Gordon College - Microbiology Exam 2
1/90
Earn XP
Description and Tags
Genetics, genomics, antibiotic resistance, viruses, and Student presentations
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
91 Terms
Genome
Entire complement of genetic information (genes that encode proteins, RNAs, regulatory sequences + non-coding DNA)
Genomics
•Mapping, sequencing, analyzing, and comparing genomes
•Requires molecular biology + computing power
•Analysis is difficult and time consuming, but incredibly informative
mutations
change in nucleotide sequence (genotype), may alter phenotype
ways other than a mutation by which genomes can change
horizontal gene transfer, gene duplicaitons, transposable elements, population changes
silent mutation
doesn’t affect phenotype of cell
always in 3rd base codon bc of genetic code degeneracy
missense mutation
informational “sense” in polypeptide has changed
if at critical location in polypeptide chain, protein could be inactive
nonsense mutation
premature termination of translation leading to incomplete polypeptide
change is from sense (coding) codon to a nonsense (stop) codon
frameshift
insertion/deletion of nucleotide base in numbers that aren’t multiples of 3
transposable elements
Genes that can move throughout genome. shape genomes through their mobilization
gene duplication
Become functionally diploid.
One remains functional and the other can accumulate mutations.
horizontal gene transfer
plasmids between bacteria.
movement of genetic information between organisms
insertion sequence (IS)
a portion of DNA that codes for transposes and has terminal repeats
transposon
like an insertion sequence, but has addition genes between terminal repeats
IS and transposons are transposable elements T/F
T
inverted terminal repeats
parts recognized by enzymes when cutting, also sites of recombination, 3’ and 5’ ends of same strand can also connect with each other
How transposable elements can change genomes
replicative transposition and non-replicative transposition
replicative transposition
transposes cuts at terminal repeats
Cut ends of transposable element attacks target sequence at 5’ ends
DNA polymerase fills in the gaps
DNA ligase seals the cuts
Recombination results in original TE staying the same but copy is inserted in between two copies of the target sequence
non-replicative transposition
Transposase brings inverted repeats together making hairpin structure
Carrier DNA is ejected and the elements attack target DNA at 5’ ends
DNA polymerase fills in gaps at 3’ ends (3→5)
DNA ligase seals cuts
Result is transposable element removed from original seuqnece and inserted between two copies of target sequence
types of base-change mutations
insertion, deletion, substitution
insertion
nucleotides added causing frameshift
deletion
nucleotides removed causing frameshift
Type of base-change mutation: substitution
nucleotides switched, can be silent, cause missense or nonsense
Base pair substitutions: effects of silent
mutated codon codes for same amino acid, no change in protein
Base pair substitutions: effects of missense
mutated codon codes for different amino acids, changes protein structure
effects of nonsense
mutated codon codes for stop codon, curing polypeptide short (stop codons: UAA, UGA, UAG)
effects of transposable elements of the genome
can change reading frame causing different kinds of mutations and regulation of gene expression
horizontal gene transfer concept
method of integrating DNA sequence from one bacterial genome into another bacterial genome
conjugation, transformation, transduction
vertical gene transfer concept
genome replication and cell division
conjugation
cell-cell contact (pili) allows DNA transfer following rolling circle replication
transformation
free DNA taken up by competent cell
transduction
how a bacteriophage transfers
mechanism of conjugation: steps
Pili connects to other cell
Pulls cell close
Circular plasmid gets nicked in one strand
One strand goes through pili into other cell
Complementary strand is synthesized in recipient cell, transferred strand is replaced in donor cell by complementary strand synthesis
Cells separate
mechanism of transformation: steps
Free DNA binds to DNA-binding protein on recipient cell
Proteins inside facilitate single strand movement through cytoplasm into bacterial genome
New strand is added to the genome via recombination
Forms heteroduplex —-------->>>>
Genome replaces other strand to fix circle
mechanism of transduction
Via bacteriophage
Transfers DNA from donor to recipient
When bacteriophage is repackaged, some host genome gets packaged
When bacteriophage goes to infect new cell, injects genome (with hosts genome) into new recipient cell
importance of conjugation and horizontal gene transfer in biotech
We can add genes to certain bacteria to make then produce certain proteins for us
Transformation
Steps to clone gene into many bacteria
Cut DNA with restriction enzyme
Add vector cut with same restriction enzyme
Add DNA ligase to form recombinant molecules
Introduce recombinant vector into host
Hallmarks of horizontal gene transfer
Gene found in distantly related species
Flanked by inverted repeats (hints has transposition)
Gene clusters
Causes of population-level changes in genomes
Selection, Genetic drift, and Bottleneck
Cause of population-level changes in genomes: selection
mutation helps organism survive, reproduce
Cause of population-level changes in genomes: Genetic drift
some have more offspring than others (random)
Cause of population-level changes in genomes: Bottleneck
populations size is reduced greatly, removes some phenotypes
Information that can be revealed from the genome
Evolutionary history
Essential genes (use comparative genomics)
How an organism functions
Virulent, metabolism, niche role, etc.
core genome
portion shared by all strains
accessory genome
optional genes in some strains
pan genome
core + all optional genes (not all in strains)
genomic or chromosomal island
gene clusters for specialized function (pan)
pathogenicity island
genomic island that codes for virulence factors
usefulness of transcriptomic information
insights into evolutionary history
uses mRNA transcripts (rather than genome) to analyze genome
molecular phylogeny
evolutionary history of organisms inferred from homologous genes
sequence similarity
% of nucleotide positions shared by any to sequences (shared/total)
homology
genes inherited from common ancestor (binary trait, either inherited or not)
suitability of 16S rRNA sequences for constructing phylogenetic trees
Encodes a small subunit of ribosomal RNA molecule
16S rRNA sequence found in all bacteria and archaea, so can compare any species
Has 9 hypervariable regions (V1-V9)
importance of gaps in sequence alignments
Not all DNA changes are point mutations
Establishes positional homology
phylogenetic tree
depicts evolutionary history of an organism
branch tips
species that exist today
node
where ancestor diverged into 2 new lineages
how to interpret phylogenetic trees
the closer the branches (connection wise not proximity), the more related
factors that may confound construction of phylogenetic trees
Choosing one of two equally supported trees
Convergent evolution
Horizontal gene transfer
convergent evolution
two species having a shared sequence not due to common ancestry
categories of antibiotic resistance
Development of resistant biochemical pathway
Ex. dihydrofolate synthase mutated to bind less efficiently to sulfonamide (antibiotic)
Antibiotic inactivation
Ex. beta-lactamasde splits beta-lactam ring of penicillin
Reduced membrane permeability
Ex. reduce porB, a porin that allows entry of penicillin
Efflux
Ex. tetA, a tetracycline (antibiotic) pump to pump out of cell
Events leading to genome-altering mutations
DNA replication errors
size of virus genome and reasons for its size
size: 2-350 genes; 2000-1.2 million base pairs
reason: crucial niche selection and specified transmission routes in the susceptible host
diverse structures of viruses
helical (rod-shaped), icosahedral (polyhedral), or complex
factors contributing: capsid, envelope, and genome type
components of viruses
nucleic acid genome (DNA or RNA) enclosed within a protein coat called a capsid
similarities and differences between these components and their cellular counterparts
similarities: genetic material, evolution, interaction with other organisms
differences: cellular structure, reproduction, metabolism, size, host dependence, genetic material, infection mechanism
5 main stages of viral infection (using T4 as example
T4 uses lytic life cycle: lysis of the host cell and release of the new viral progenies
attachment
entry
uncoating
replication
maturation
release
Attachment
controls host specificity
surface factors interact w/ host receptor
surface factor = protein/lipid w/ other function
outcomes of viral infection in bacterial and animal cells
direct cell damage and death
differences between viral and bacterial growth curves
occurs when visions are released from the lysed host cell at the same time
sense vs antisense strand terminology
two strands of DNA that make up a double-stranded DNA molecule
Baltimore scheme of classifying viruses
classifying viruses based on the type of genome and its replication strategy
examples of viruses from groups I, IV, V, VII of the Baltimore classification scheme
group I (dsDNA): HSV, adenovirus, poxvirus
group IV (+ssRNA): Poliovirus, corona, rotavirus
group V(-ssRNA): Influenza virus, rabies virus
group VII (dsDNA-RT): hepatitis B virus
Baltimore classification scheme: how each of these viruses replicate
group 1: Double-stranded DNA (dsDNA) viruses
group 2: single-stranded DNA (ssDNA) viruses
group 3: double-stranded RNA (dsRNA) viruses
group 4: positive-sense single-strand (+ssRNA) viruses
group 5: negative-sense single-stranded RNA (-ssRNA) viruses
Baltimore classification scheme: proteins encoded by these viruses/unique features of each virus
group 1: DNA polymerase, thymidine kinase, capsid proteins, immediate early proteins, latency-associated proteins
group 2: replication initiator proteins, capsid proteins, nonstructural proteins
group 3: RNA-dependent RNA polymerase, structural proteins, outer shell proteins
group 4: RNA-dependent RNA polymerase, Protease, Capsid proteins, envelope glycoproteins, accessory proteins
group 5: RNA-dependent RNA polymerase, Nucleocapsid proteins, envelope glycoproteins, matrix proteins, accessory proteins
group 6: Reverse Transcriptase, integrate, protease, capsid proteins, envelope glycoproteins, regulatory proteins
group 7: reverse transcriptase, capsid proteins, surface glycoproteins, x proteins
SARS-CoV2 structure
positive sense RNA virus.
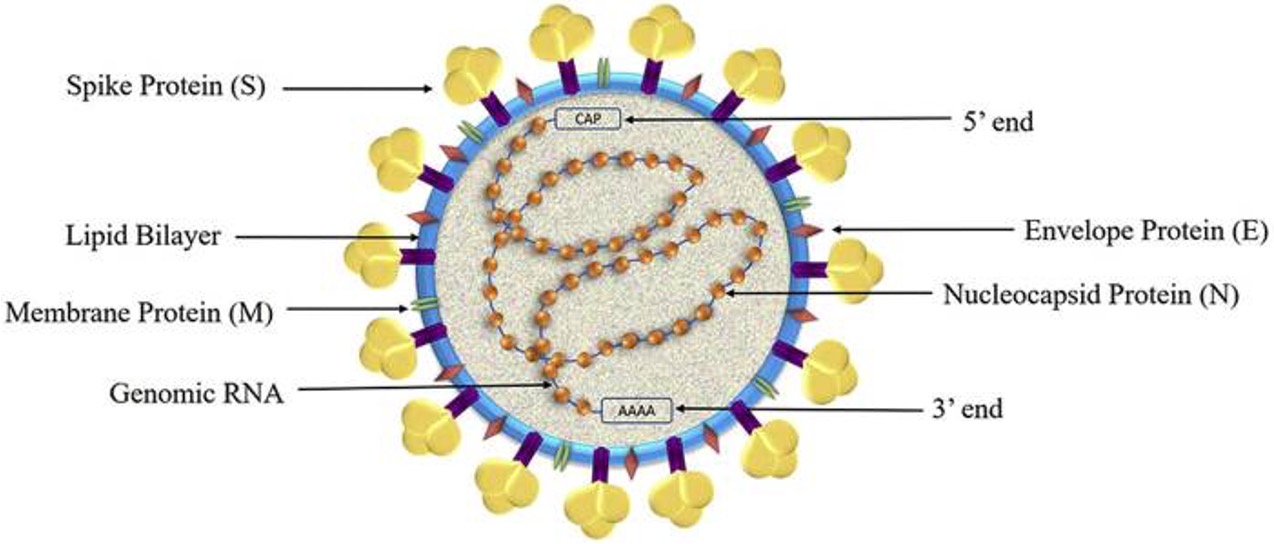
structural proteins of SARS-CoV2
envelope, membrane, spike, and nucleocapsid
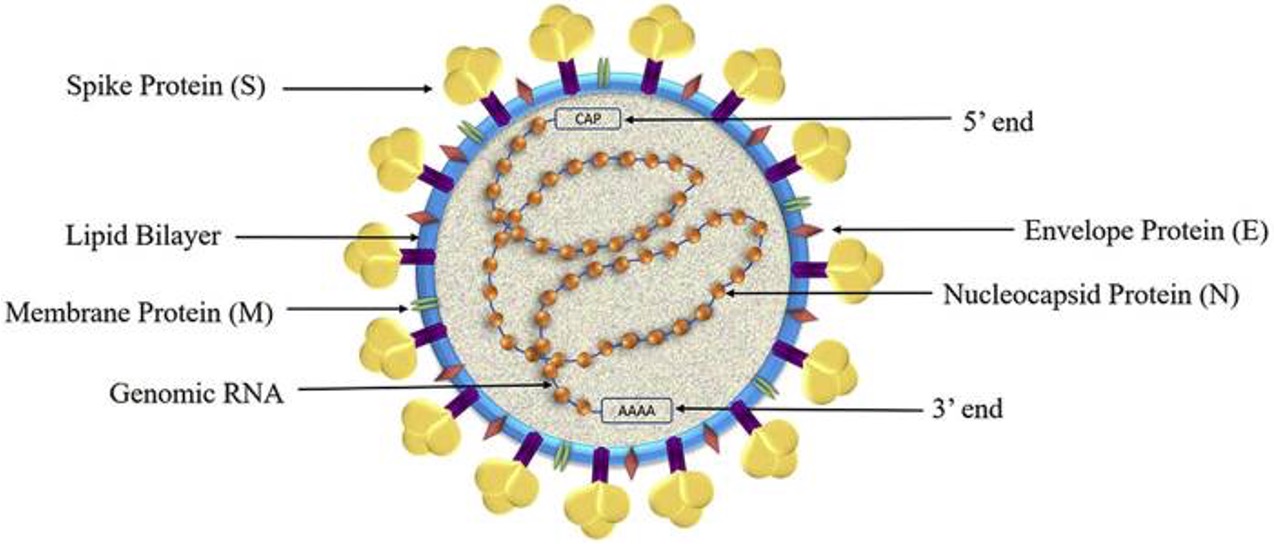
Non-structural proteins of SARS-CoV2
NSP1 to NSP11 and NSP12 to NSP16
accessory proteins of SARS-CoV2 and their functions
ORF3a, ORF3b, ORF6, ORF7a, ORF7b, ORF8, ORF9b, and ORF10, essential for controlling the hosts response to COVID infection
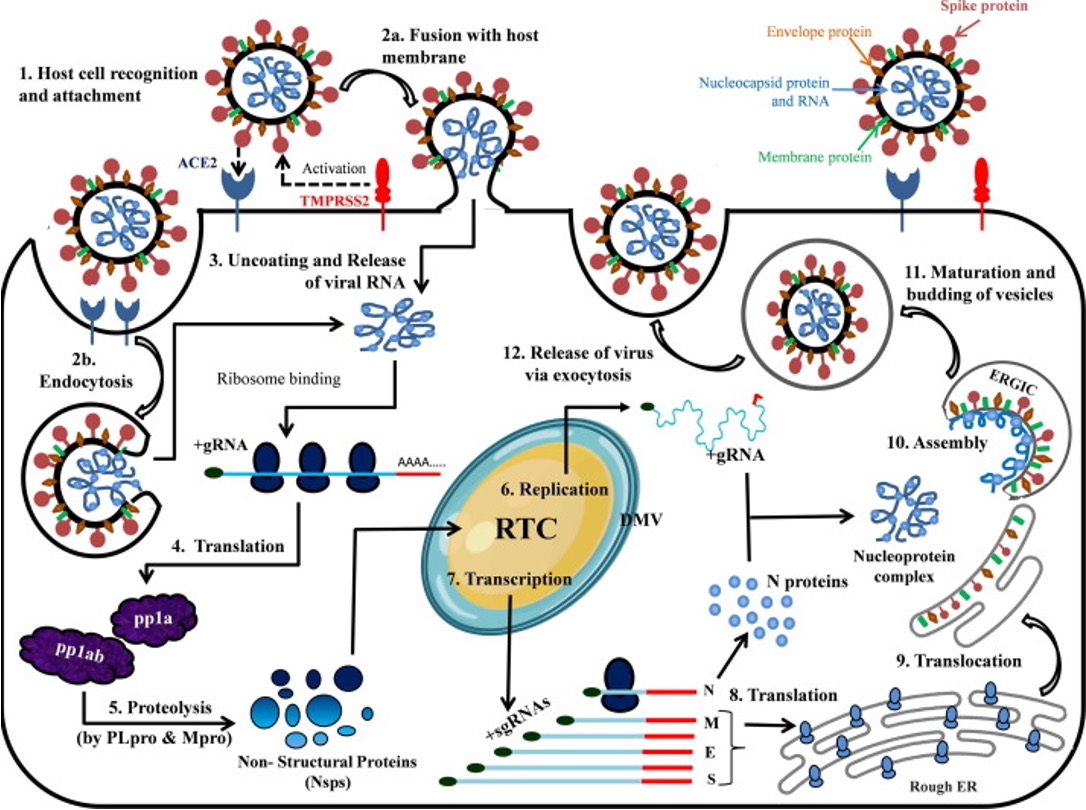
Replication cycle of SARS-CoV2
opening double helix and separation of DNA strands, priming of the template strand, and assembly of the new DNA segment
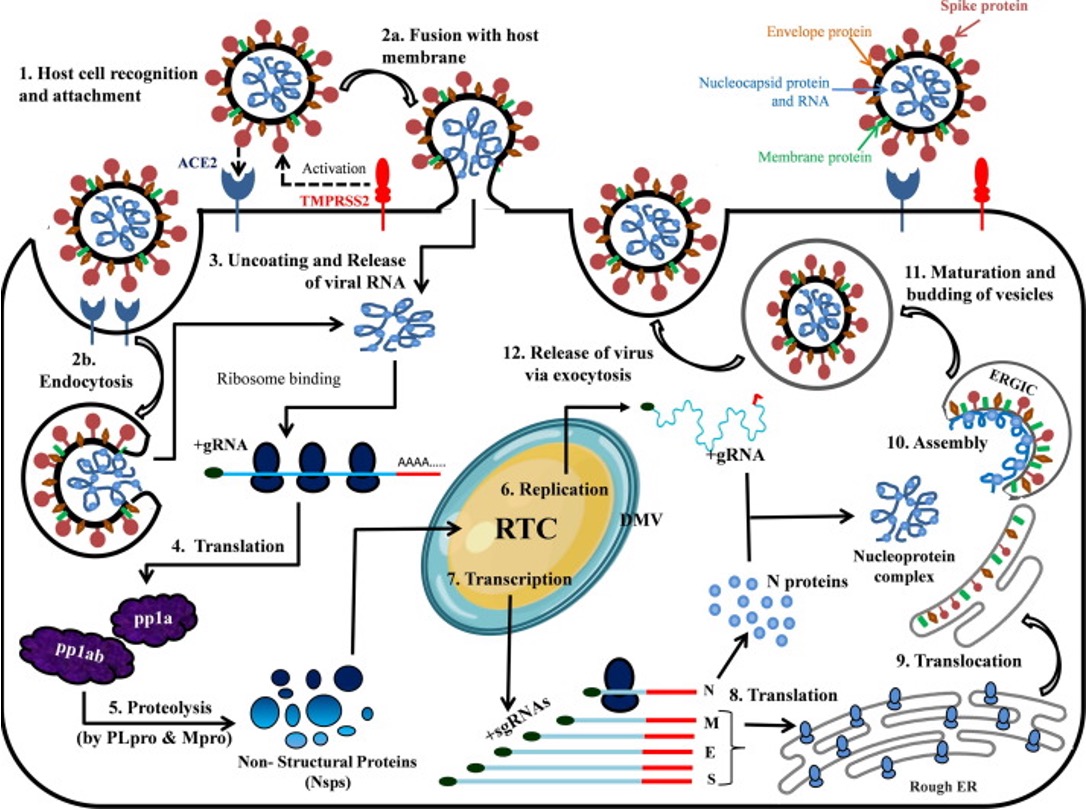
Replication cycle of SARS-CoV2: Where and how each of the 5 main stages of the viral life cycle is accomplished by SARS-CoV2
virus entry
translation of viral replication machinery
replication
translation of viral structure proteins
virion assembly
release of virus
Penetration
bacteria: entry of nucleic acid + specific viral proteins
animal: entry of full virion
synthesis
using host cell machinery, but directed by virus; early proteins, middle proteins, late proteins
assembly
package genome into preassembled capsids
release
package genome into preassembled capsids (late proteins breach host cytoplasmic membrane and peptidoglycan layer
sense (coding strand)
a strand of DNA that carries the code for making a protein
anitsense strand
non-coding DNA strand of gene. serves as template for producing mRNA, directs the synthesis of a protein
Which of the following events is an example of a mutation?
A. Phosphorylation of an amino acid that is part of an enzyme increases the efficiency of that enzyme
B. A mistake is made by DNA polymerase during DNA replication resulting in the change of an ‘A’ to a ‘C’ in the genome of a cell
C. An mRNA molecule is degraded by nucleases
D. A protein changes its conformation upon binding to another protein
B
which of the following mutations is likely to have the smallest impact on a cell?Explain your answer
A. Insertion
B. Deletion
C. Substitution
D. Frameshift
C. because it changes just one base in the DNA, which may not alter the protein if the new codon codes for the same amino acid.
In which of the following examples will there be two copies of the gene present at the end of the process. Select all that apply (1) Explain your answer (1)
A. Conjugation
B. Conservative transposition
C. Generalized transduction
D. Transformation
A. Conjugation and C. Generalized transduction result in two copies of the gene because genetic material is transferred while the donor retains its original copy. In B. Conservative transposition and D. Transformation, only one copy of the gene is present at the end.
Transcriptomics gives us information about the sequence of which molecule?
A. RNA
B. DNA
C. Proteins
D. Carbohydrates
A. RNA because Transcriptomics studies the RNA molecules made from DNA, giving information about gene expression.
Which of the following features is not identified as part of a typical bioinformatics workflow used to generate genomic information (1)
A. Start codons
B. Stop codons
C. Ribosome binding sites
D. Post-translational modifications
D. Post-translational modifications. This feature occurs after protein synthesis and is not part of the genomic information workflow, unlike start codons, stop codons, and ribosome binding sites.
At which stage of the viral life cycle would you expect to detect the largest amount of virions in the infected tissue of a patient? (1) Explain your answer (1)
A. Attachment
B. Penetration
C. Synthesis
D. Release
D. Release. This is when newly formed virions are expelled from infected cells, resulting in the highest concentration of viruses in the tissue.