Exam 2. Biochemistry.
1/41
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
42 Terms
Relate and define the terms “enzyme” and “catalyst”, recognize that an enzyme is a binding protein that also catalyzes a reaction.
catalyst: a substance that participates in a reaction wiout being consumed (emerges in its original form)
enzyme: a catalyst (typically a protein) that increases the rate of chemical reactions in living systems
Differentiate between cofactor, coenzyme and prosthetic group
coenzyme: a small organic molecule that is required for the catalytic activity of an enzyme
cofactor: a small organic molecule (coenzyme) or metal ion that is required for the catalytic activity of an enzyme
prosthetic group: an organic group (like a coenzyme) that is permanently associeated with a protein
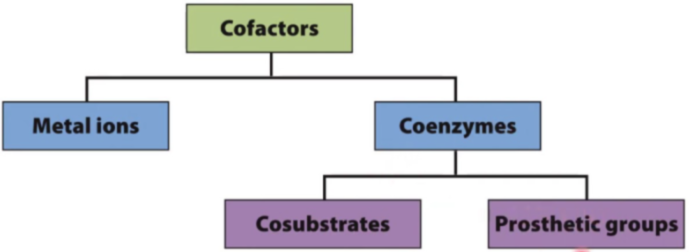
List and describe the six internationally recognized enzymes types.
oxidoreductases: transfers e- or H+ from one molecule to another (oxidation-reduction reactions)
transferases: moving functional group from one molecule to another (group transfer)
hydrolases: transfer of functional group to water; catabolic hydrolysis (hydrolysis reactions)
lyases: group elimination to form double bonds
isomerases: rearrangement of atoms within a molecule (isomerization reactions)
ligases: bond formatio coupled with ATP hyrolysis (anabolic)
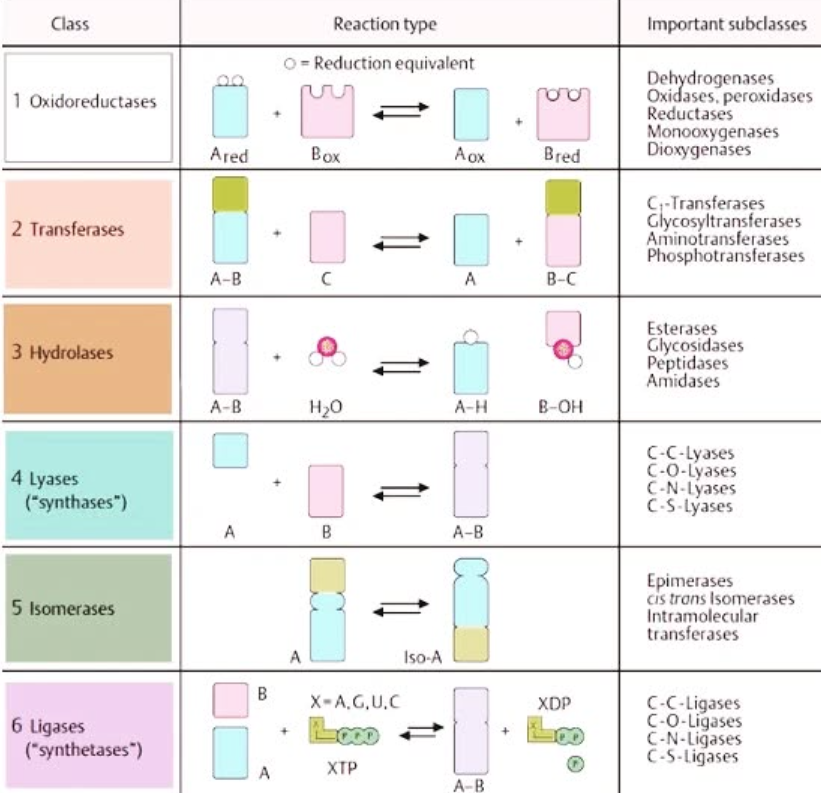
Describe the lock and key model and the induced fit model of substrate binding and differentiate between them.
lock-and-key model: substrates fit enzymes like a key in a lock
induced fit model: substrates actually fit enzymes like a hand in a glove
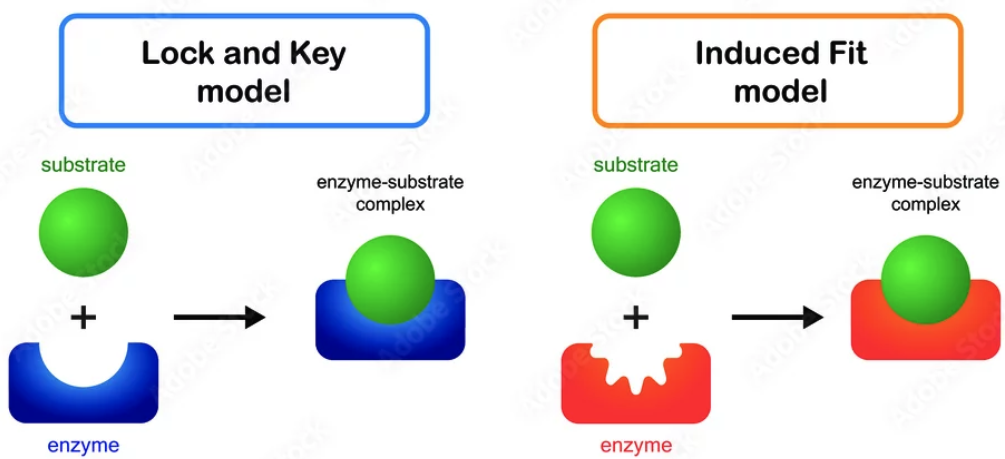
Describe how enzymes catalyze reactions by stabilizing transition states (lowering activation energy) both in words and in free-energy diagrams.
the enzyme binds to the transition state of a substrate, stabilizing it, ensuring it is in the right position to react, and bringing reacting parts closer together to increase collisions. This lowers the activation energy as seen in free-energy diagrams.
Describe the mechanisms enzymes use to stabilize transition states (acid-base catalysis, covalent catalysis, metal ion catalysis)
acid-base catalysis: a H+ is transfered between the enzyme and substrate to form transition state(catalyst acts as an acid/base)
covalent catalysis: cobalent bond forms between catalyst and substrate during transition state formation (catalyst acts as a nucleophile, electron-rich group wanting an electrophile)
metal ion catalysis: metal ions participate in enzymatic reactions
Understand that amino acid side-chains at the active site also impact enzyme catalysis, and determine which amino acid side-chains can participate in catalysis.
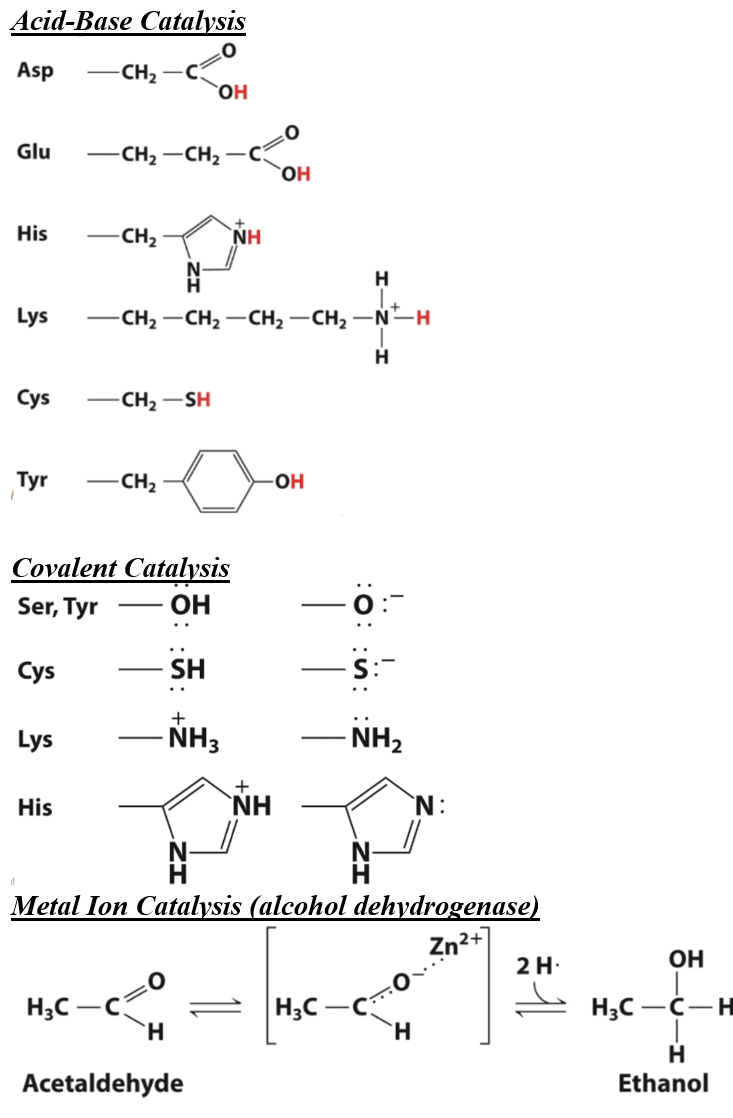
Define active site and describe its properties
active site: the region of an enzyme in which catalysis takes place (typically found in a cleft with two domains (a pocket))
Outline the reaction catalyzed by chymotrypsin, explain the operation of chymotrypsin’s catalytic triad, define the oxyanion hole and its role in chymotrypsin catalysis
catalytic triad: ser195, his57, asp102 work together to cleave peptide bonds (highly efficient nucleophilic attack mechanism and stablize reaction intermediates)
oxyanion hole: backbone of amino groups that form hydrogen bonds with negatively charged oxygen atom in the intermediate, structural feature in active site of chymotrypsin
Define the term specificity pocket and describe how enzymes use specificity pockets to differentiate between potential substrates
specificity pocket: a cabity on te enzyme surace at the active site that accommodates the residue on the N-terminal side of the scissile peptide bond
Differentiates between substrates based on size, shape, and atom positions
Describe the mechanisms cells use to regulate protease activity
protease inhibitors (the inhibitors pose as protease substrates but are not completely hydrolyzed)
Describe how blood coagulation operates as a protease cascade
one protease is required to act as a substrate to make another, repeat several times before blood coagulatino occurs
Describe how the rate (velocity) of an enzymatic reaction is measured/calculated
velocity = (concentration of substrate or product) / (reaction time)
Describe and calculate “rate enhancement”.
rate enhancement: the increase in rate of a chemical reaction due to presence of a catalyst
calculation: rate enhancement = (rate of catalyzed rxn)/(rate of uncatalyzed rxn)
Be able to define the rate equation for unimolecular and bimolecular reactions.
unimolecular: rate = k[A] units: k = sec-1
bimolecular: rate = k[A][B] units: k = M-1 * sec-1
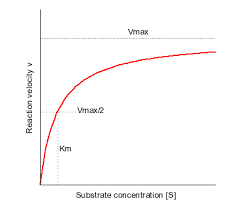
Know the Michaelis-Menten equation and relate it to the Michaelis-Menten plot, use the Michaelis-Menten equation to calculate kinetic parameters.
V0 = (Vmax [S]) / (KM + [S])
![<p>V<sub>0 </sub>= (V<sub>max </sub>[S]) / (K<sub>M </sub>+ [S])</p>](https://knowt-user-attachments.s3.amazonaws.com/997f948f-6f87-4db4-8120-042de8b3c78f.png)
Derive key kinetic parameters (Vmax and KM) from Michaelis-Menten plots.
Define the meaning of the “steady state assumption” in Michaelis-Menten kinetics
in a steady state assumption, [ES] remains constant until almost all substrate molecules have converted to product
[ES] , [E] , [P], [S]
![<p>in a steady state assumption, [ES] remains constant until almost all substrate molecules have converted to product <br><span style="color: green">[ES]</span><span> , </span><span style="color: purple">[E]</span><span> , </span><span style="color: red">[P]</span><span>, </span><span style="color: #4ce5af">[S]</span></p>](https://knowt-user-attachments.s3.amazonaws.com/6f74b344-759d-4edb-9a5d-51d30271809e.png)
Define what we mean when we say an enzyme is “saturated” and describe the circumstances under which an enzyme is saturated.
an enzyme is saturated when there is so much substrate that all of the enzyme’s active sites are full and the enzyme is opporating at max capacity (Vmax)
Explain the meaning and physiological significance of Vmax and KM
Vmax happens when an enzyme is opporating at maximum capacity. This is significant because physiological processess that need to occur rapidly (production of energy, synthesis of essential molecues, etc.) need a high Vmax
KM measures the affinity for a substrate, or measures how easily the enzyme can form the ES complex (low KM has high affinity, high KM has low affinity). This is significant because in physiological processes, if KM is low, enzymes can function effectively even in low substrate concentrations (enzymes in bloodstream) and if KM is high, the substrate concentration needs to be high for rapid processing (liver or muscle cell enzymes in high-energy processes)
Describe and calculate kcat (turnover number), describe what it can tell you about an enzyme.
kcat = (Vmax) / ([E])
it tells you the number of substrate molecules converted to product per enzyme molecule in a unit of time
Explain the use of kcat/KM (specificity constant) to measure the efficiency of an enzyme.
it tells us how efficiently an enzyme can convert substrate to product (low = less efficient; higher = more efficient)
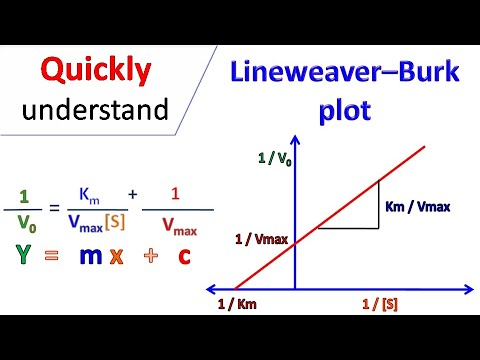
Describe the relationship between the Michaelis-Menten equation and the Lineweaver-Burke equation.
Lineweaver-Burke is the Michaelis-Menten in linear form (y=mx+b)
Be able to use the Lineweaver-Burk plot to determine key kinetic parameters (Vmax and KM).
y int is Vmax -1 and x int is Km-1
Describe the circumstances under which Michaelis-Menten kinetics are not applicable
allosteric (alters enzyme structure, which affect affinity); substrate is supersaturated, slow reaction, multiple substrates
Describe and differentiate between competitive and non-competitive, reversible and irreversible enzyme inhibition as well as feedback inhibition.
Understand how KM and Vmax change based on the nature of inhibitors.
Define “transition state analog” and describe why they are effective inhibitors/drugs.
Define/describe allosteric regulation the terms “positive effector” and “negative effector”
positive increases activity, negative decreases activity
Relate lipid structure to amphipathic behavior, explain why amphipathic molecules tend to form micelles and bilayers when mixed with water.
Describe the general structure of a fatty acid, differentiate between saturated and unsaturated fatty acids.
Show esterification reactions between glycerol and fatty acid(s) leading to formation of either monoacylglycerol, diacylglycerol, or triacylglycerol; define the function(s) of triacylglycerols
Give the general structure of the glycerophospholipids, sphingolipids and glycolipids; compare and contrast the functions of glycerophospholipids, sphingolipids, and glycolipids
Explain how cholesterol differs in structure and function from the other membrane lipids.
Describe some of the roles of lipids in the cell, outside of the formation of lipid bilayers
Explain why lateral diffusion within a “leaflet” of a lipid bilayer is relatively easy while movement between “leaflets” in the bilayer is difficult.
Define what is meant by “membrane fluidity”, and describe the primary factors contributing to the fluidity of membranes.
Differentiate between peripheral membrane proteins, integral membrane proteins and lipid-linked proteins.
Describe the methods hydrophilic proteins use to be able to interact with the hydrophobic core of lipid bilayers.
Explain what is meant by the “fluid-mosaic model” of lipid bilayers.
Define and describe the proteins used to passively transport molecules/ions through the lipid bilayer (porins, aquaporins, ion channels, gated channels, mechanosensitive channels, transport proteins)
Describe how proteins can move ions/molecules against their concentration gradient (active transport); describe the specific mechanism of the Na/K-ATPase