/genomic library with a probe → identify and isolate clone
Via restriction digest: if restriction sites flank gene → cut with enzymes → purify via gel electrophoresis
Vector
Piece of “engineered” DNA that functions as a vehicle to carry foreign DNA molecules into another cell
Can be designed specifically for human cells (could contain human promoters)
Insert healthy gene into human cells using a viral vector (contains necessary genes to insert own DNA into host cell DNA)
Recombinant proteins
Host selection considerations crucial for amount of protein & correctly folded modified protein
Bacteria: produce more protein, less proper modification
Eukaryotic cells: produce less protein, more properly modified
Require: transcription start site, translation start site, ORI site
Inducible expression is advantageous so include an operon in bacterial systems
Isolating recombinant proteins
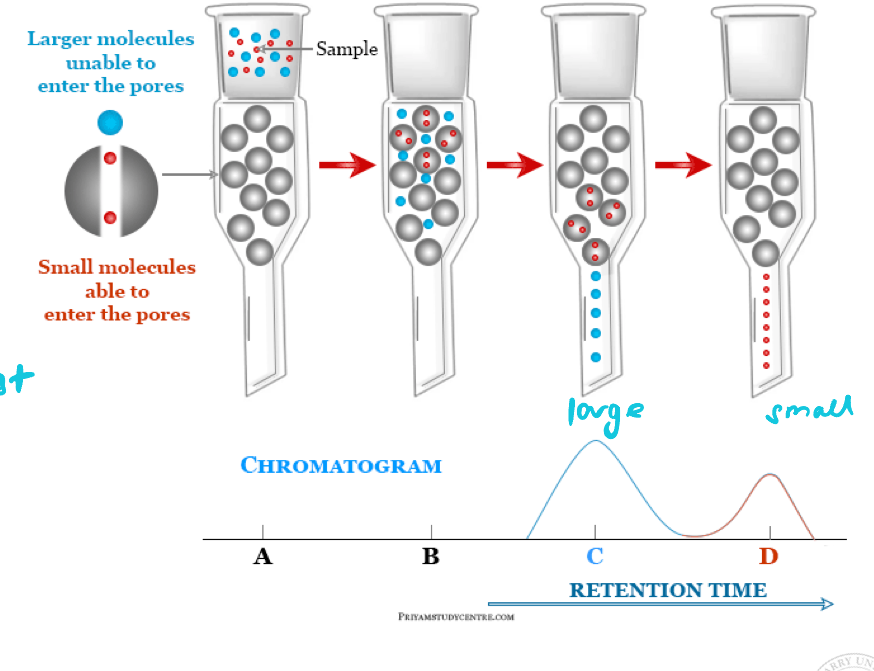
Column-based chromatography
Size exclusion: most common method to separate proteins
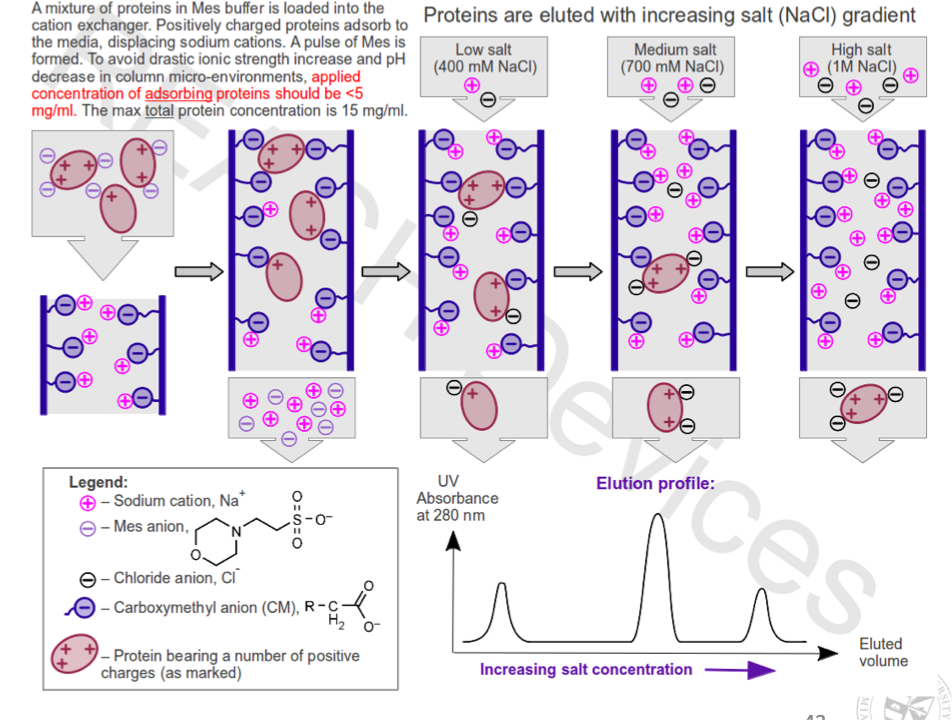
Ion exchange
Affinity
Electrophoresis: separates proteins by size/charge
SDS-PAGE for size-based separation
Size exclusion chromatography
Column contains a stationary phase made of beads with tiny pores
More interaction between small proteins and pores → slower progress of small proteins through column
Larger proteins elute first
Fractions collected separately
Molecular weight of proteins in each fraction investigated by SDS-PAGE
Ion exchange chromatography
Column contains stationary phase made of beads with charge
More interaction between opposite charged proteins and stationary phase → proteins retained while others pass through
To remove retained proteins wash with salt or pH gradient buffer
Salt (NaCl) competes with proteins for binding sites on resin → increasing concentration causes more proteins to elute
Downside: salt and pH can alter protein structure
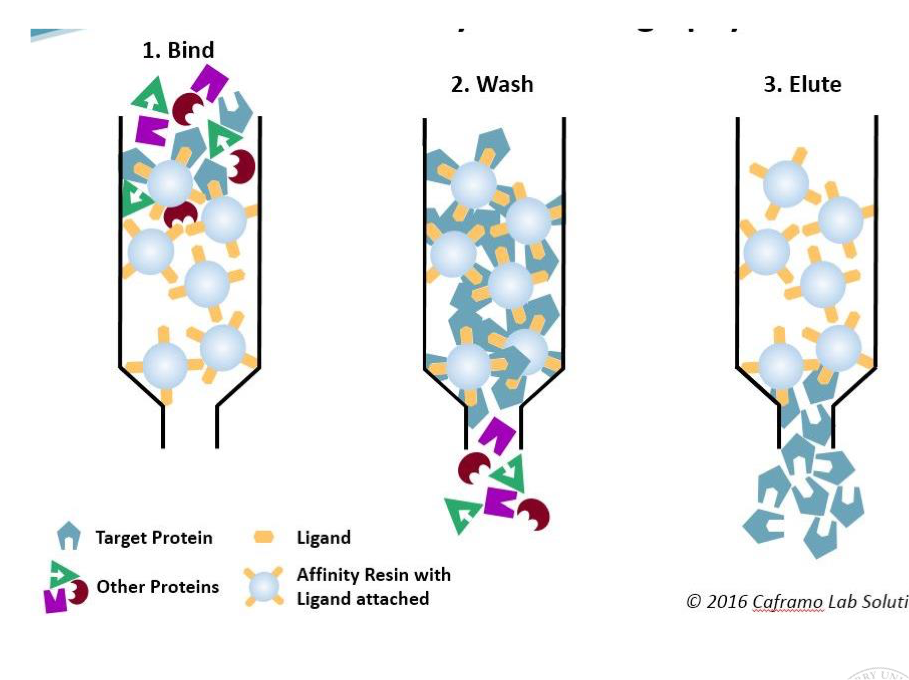
Affinity chromatography
Column contains a stationary phase made of beads with a molecule that protein of interest is known to bind e.g. ligand, antigen, antibody
Protein is retained in stationary phase by binding to specific binding molecule
To remove retained proteins, need to outcompete stationary phase for binding to your protein → add competing molecules
high concentration of ligand (to compete with the bead-bound one)
add salt or change pH gradient (to weaken binding)
Eluted proteins may contain ligand when eluted
Downside: salt and pH gradient can alter protein structure
PAGE electrophoresis
Poly-acrylmide gel electrophoresis
Used to perform analysis of proteins (know if protein is correct)
Separate proteins based on size and charge
Small, charged molecules move faster through gel than larger uncharged ones