19. mRNA processing
1/21
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
22 Terms
in e.coli transcription termination - 2 mechanisms
Rho-independent: RNA polymerase only
Rho-dependent: RNA polymerase + protein RHO
Premature termination = attentuation
rho dependent termination
rho binds to rut site n mRNA
RNAP pauses at termination site → provides time for rho to catch up (hairpin required)
rho has no affinity for RNAP, tracks along mRNA until it catches up
rho unwinds DNA-RNA hybrid (requires ATP)
mRNA, rho, RNAP all released
RHO independent termination sites
GC rich self-complementary region with several intervening nucleotides, when transcribed - forms stem loop structure
followed by series of U resides
Rho independent termination
a rho independent terminator contains an inverted repeat followed by a string of approximately 6 adenine nucleotides
the inverted repeats are transcribe into RNA
inverted repeats in RNA fold into a hairpin loop, which causes RNA polymerase to pause
RNA transcript separates from the template, terminating transcription
DNA sequence where RNA polymerase either terminates or continues with transcription
not a termination site (t site)
attenuation in the trp operon
regulatory mechanism where transcription stops early if trp levels are high, and continues if trp levels are low
expression of trp operon controlled at two levels
trpR gene repressor, in the presence of tryptophan binds the operator (o)
attenuator sequence that prematurely terminates transcription when high levels of trp are present (attenuated RNA consists of only a short leader sequence)
RNAP I requires a
polymerase-specific termination factor that binds to a specific DNA sequence
RNAP III terminates after
transcribing a series of U resides but does not require a stem-loop structure
RNAP II terminates due to
3’ processing of the transcript - polyadenylation by protein complex that is carried by the phosphorylated CTD tail and binds to elements in 3’ UTR of mRNA
heterogenous nuclear RNA
transcripts made by RNAPII before they are fully processed
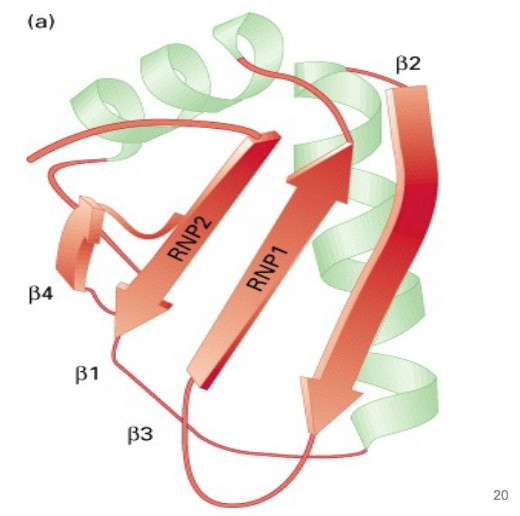
RNP motif - RBD (Rna Binding Domain) most common
~ 80 residues, two highly conserved regions - 4 beta sheets flanked with 2 alpha helices
premRNa processing
5’ capping CTD tail
RNA splicing CTD tail
Polyadenylation CTD tail
spatial separation fo transcription and translation
protects the cell from making faulted proteins
cotranscriptional modifications
7-met-guanosine coupled to 5’ end
methylation of ribose
N-methylation of adenine
phosphorylated CTD recruits cappping enzymes
RNA 5’ triphosphate removes the 5’ phosphate
guanylyl transferase attaches GMP
7-methyltransferase modifies the terminal guanosine
mRNA 5’cap released from CTD tail
Poly a tail
increase transalation efficiency by aiding ribosome recycling and mRNA circularization
polyadenylation requires
nuclear poly A binding protein Ii PABII 200-250 nucleotides
splice sites can be determined by comparing genomic dna with
cDNA or mRNA
Group I and Ii introns
self-splicing introns - RNA is plicing itself (no proteins invovled) RNA = ribozyme
most eukaryotic cells - spliceosomal splicing
splicing assisted by at least 5 small nuclear RNAs (snRNAs)
U1, U2, U4, U5, U6
spliceosome protein complexes associated with phosphorylated CTD tail
factors invovled in splicing: helicases
assist snRNAs and Sm protiens in: spliceosomal protein-protein interaction