FMLec | M2 Non-Culture-Dependent Techniques
1/68
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
69 Terms
3 types of non-culture-dependent techniques
Direct microscopic counts
Rapid end-detection methods
Nucleic acid- and protein-based methods
Non-culture-dependent technique
Total count
Commonly used for milk and other dairy products (also known as Breed count)
A measured volume (0.01 mL) of bacterial suspension is placed within a defined area (1 cm2) on a microscope slide
Steps:
Smear
Stain
Count
Direct microscopic counts
3 steps to direct microscopic counts
Smear
Stain
Count
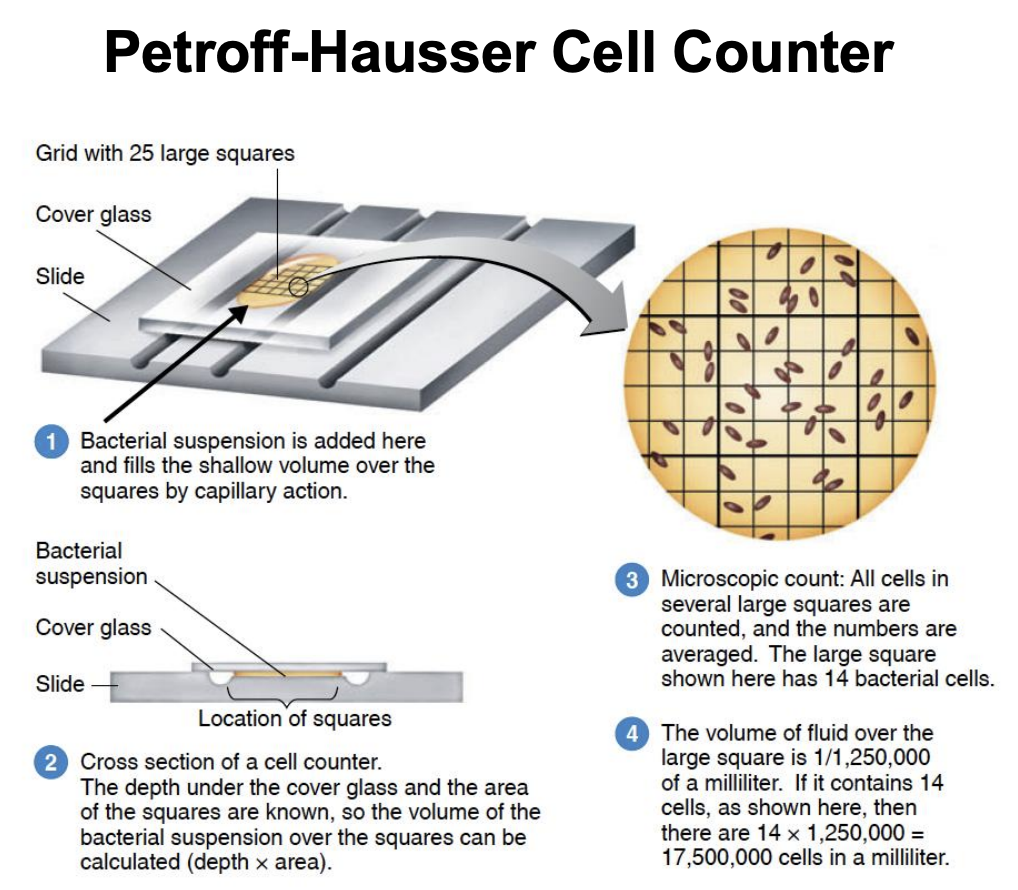
Explain Petroff-Hausser Cell Counter procedure
Grid with 25 large squares, cover glass slide, bacterial suspension is then added and fills shallow volume over squares by capillary action
Depth under cover glass and area of squares are known, so volume of bacteria suspension over squares can be calculated (depth x area)
Microscopic count: All cells in several large squares are counted, and numbers are averaged. Large square has 14 bacterial cells.
Volume of fluid over large square = 1 / 1,250,000
14 cells x 1,250,000 mL = 17,500,000 cells/mL
Advantages and Disadvantages of Petroff-Hausser Cell Counter
Advantages rscs
Rapid
Simple
Cell morphology can be assessed
Suited for samples with low numbers of bacteria
Disadvantages enfihm
Exhausting to analyst
Nonviable cells are enumerated
Food particles are not always distinguishable from microorganisms
Invariably higher than SPC counts
High limit of detection (1.0×107)
Motile bacteria are difficult to count
Explain Viability Staining Protocol
This allows differentiation between viable and nonviable cells because
Propidium iodide can only penetrate membrane of nonviable lysed cells, turning these into red
5 rapid end-detection methods
erfba
Enzyme-Linked Immunosorbent Assay (ELISA)
Reversed passive latex agglutination
Flow cytometer
Biosensor
ATP bioluminescence technique
ELISA limit of detection
104 - 106 CFU/mL
May require pre-enrichment / selective enrichment
Petroff-Hausser Cell Counter limit of detection
1.0×107 (high)
Rapid end-detection technique
Antigen-antibody based detection system
May require pre-enrichment or selective enrichment
Commercially available kits: Campylobacter, Salmonella,
L. monocytogenes, E. coli O157:H7
ELISA
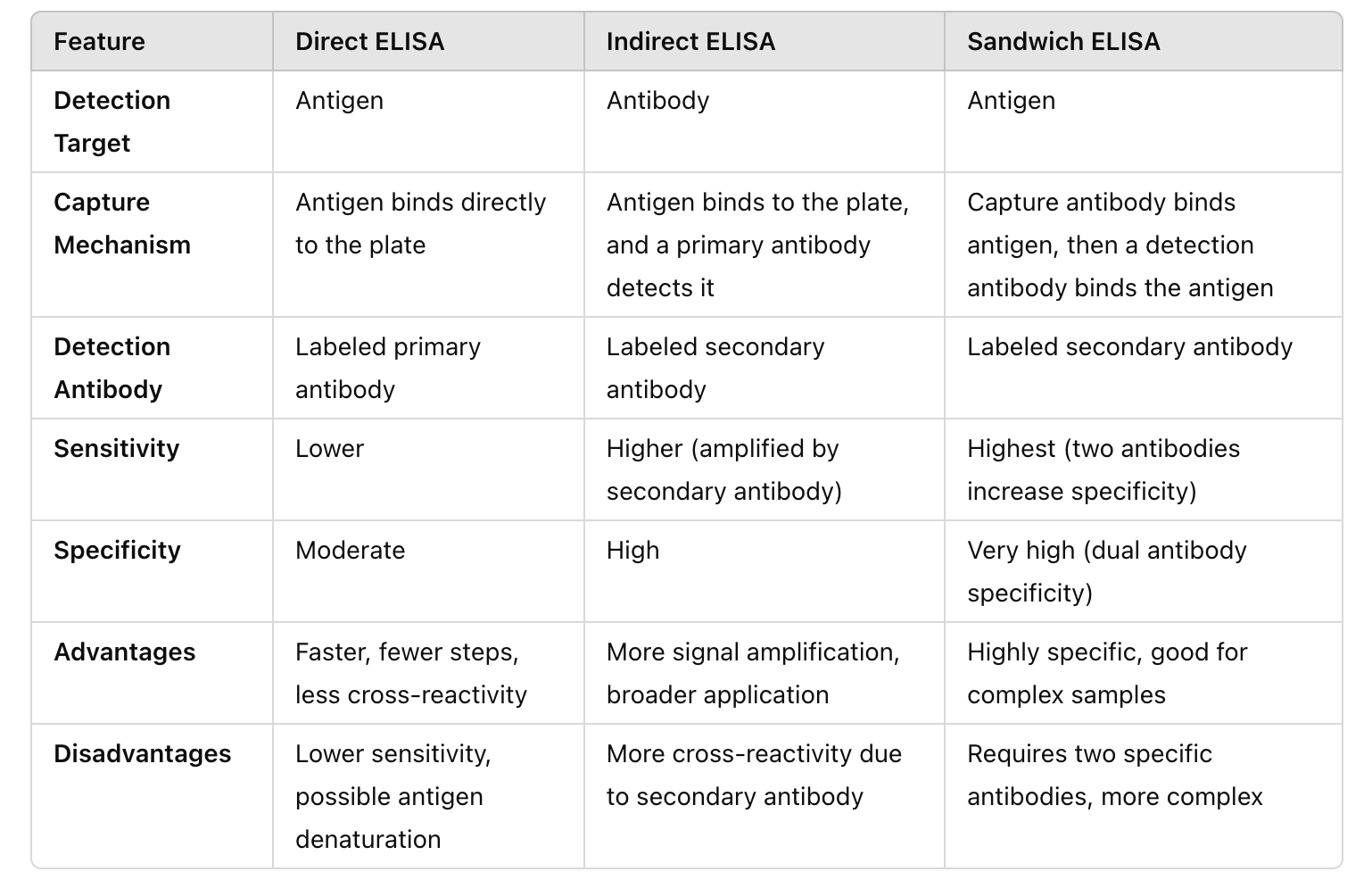
Direct vs. Indirect vs. Sandwich ELISA
Direct ELISA for rapid antigen detection
Indirect if high sensitivity is needed (since this uses secondary antibodies that amplify signals)
Sandwich if high specificity is required for complex samples, e.g., blood
T/F: Sandwich ELISA cannot be used if the antigen has only one epitope
TRUE
It requires two antibodies binding to different sites on the same antigen; if there’s only one epitope, this wouldn’t work
T/F: The signal intensity in an Indirect ELISA is lower than in a Direct ELISA
FALSE
Indirect ELISA has a higher signal because of the amplification effect from the secondary antibody
T/F: Sandwich ELISA is useful for detecting large, complex antigens with multiple epitopes
TRUE
Since two antibodies target different epitopes, it works well for large, multi-epitope molecules
Rapid end-detection technique
Used for detection of microbial toxins, e.g., Shiga toxin of Shigella dysenteriae
Reversed passive latex agglutination
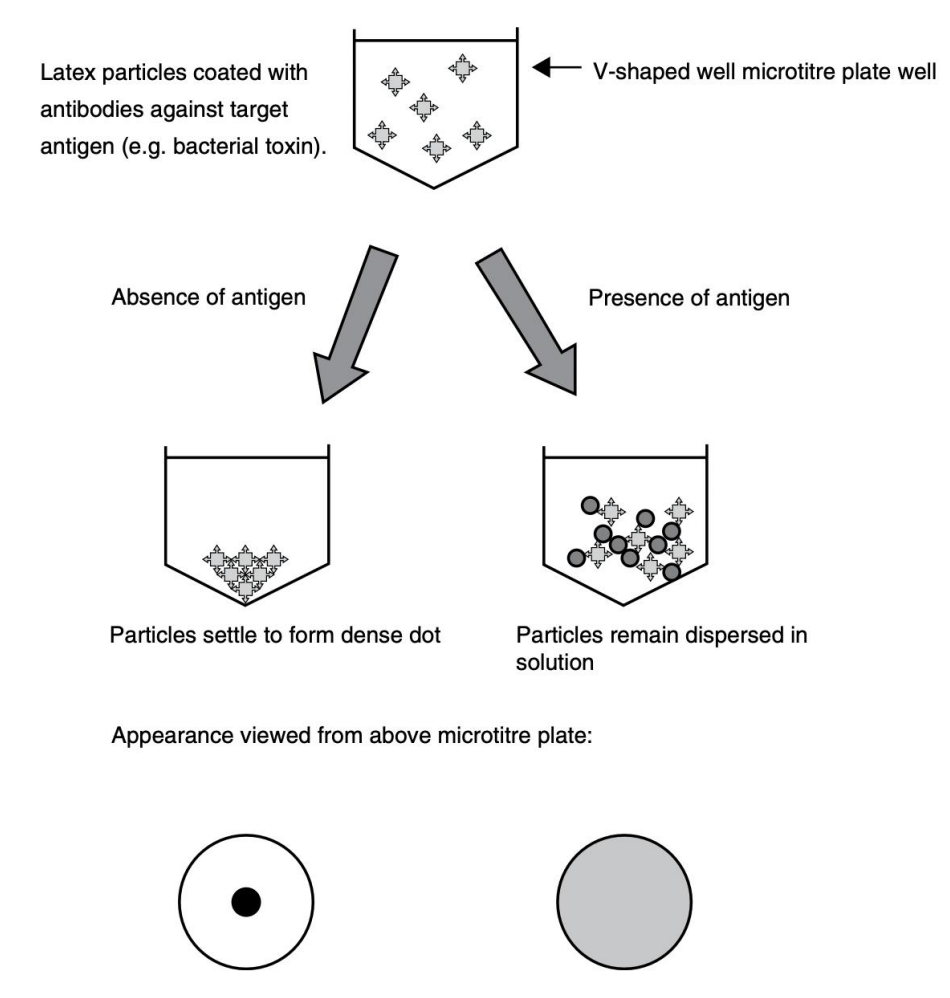
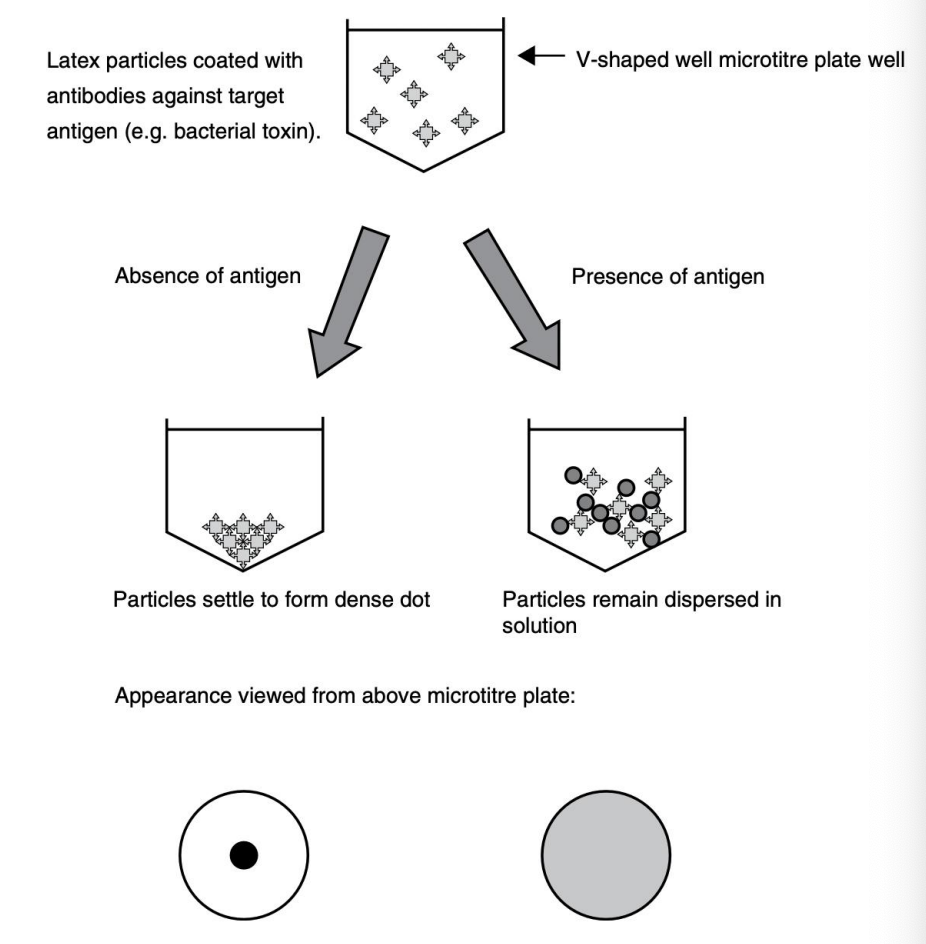
Explain principle behind reversed passive latex agglutination
Latex particles coated with antibodies against target antigen (V-shaped well in microtiter plate)
Absence of antigen = particles form a dense dot
Presence of antigen = particles remain dispersed in solution
Flow cytometer limit of detection
104 CFU/mL
Rapid end-detection technique
Detects microorganisms based on light scattering by cells and fluorescent labels
Sort cells based on size, shape, complexity, other properties
Fluorescence‐labeled antibodies or nucleic acid
Flow cytometry
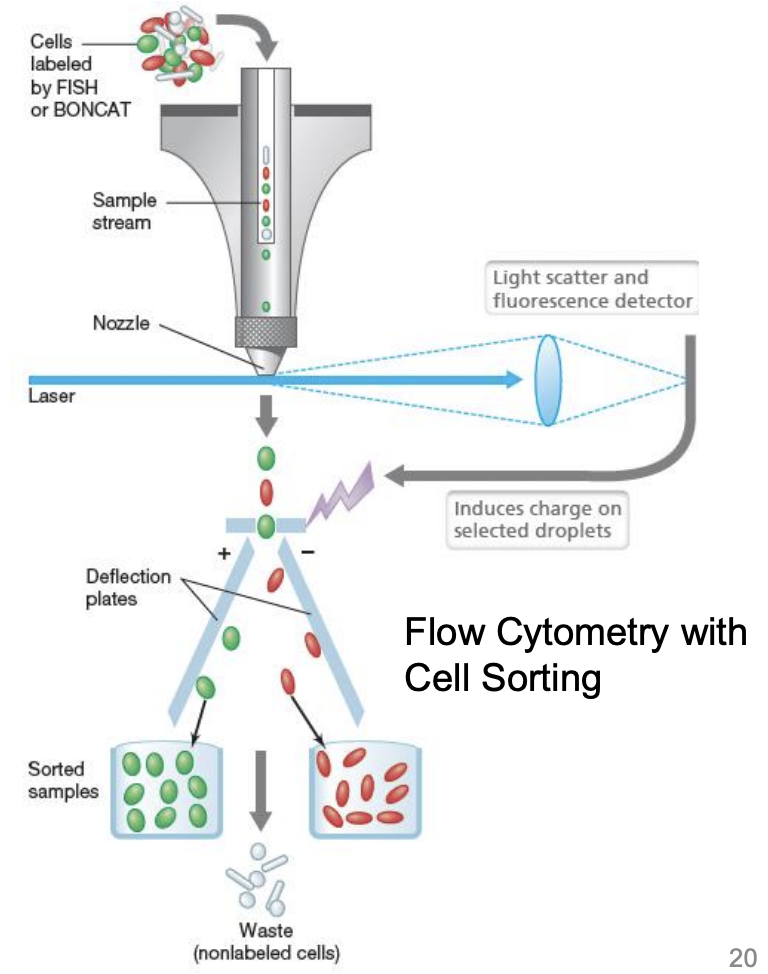
Rapid end-detection technique
_ refers to analytical device that integrates biological material for target recognition with a physicochemical transducer
Use either _ to give high specificity
Biosensors
DNA probes or specific antibodies
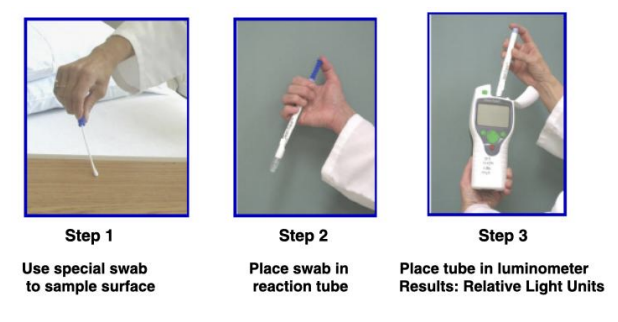
ATP bioluminescence technique limit of detection
1 pg (1000 bacterial cells)
Rapid end-detection technique
_ has ATP as indicator of living bacterial cells
Limit of detection = 1 pg = 1000 bacterial cells
Primarily used as hygiene or cleaning regime monitoring method
ATP bioluminescence technique
T/F: Indirect ELISA is the preferred method when detecting small molecules due to its ability to amplify the signal through multiple secondary antibodies
TRUE
When performing cell sorting via flow cytometry, _ reflects cell size while side scatter (SSC) reflects shape; SSC and fluorescence can also be indicators of viability
forward scatter (FSC)
T/F: Flow cytometry is more sensitive than ATP bioluminescence in detecting bacterial contamination because it does not require cell enrichment
FALSE
ATP bioluminescence can detect as low as 1000 cells, whereas flow cytometry typically requires at least 10⁴ CFU/mL
T/F: ATP bioluminescence can be used to quantify both bacterial and fungal contamination, but it cannot distinguish between them
TRUE
ATP is a universal indicator of living cells, but it does not differentiate between bacteria and fungi
T/F: The ATP bioluminescence technique is limited to detecting only culturable bacterial cells
FALSE
It detects ATP from all living cells, regardless of culturability
T/F: ATP bioluminescence can be used to confirm the effectiveness of antibiotic treatment by measuring a decrease in ATP levels over time
TRUE
T/F: Reversed passive latex agglutination can be used to detect bacterial toxins but cannot differentiate between viable and non-viable bacterial cells
TRUE
10 nucleic acid- and protein-based methods
polpra rvrm
Polymerase chain reaction (PCR)
Oligonucleotide DNA microarrays
Loop-mediated isothermal amplification (LAMP)
Pulsed-field gel electrophoresis (PFGE)
Restriction fragment length polymorphism (RFLP)
Amplified fragment length polymorphism (AFLP)
Random amplification of polymorphic DNA (RAPD)
Multi-locus variable number tandem repeats (VNTR) analysis
Ribotyping
Matrix-assisted Laser Desorption Ionization Time-of-Flight Mass Spectrometry
Nucleic-acid and protein-based method
Amplifies DNA to levels that can be detected
Repetitive cycle of heating and cooling to amplify the target DNA
Heat-stable DNA Polymerase and specific primers
PCR
Advantages vs. Disadvantages of PCR
Advantages hssq
High specificity
High sensitivity
Quick turnaround time to obtain results
Disadvantages etcp
Expensive and high-maintenance equipment
Trained personnel
Need for extensive DNA clean-up to avoid cross-contamination
Need to pre-incubate
Limit of detection: ~100 cells
Allows to differentiate live and dead cells
PCR limit of detection
~100 cells
The presence of DNA amplification in LAMP can be detected by a _
color change
5 PCR-based methods
em 3r
EMA/PMA-PCR
Multiplex PCR
Real-time or quantitative PCR (qPCR)
Reverse transcriptase PCR (RT-PCR)
Rep-PCR
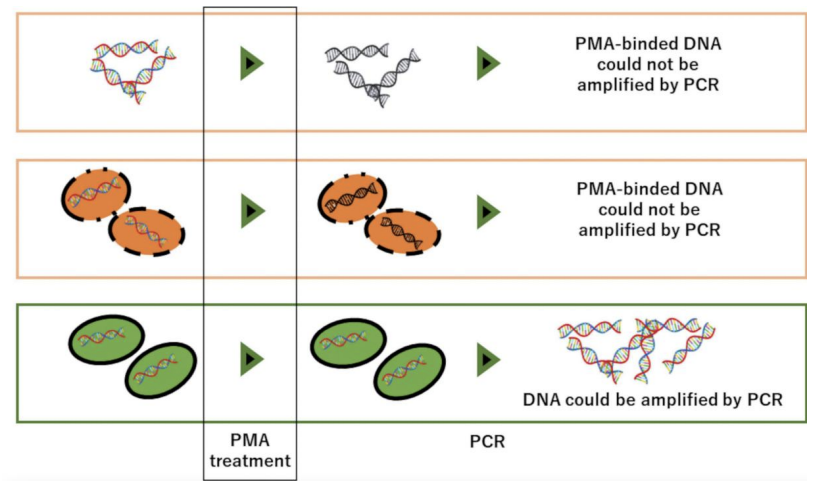
Explain EMA/PMA-PCR procedure
Sample pretreatment with Ethidium monoazide (EMA) and Propidium monoazide (PMA), which can only enter bacterial cells with damaged cell membranes and intercalate / cross-link their DNA after exposure to strong visible light, preventing DNA amplification
Low amplicons = high amount of dead bacterial cells (whose DNA amplification has been inhibited)
PCR-based method
Different primers are used to amplify different DNA
regions simultaneously
Multiplex PCR
PCR-based method
Monitors [DNA] as amplification progresses through fluorescence
Real‐time or quantitative PCR (qPCR)
PCR-based method
Detection and quantification of RNA expression
Reverse transcriptase PCR (RT-PCR)
PCR-based method
Amplifies noncoding (introns) intergenic repetitive sequences
Number of amplicons differs between unrelated, non-clonal strains
Repetitive-element PCR
_ can simultaneously detect different sequences in mixed DNA samples
Multipathogen microarray
T/F: LAMP amplification results in linear DNA fragments, similar to PCR
FALSE
LAMP produces long, looped, and cauliflower-like DNA structures instead of linear fragments
LAMP can be performed in a _ instead of a PCR machine.
water bath or simple heat block
T/F: Loop primers in LAMP help accelerate amplification by facilitating strand displacement
TRUE
The loop primers create faster strand displacement and rapid DNA amplification
Nucleic acid-based method
Consist of hundreds to thousands of specific oligonucleotides on a solid support
Oligonucleotides (25–80bp)
PCR amplified
Positions (in a grid pattern) are recorded by spot location
Oligonucleotide DNA microarrays
_ allows autocycling amplification under constant temperature (60-65C)
Bst DNA polymerase (Geobacillus stearothermophilus)
_ is an alternative to PCR
Amplification at one fixed temperature
Colorimetry, fluorescence, bioluminescence
Loop-mediated isothermal amplification (LAMP) technique
LAMP vs. PCR
ttpda | LAMP | PCR |
Temperature | 65 C only | Varies (cycling) |
Time | 20-40 mins (shorter) | 1.5+ hr |
Primers | 4-6 primers | 2 primers |
Detection | Fluorescence (could be real-time or endpoint), color, turbidity | Fluorescence (could be real-time or endpoint) |
Amplicon | Concatemeric | Discrete |
Explain important role of Bst DNA polymerase in LAMP
Bst DNA polymerase has a strong strand displacement activity that eliminates the need for heat-induced denaturation of strands and thus allows continuous amplification at constant temperature
T/F: Loop primers in LAMP accelerate amplification by providing additional binding sites for DNA synthesis.
TRUE
Loop primers reduce reaction time by creating more initiation points for DNA extension, leading to faster amplification
Nonculture-dependent technique
Used to separate or resolve very large DNA fragments
RE digestion of gDNA
Size-dependent net migration of DNA fragments
Periodic reversal of polarity of electric field that reorients DNA to move in different direction or travel across the gel matrix
Pulsed-Field Gel Electrophoresis
Nonculture-dependent technique
Detects variations in restriction sites using DNA probes
Fragment size = determined by position of restriction sites on gel matrix (identifier)
Number of bands = determined by number of restriction sites
Restriction Fragment Length Polymorphism (RFLP)
T/F: The pulsed electric field in PFGE is applied at random intervals to maximize DNA movement
FALSE
The pulses are carefully controlled and alternated at specific angles to optimize the separation of large DNA fragments
Nonculture-dependent technique
Specific co-amplification of restriction fragments formed after RE digestion (50-100 bp)
Amplified fragment length polymorphism (AFLP)
Explain RFLP vs. AFLP
RFLP = DNA is cut using restriction enzymes, then resolved via southern blot
AFLP = Amplifies a subset of restriction fragments produced from RE digestion; gel electrophoresis
Nonculture-dependent technique
Parallel amplification of a set of fragments using short arbitrary primers (10 bp long)
Differentiates bacteria based on number and length of PCR amplicons
Random amplification of polymorphic DNA (RAPD)
Explain how RAPD vs. RFLP, AFLP
RAPD
No RE digestion step
Random regions were amplified due to the use of short arbitrary primers
But it is a quick way to detect genetic variation without prior knowledge of genome
_ is unique for each strain and can be used to generate profile for genotyping
Number of variable number tandem repeats (VNTR) copies
Nonculture-dependent technique
Amplifies VNTR regions
Allows microorganism identification because the number of VNTR copies is unique for each strain and this can be used to generate profile for genotyping
Multiple-locus Variable Number Tandem Repeats (VNTRs) analysis
Nonculture-dependent technique
Bacterial identification and characterization based on rRNA genes (DNA is being analyzed)
Steps ggp
gDNA RE digestion
Gel electrophoresis
Probing with 16S or 23S rRNA sequences (southern blotting; fluorescence)
Ribotyping
Nonculture-dependent technique
Measures masses of bacterial proteins and/or lipids
Effective and rapid tool for genus- and species-level identification
Requires single colony or pure culture
MALDI-TOF MS
T/F: Microarrays can be used to detect SNPs (single nucleotide polymorphisms), but they are less effective for detecting large structural variants in the genome
TRUE
T/F: Southern blotting is commonly used in RFLP analysis to detect specific DNA fragments after electrophoresis
TRUE
T/F: VNTR analysis is more discriminatory than SNP-based genotyping for closely related bacterial strains
TRUE