MCB--> DNA Structure/Properties and Chromosome Structure (Gochin)
1/107
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
108 Terms
Describe the formation of DNA structure from two chains of deoxynucleotides.
1) Two chains of deoxynucleotides connected by phosphodiester bonds, to form a polynucleotide chain
2) Two anti-parallel polynucleotide chains held together by complementary hydrogen bonding between bases and hydrophobic interactions
3) The two strands twist into a right-handed helix with purine and pyrimidine bases shielded from water on the inside and negatively charged phosphates outside
What groups are on the 5' end and 3' end of a strand of DNA? What is the significance of the group on the 5' end?
5' - "free" phosphate group, which contributes to the negative charges of DNA that project outwards from the helix to interact with water or positively charged histone proteins
3' - free -OH group, which attaches to the phosphate of the phosphate group to form a phosphodiester bond
Coding Strand
Strand in which the protein sequence read from 5' to 3'
Template Strand
Strand in which a new daughter strand is assembled
How are the Watson-Crick hydrogen bonded base pairs G-C and A-T formed?
The packing of hydrophobic bases in pairs in stacked arrangement in the inside of the double helix, allowing the aromatic rings of bases to interact (hydrophobically) in this position
Are G-C or A-T base pairs more stable? Why?
G-C has 3 hydrogen bonds, whereas A-T has 2 hydrogen bonds
What does the denaturation of DNA into single strands depend on?
A melting temperature Tm dependent upon:
1) length of the DNA
2) number of G-C bases
A hyperchromic shift (increase in OD(260) occurs with denaturation)
Note: single strands will re-anneal upon cooling by the process of hybridization
How do single strands of DNA re-anneal?
Hybridization: re-anneal upon cooling; critical for PCR
What compounds compose DNA?
1) Pentose sugar: deoxyribose
2) Inorganic phosphate
3) Nitrogenous bases: purines (A, G) and pyrimidines (T, C)
What is the basic unit of DNA?
Deoxynucleotides
What is the difference between ribonucleotides and deoxyribonucleotides?
Deoxyribonucleotides: only for DNA synthesis; -OH group at the 2' position of the ribose ring is replaced with hydrogen
Ribonucleotides: has an -OH group at the 2' position of the ribose ring; structure allows for different hydrogen bonding patterns and backbone flexibility of RNA compared to DNA
"Genes on a chip" Technology
By DNA hybridization, used to examine mutations in genomic DNA or changes in expression patterns (cDNA)
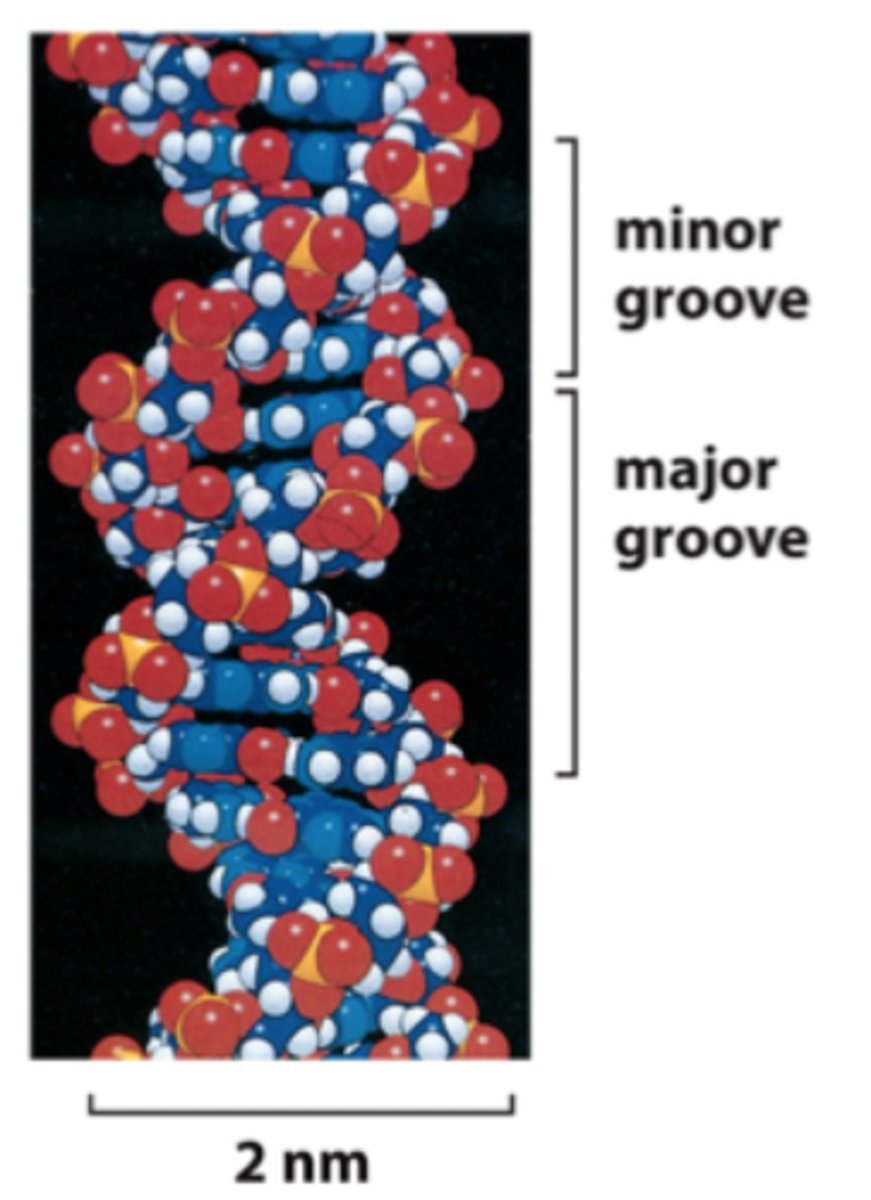
What is the significance of the narrow and wider grooves of a DNA double helix?
Recognition by proteins to alter DNA structure or to regulate transcription/replication
Also recognized by drugs
CpG Islands
Tracts of DNA sequence containing a lot of CpG on the same strand
Methylated - gene silencing (associated with aging/cancer)
Associated with 5' end of many housekeeping genes and some regulatory genes
A & C contain one (amino/keto) group and G & T contain one (amino/keto) group.
Amino; keto
Note: G & T are not susceptible to deamination b/c they do not have amino groups
How is DNA with mismatched base pairs used in assays?
DNA with mismatched base pairs has a lower annealing temperature, so can be used in assays to recognize specific base mutations
Is ATP/GTP/UTP/CTP/TTP always in the ribose form or deoxyribose form?
Ribose form
Deoxyribose form is ONLY for DNA synthesis or for enzymes making deoxyribonucleotides
Glycosidic Bond
Holds the nitrogenous base onto the pentose sugar of DNA
Chargaff's Rules
1) [A] = [T], [G] = [C]
2) [purines] = [pyrimidines]
3) [amino bases] = [keto bases]
Chemical analysis reveals that the DNA extracted from a sample is 30% cytosine. What percentage is adenine?
A. 20%
B. 30%
C. 40%
D. 70%
Answer: A
[C] = [G], so 30% C + 30% G = 60%
[A] = [T], and 100% - 60% = 40%, so 20% A and 20% T
Why does the structure of DNA provide a mechanism for heredity?
From the complementary sequence:
1) DNA is a template for its own duplication
2) DNA encodes information for producing proteins
T/F: Two polynucleotide chains interact by covalent forces.
False; interact by non-covalent forces (hydrogen bonding, hydrophobic bonding)
Why don't the two polynucleotide chains of DNA run parallel?
The bases will not be projected toward each other in a manner permitting hydrogen bonding if they run in the same direction
What is the most common structural form of DNA?
B-DNA: aka the Watson-Crick Model, which has major & minor grooves
Proteins typically bind in the major grooves
Most stable under physiological conditions
A-DNA
Structural form of DNA that is also a right-handed helix, but is more compact
Favored in solutions devoid of water to allow DNA crystallization
AKA duplex RNA
Z-DNA
Structural form of DNA that is a left-handed helix that forms in regions rich in alternating purine and pyrimidine bases so that the backbone is more extended and looks zigzag in appearance
With its characteristic high salt or methylation, this form may play a role in regulation of gene expression
Why are there multiple structural forms of DNA?
Regulation of gene expression
What are the physiochemical properties of DNA?
1) Chemically inert at high temperatures (100 C) and long times
2) RNA is much more sensitive to degradation by RNAses found commonly on the skin
3) Duplex DNA denatures by strand separation
What happens when the secondary structure of DNA is denatured?
1) Disrupts the H-bonds and hydrophobic interactions of the bases
2) Helix unwinds and the strands separate
What is a hyperchromic shift and why does it occur?
Hyperchromic shift: increase in the absorption of ultraviolet light when DNA is denatured and unwound with heat
When the shielding of the aromatic bases inside the native helix is removed, the aromatic bases can absorb more UV light
As the helix structure of DNA is disrupted by heating, the UV absorption of the DNA:
A) Does not change
B) Significantly decreases
C) Depends on the ionic composition of the solution
D) Moves to a higher wavelength
E) Significantly increases
E; a hyperchromic shift occurs
What is the melting temperature of a melting curve?
Midpoint of a DNA melting curve in which DNA transitions from a double strand to a single strand
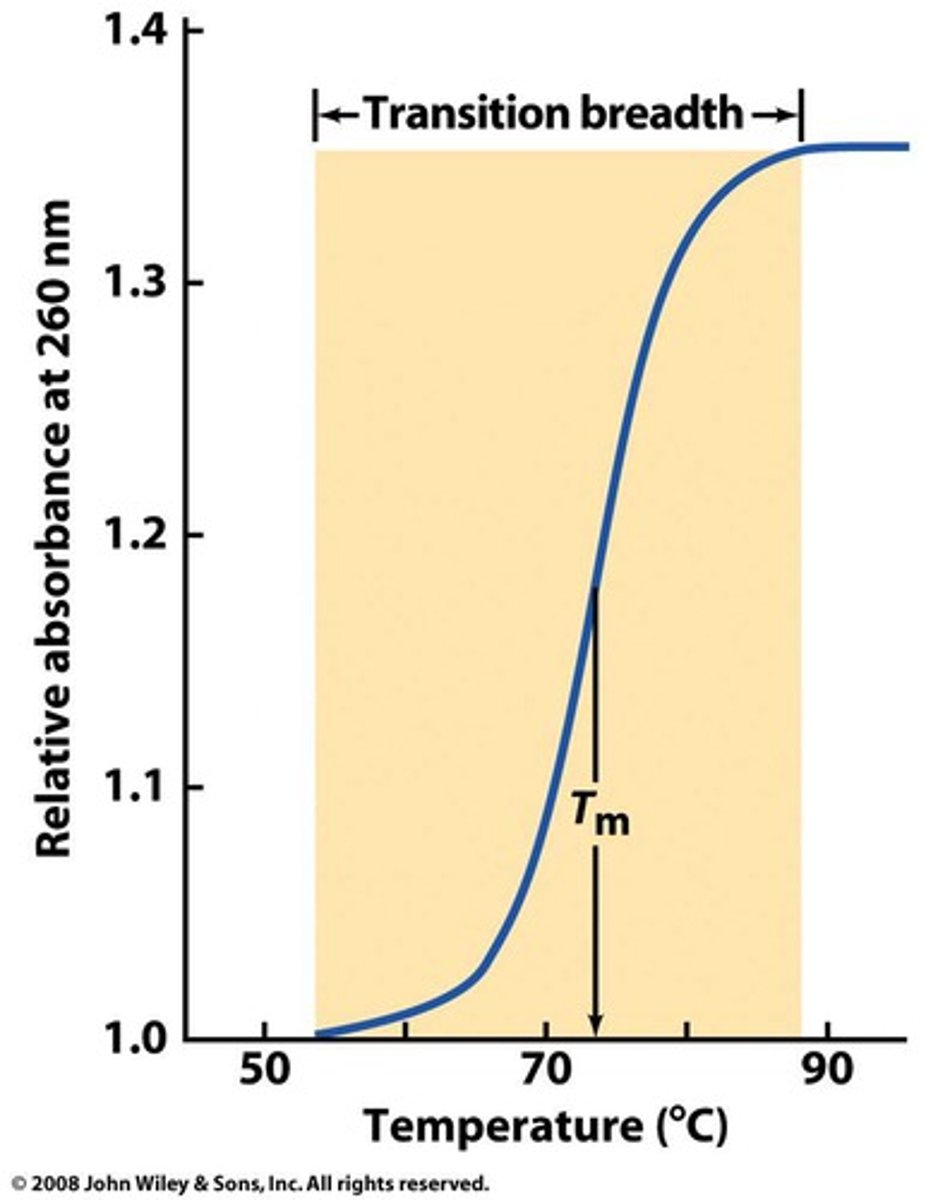
What is the relationship between Tm and GC content?
The more G & C, the higher the Tm
The temperature needs to be higher in order to break 3 hydrogen bonds in each G-C pair
What is more energetically favorable to initiate DNA replication - high GC content or high AT content?
High AT content - less energy required
A-T pairs only have 2 hydrogen bonds, as opposed to G-C having 3 hydrogen bonds. Because DNA replication is initiated by separating the two strands, DNA with high AT content is ideal
How do you determine Single Nucleotide Polymorphisms (SNPs) using DNA denaturation?
1) Denature template DNA and probes (affected DNA)
2) Compare absorbance v. temperature curve or template DNA to that of probes
3) Determine if a curve represents homozygous normal, heterozygote, or homozygous recessive
What are the characteristics of an SNP homozygous recessive curve?
Decreased Tm due to decreased H-bonding as a result of a mismatched base pair (SNP). With decreased H-bonding, the homozygous recessive DNA doesn't need a high temperature to denature, so the curve shifts to the left
What are the characteristics of an SNP heterozygote curve?
Half of the curve follows the normal template DNA denaturation curve (1/2 of the sample is normal), whereas the other half of the curve follows the SNP homozygous recessive curve (1/2 of the sample contains missing H-bond)
What is the use of cloning DNA in diagnosis/treatment?
To produce billions of identical copies for forensics, mutation analysis, and human protein production
What is the use of cleaving DNA in diagnosis/treatment?
Restriction digest using restriction endonucleases to break down cellular DNA into manageable fragments Used for mutation analysis of a gene or selected regions of a genome (DNA "fingerprint), and library of clones
What is the use of hybridizing DNA in diagnosis/treatment?
Use a probe oligonucleotide to locate allele-specific mutations, gene locus, and translocations
What is the use of rapid sequencing of DNA in diagnosis/treatment?
ID a gene or protein for detection of unusual mutations
What is the use of monitoring the expression level of the gene in diagnosis/treatment?
Detect cancer and disease
What are the ingredients for synthesizing DNA in a test tube?
1) DNA template
2) dNTPs
3) Primer with free 3' -OH group
4) DNA polymerase I
5) Mg2+
Note: experiments determining these ingredients also led to the discovery of the stability of DNA polymerase at high temperatures
Southern Blot
Separate genomic DNA on an electrophoretic gel and identify using a radio-labeled or fluorescent-labeled DNA probe that hybridizes a specific sequence (probe anneals to the DNA fragment of interest)
Northern Blot
Separate RNA on an electrophoretic gel and identify using a radio-labeled or fluorescent-labeled RNA probe that hybridizes a specific sequence
Used to analyze gene expression
cDNA microarray
More current method for gene expression profiles (hybridization patterns for patient vs. control DNA) cDNA (complementary to expressed mRNA) is hybridized to oligonucleotides on a chip selected to represent a particular disease state
Recombinant DNA
Produced by PCR/cloning
Combine two or more separate DNA strands that were cleaved using restriction enzymes to create sticky ends that can be annealed together
Note: the production of recombinant proteins are important sources of protein drugs (i.e. insulin)
Western Blot
Perform gel separation analysis on proteins
Used along with ELISA to test for the presence of antibodies or antigens in a patient's serum
Restriction Fragment Length Polymorphism (RFLP)
Separate DNA (cut by restriction enzymes) using agarose gel electrophoresis or pulsed-field gradient electrophoresis
DNA (negatively charged) travels to the positive pole of the gel, with smaller pieces moving faster
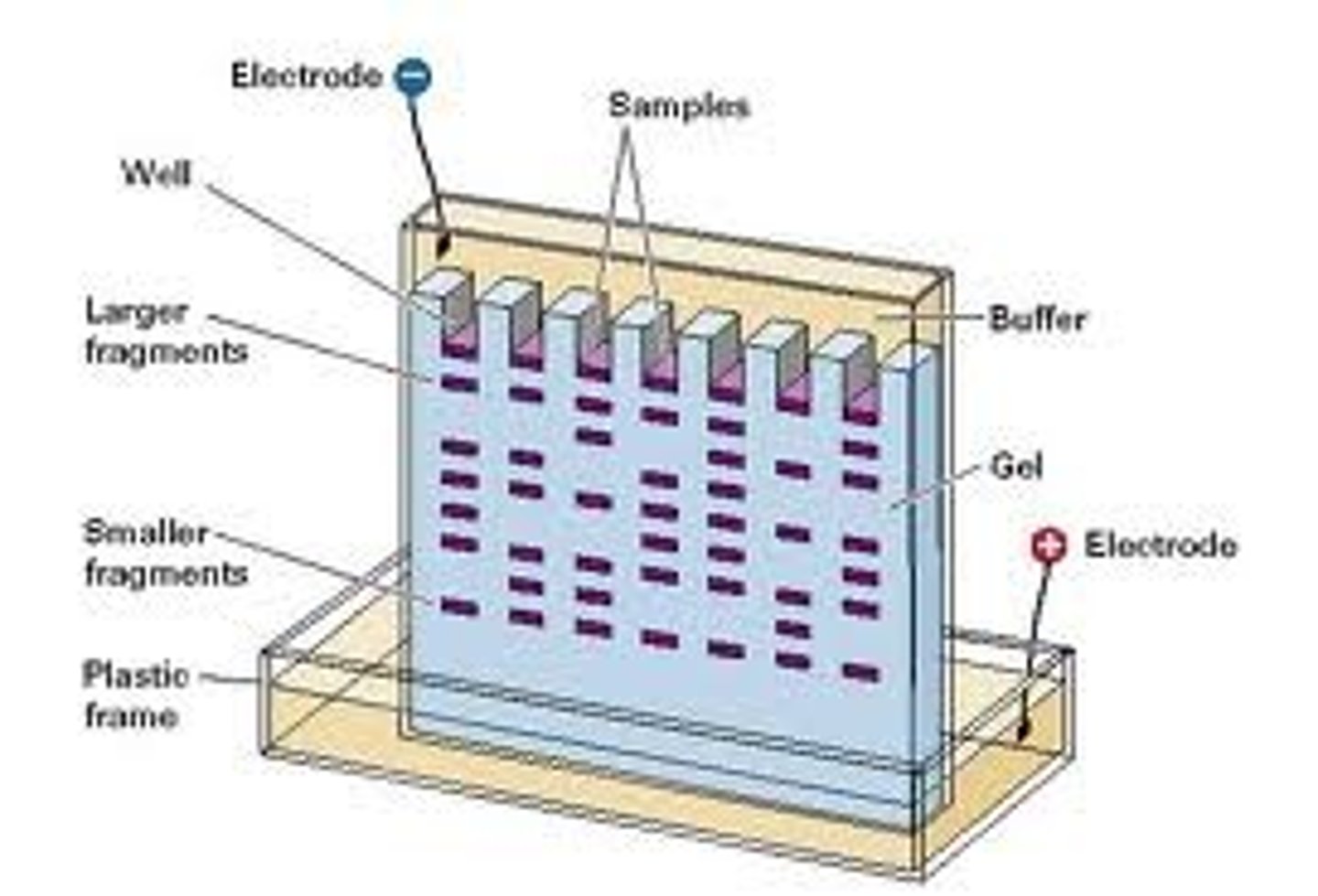
DNA probe
Short piece of single-stranded DNA or RNA that is fluorescent/radio-labeled to hybridize to the DNA of interest (i.e. region of mutation, disease-causing repeat sequence, or marker)
Need to select DNA of interest after a restriction digest of genomic DNA that produces millions of fragments
What indicates anticipation in RFLP diagnosis of multiple repeat polymorphisms?
The size of fragments increase from generation to generation on a southern blot
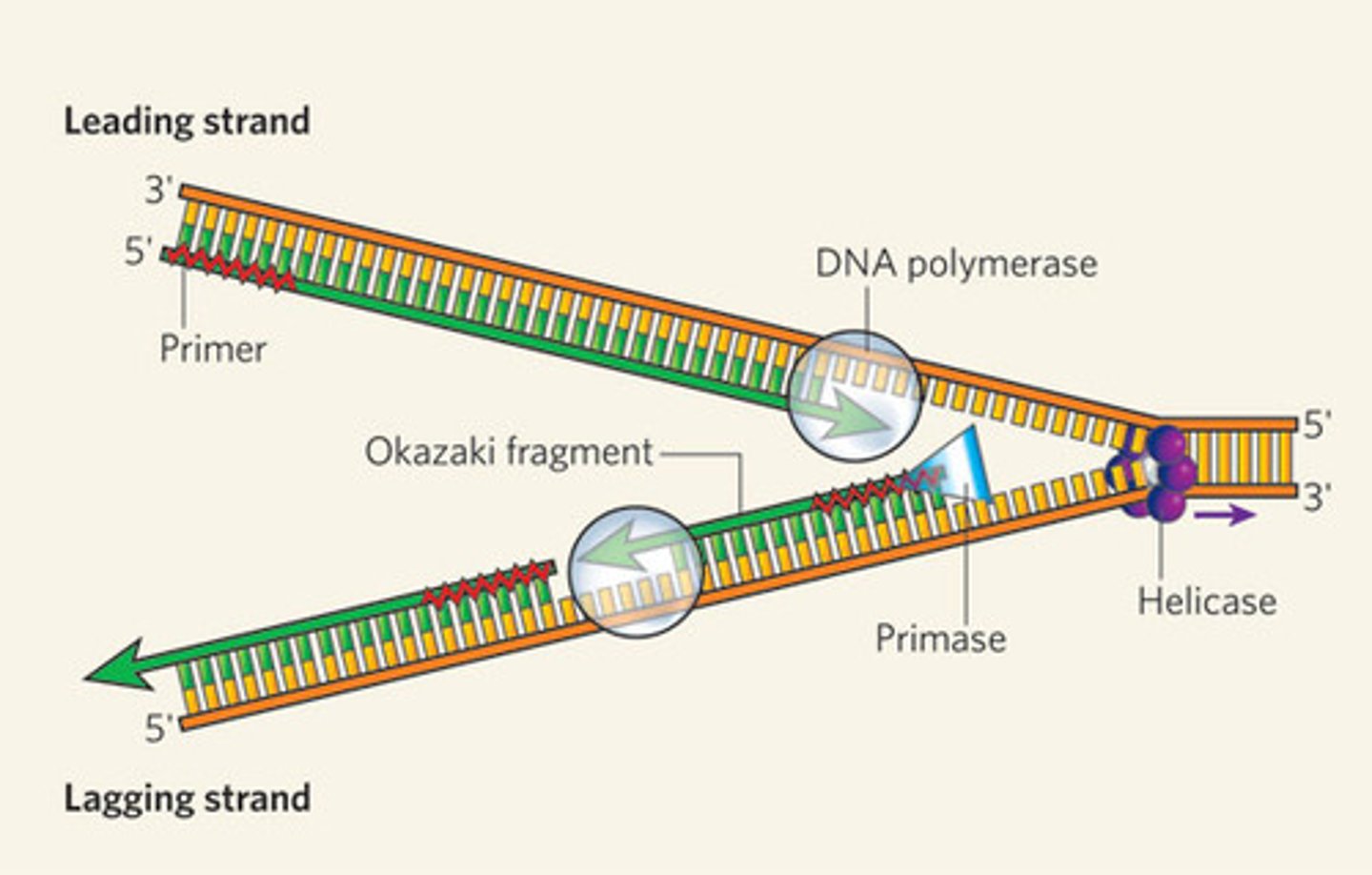
Polymerase Chain Reaction (PCR)
1) Melt DNA duplex at a high temperature
2) Primers anneal to bracket a region of DNA of interest by reducing temperature
3) DNA polymerase copies the region between the primers and create new DNA strands
4) Repeat steps until millions of copies are made
What is DNA polymerization and in what direction does DNA polymerase move?
Enzyme (requires a primer - short DNA/RNA segments) that acts on the template strand for DNA elongation
Moves 5' to 3' (copies the template from 3' to 5')
What is the significance of dideoxynucleotides (ddNTPs) in DNA sequencing?
AKA Sanger method
With 2' AND 3' H-bonds, unlike dNTPs (larger amounts) that have only 2' H-bonds, ddNTPs (small amounts) do not have an -OH group on the 3' that would have been used to form a phosphodiester bond. Thus, ddNTPs act as chain-elongating inhibitors of DNA polymerase to terminate strand synthesis
Capillary Electrophoresis
After separating fragments through the Sanger method, ddNTPs will fluoresce a different color when illuminated by a laser beam and can be arranged from longest to shortest to form the complementary DNA strand
Functional Genomics
1) Changes in gene expression levels give a molecular fingerprint of cell transformation, neoplasia, abnormal pathology, infectious organisms, et.
2) mRNA is amplified by PCR and converted to cDNA
3) cDNA is evaluated with a northern blot using a probe, or a cDNA microarray with preselected probes for different genes
Chromosome
Single DNA molecule compacted into chromatin
Heterochromatin
Highly condensed chromatin, not transcriptionally active
Spirals of chromatin filaments = condensed chromatin, which are in metaphase
Euchromatin
Loosely condensed, transcriptionally active
Chromatin Composition
Nucleosomes (histones + DNA) wrapped into solenoid fibers and loops in a hierarchical assembly
Loops = chromatin filaments + nuclear scaffold
Histones
Positively charged molecules (due to lysine and arginine) that dissociate from DNA for replication and transcription when the positive charge is reduced by acetylation
Types: H1, H2A, H2B, H3, H4
Dyad
Replicated chromosomes (two chromatids) held together by a centromere
Telomeres
At the ends of chromosomes to distinguish them from DNA breaks
Contain hexameric repeat TTAGGG to protect the ends of the chromosomes (minisatellite DNA)
Enables DNA replication to avoid a sticky end
Site for recognition of telomerase
Telomerase
Reverse transcriptase that uses an RNA template to extend the length of the telomere
Active only in germ, stem, and cancer cells, immortalizing the cell line
Telomere shortening
Associated with aging
Progressive loss of telomeres may be responsible in eukaryotes for the Hayflick limit
Hayflick limit
Limit to the number of times a cell can divide
Solenoid
Chromatin filaments composed of strings of nucleosomes and H1
Nucleosome
H2A + H2B + H3 + H4 + DNA
146 base pairs of core DNA wrapped around a histone octamer
How much DNA is between histone octamers?
8-114 bp; aka linker DNA
How many base pairs are between successive nucleosomes?
~40
H1
May reach from one nucleosome to the next to help stabilize them in higher orders of organization
Packing requires one H1 per nucleosome
Has been implicated in controlling gene expression
In inactive DNA, H1 is (tightly/loosely) bound to DNA, whereas in active DNA, H1 is (tightly/loosely) bound to DNA.
Tightly; loosely
"Tails" of histone proteins
Extend beyond DNA
May play a role in making contact with adjacent nucleosomes as the chromatin folds back on itself to form the higher order structure to compact DNA further
What post-translational modifications are related to the organization of the higher order chromatin structure?
Acetylation, phosphorylation, methylation
Tandem repeated DNA
Sequence of bases repeated over and over linearly along the chromosome; highly repetitive DNA
Can be satellite DNA, minisatellite DNA or microsatellite DNA (for forensic medicine, establish paternity, and crime forensics), with repeats of >100bp, 5-10bp, and 1-4bp respectively
Satellite DNA
High number of tandem repeats clustered at the centromere
Forms heterochromatin
5-171bp long
Minisatellite DNA
High number of tandem repeats clustered at telomeres
6-24bp long
Some of minisatellite DNA is transcribed
Sometimes used fro DNA fingerprinting or VNTR (Variable Number of Tandem Repeats) analysis
Minisatellite DNA for profiling - what is the probability that one band matches in the general population as opposed to 10 bands matching?
1 band: 1/4
10 bands: 1/1,000,000
Chances of a random match are reduced to close to nil
Microsatellite DNA
High number of short tandem repeats (STRs) (1-4bp) throughout non-coding DNA
Polymorphic (highly variable number of repeats between individuals) and intergenic (Variable Number of Tandem Repeats - VNTR - used in DNA fingerprinting)
Used in forensics
Also involve CpG islands
What are common STRs?
AATG, GA, TATG
Interspersed repetitive DNA
Degenerate copies of DNA segments repeated in various places in the genome
I.e. LINES, SINES, DNA transposons and retrotransposons, mobile genetic elements that multiplied in the human genome by replicating themselves and inserting new copies in different positions
LTR repeats
Associated with retroelements (decayed relics of retroviruses)
"Processed" Psudeogenes
Lack sequences corresponding to introns, indicating that they may have arisen from reverse transcription of mature mRNA
Unique Sequences: single copy
AKA non-repetitive
gene/sequences for proteins/enzymes
Note: 40% of base pairs are gene-related and only 3% are protein-coding regions; 100,000 proteins, but only 25,000 genes
Unique Sequences: mid-range copy numbers
AKA non-repetitive; multiple copies to code for
- nucleotide sequence in tRNA, rRNA (1,000 copies)
- histones (20-50 copies)
- immunoglobulins (copy number varies
- collagen, actin
Note: 40% of base pairs are gene-related and only 3% are protein-coding regions (exons); 100,000 proteins, but only 25,000 genes
Moderately repetitive DNA
1) Regulatory regions for transcription factor binding (CpG repeats - control housekeeping genes); cis/trans acting
2) LINES (L1): code for reverse transcriptase, enabling LINES and SINES (Alu) to be copied and pasted into new regions of the genome (AKA retrotransposition)
SINES
Short interspersed elements
Along with pseudogenes, are non-functional relics of genes
LINES
Long interspersed elements (RT-reverse transcription)
Code for reverse transcriptase required for retrotransposition
What is the significance of LINES/SINES?
Evolution; some genes that are beneficial may be copied and pasted
Retrotransposons
Mobile genetic elements that can copy and paste themselves into new loci
"Jumping gene"
Requires reverse transcriptase to make a mobile copy of itself and move by this RNA intermediate through RT synthesis of DNA
I.e. LINES, SINES
Retroviral-like transposons encode reverse transcriptase
Transposition
DNA-only; move transposons into new regions with an integrase function by this cut and paste mechanism, NOT copy and paste like LINES/SINES (in yeast/bacteria)
Can contribute to drug resistance
A young boy was diagnosed with Hemophilia A. There was no history of disease in his extended family, going back three generations. Gene sequencing revealed Alu integration within exon 14 of the Factor VIII gene. In this case, a lethal Alu (SINE) insertion occurred. How did it occur?
A) By insertion of a SINE DNA sequence translocated from another locus (cut and paste)
B) By repeat copies in tandem inserted during DNA replication
C) By reverse transcription of SINE RNA from another locus (copy and paste)
D) By a splicing error
C
How are genes identified by conserved regions between species?
Exons remain invariate, introns do not
Half of the eukaryotic genes are associated with an upstream CpG island
Intron Properties
1) Its longer length (100->5,000bp) increases frequency of recombination and decreases the chance of damage to the coding exon; the farther apart two gene segments are, the greater the chance of crossing over
2) Higher mutation rate than exons
3) Begin with GT and end with AG - helps the "splicing out" of the sequences from the RNA transcript
4) Transcribed into RNA, but removed from the RNA transcript by splicing
5) RNA transcripts of introns may be regulatory signals to other genes
6) May act as ribozymes in RNA splicing
7) Enable diversity of translated proteins
Which genes lack introns?
Interferons, histones
Exon Properties
1) Typically <200 bp
2) Code for protein
3) 1 exon = 1 domain
4) Highly conserved
5) Uninterrupted genes have 1 exon; most genes in mammals are interrupted
6) May code for the different functional units/domains of a protein or code for different functions of a gene at different stages of development of an organism
7) Number of exons varies with the protein
Natural somatic mutation
Exon is added or dropped during replication, producing different proteins
Often occurs in the cells of the immune system
Describe the relationship between trinucleotide repeats and strand slippage
If strand slippage occurs in the daughter strand, heterogeneity occurs in the alleles. If strand slippage occurs in the parental strand during DNA replication, the number of repeats is reduced
Symptoms of trinucleotide repeat diseases are associated with INCREASING number of repeats (slippage of template strand during lagging strand synthesis)
Histone Acetyltransferases (HATs) and Histone Deacetylases (HDACs)
Regulate accessibility of DNA
HDAC inhibitors
Class of anticancer agents in development