C1.1: Enzymes and Metabolism
1/34
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
35 Terms
Enzyme
Biological catalysts made of proteins or rRNA, increase the rate of biochemical reactions without being changed
Without them, necessary processes for life would occur very slowly
*convert substrates to products*
Metabolism
Complex network of interdependent and interacting chemical reactions in living organisms
Arranged as a series of reactions that are catalyzed by unique enzymes at each step, can exist as wither chains or cycles
Enzyme Specificity
Enzymes are specific and catalyze only one specific reaction.
Organisms require many different enzymes to survive, and cells control rxn rate by making more/less of different enzymes
Metabolic Pathways
Most enzymes produce small changes in pathways, significant transformations happen in a sequence of small steps
Can be a linear pathway (chain) or a cyclical pathway (cycle)
Chain Pathway
A linear metabolic pathway, ex. glycolysis converts glucose to pyruvates
Cyclical Pathway
More common, a cycle in which every intermediate is a product of previous rxns and substrate of next rxn
ex. calvin and krebs cycle
What makes enzymes?
ribosomes
Enzyme Locations
Extracellular or Intracellular
Extracellular Enzymes
exoenzymes, made by attached ribosomes on ER and released from cell.
They break down larger macromolecules, ex in digestive system or fungi that secrete enzymes to absorb nutrients bc can't do endocytosis
Intracellular Enzymes
Made by free ribosomes in cytoplasm and used within cell, ex glycolysis
Some are used inside organelles like in krebs cycle in mitochondrial matrix
Types of Reactions
Catabolism and Anabolism
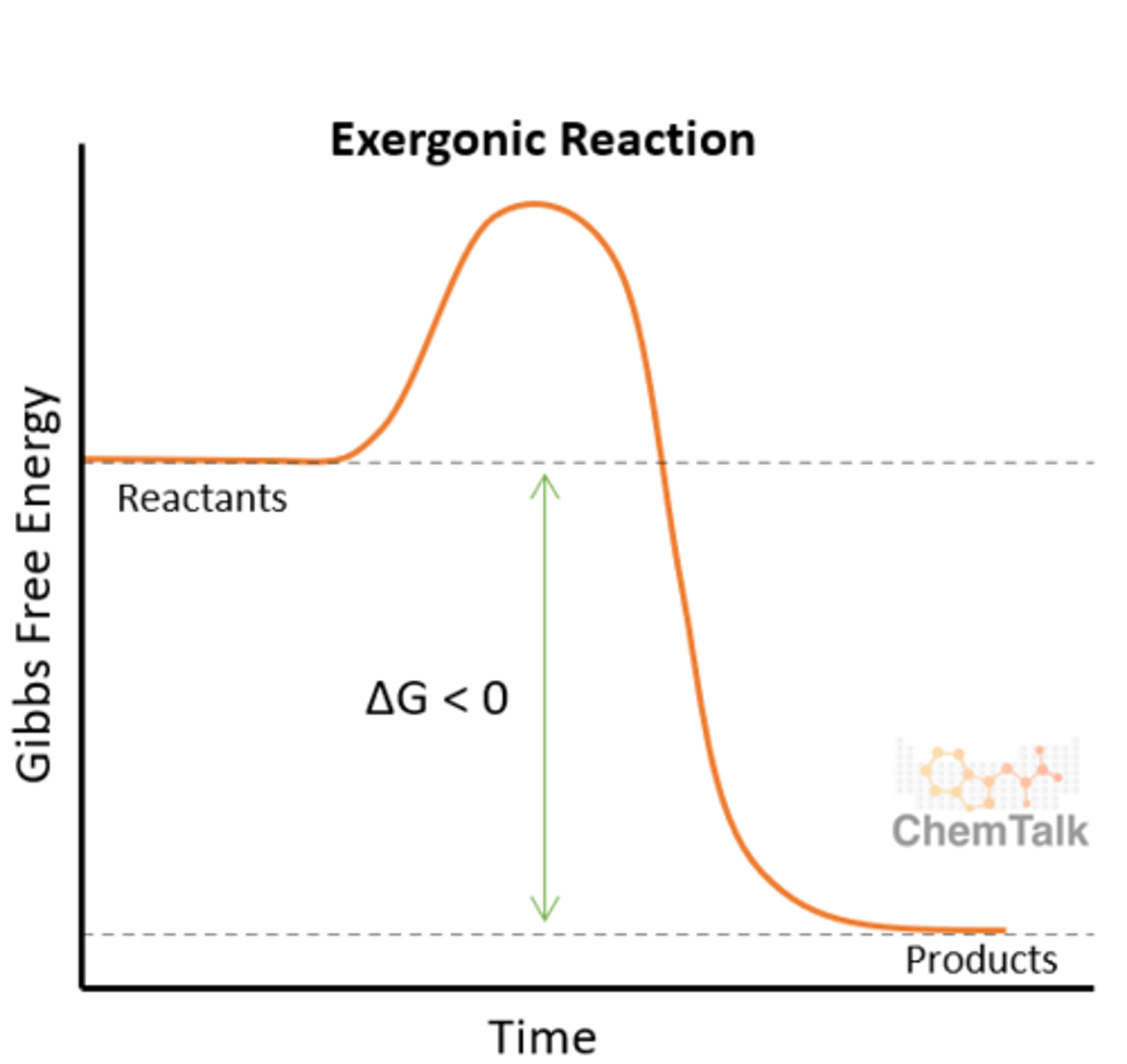
Catabolism
Breakdown, polymers to monomers via hydrolysis
Exergonic, releases energy (deltaG < 0), reactants have more energy than products.
Spontaneous
ex. digestion/cellular respiration
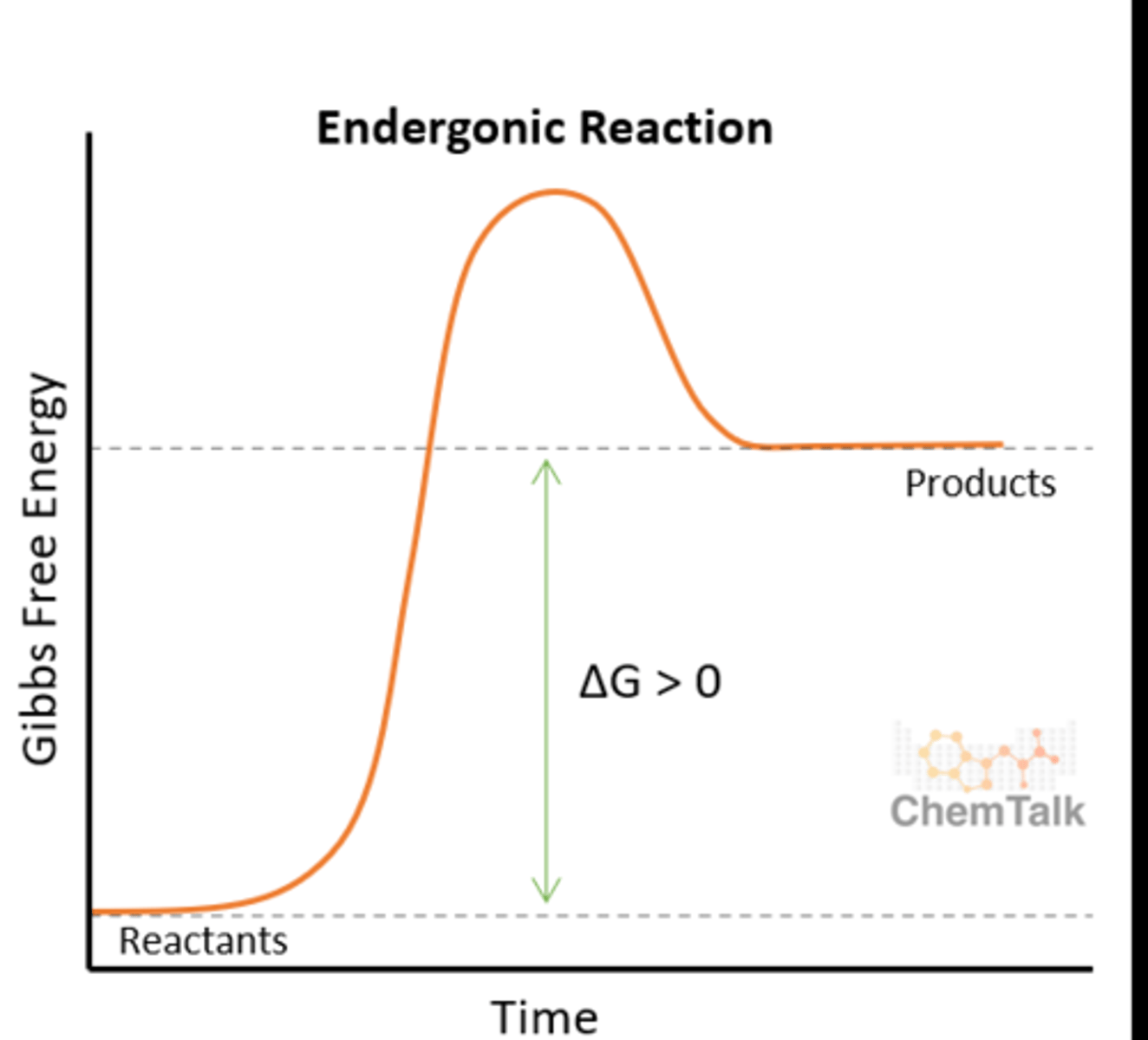
Anabolism
Synthesis, monomers to polymers via condensation reaction
endergonic, requires energy (deltaG > 0), reactants have less energy than products
Enzyme Structure
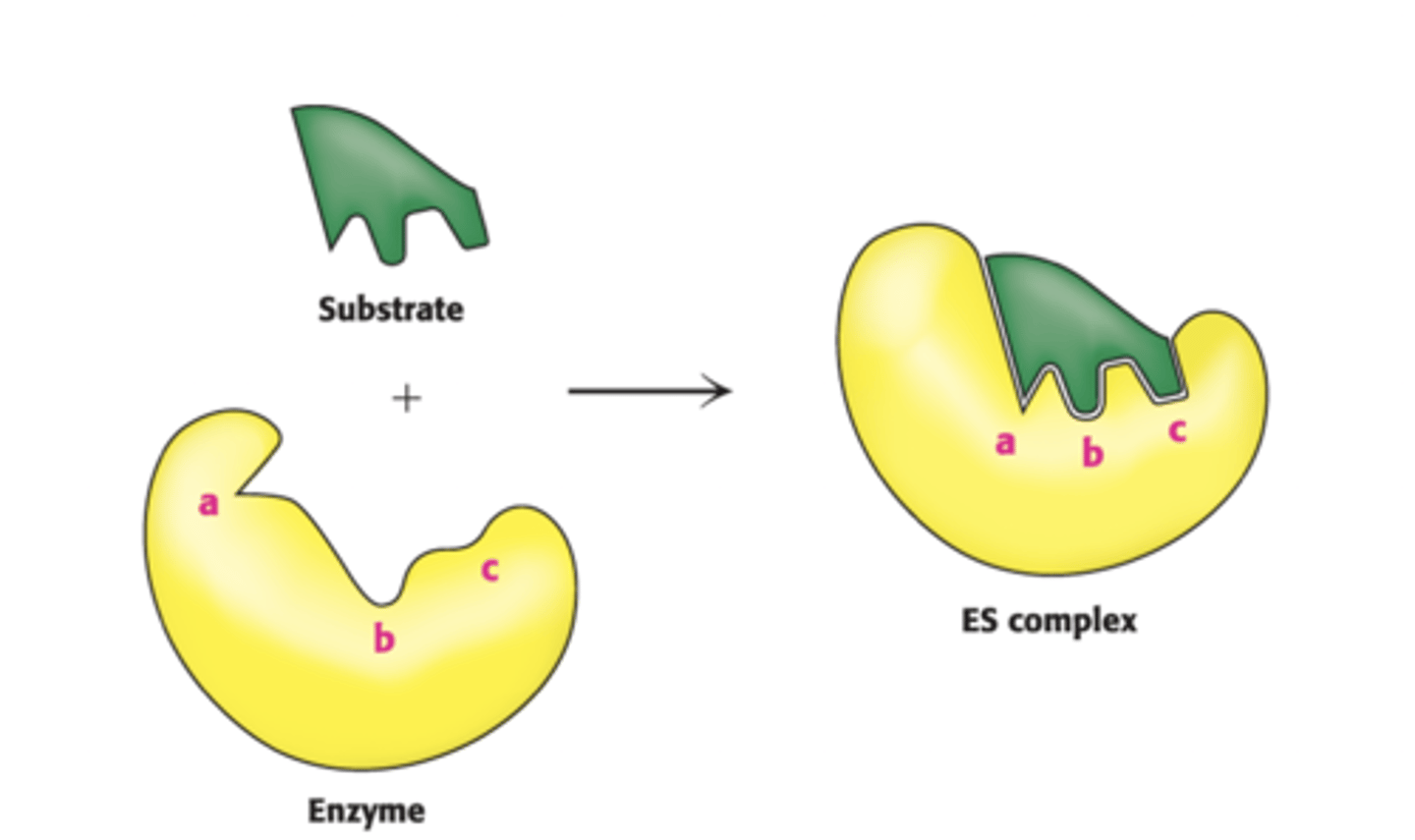
Globular protein with specific 3D shapes
Active sites display substrate specificity, the two are complimentary in shape and chemical property and once bound, substrate becomes product
Enzyme Active Sites
Active sites consist of only a few AAs, made by unique folding of polypeptide, and AAs that make up active site are not found next to each other in AA chain
Importance of Enzyme Structure
If any part of structure altered, overall shape and active site may change, and the enzyme will lose its function
Induced-Fit Binding
When substrate and enzyme are close enough, their chemical properties attract each other
This interaction causes BOTH to change in bond angles and bond length, leading to INDUCED FIT
*if there is a second substrate, it binds to another part of the active site, causing it to change shape again*
Brownian Motion
Molecules move randomly in aqueous solutions, substrate and active site must collide for rxn to occur and the two must be aligned for bonding
Attractive forces only work over short distances
**substrates typically move more than enzyme due to smaller size, membrane embedded enzymes don't move at all**
Byproduct of Metabolism
Heat
-transfer of energy never 100% efficient, therefore excess energy lost as heat
-mammals and birds use heat to maintain body temp.
If too much heat...
Body sweats (evaporative cooling)
If not enough heat...
Body shivers, involuntary muscle contractions, or body activates brown fat tissue which has many mitochondria to carry out cellular respiration
Structure Equals Function
Enzyme-substrate specificity, some enzymes are absolutely specific and bind to same substrates only, others are less specific and bind to a group of substrates
*enzyme structure determines its function*
-changes in different parts of the enzyme (even not at active site) will likely change the active site
-changes in active site prevents substrate from binding and therefore stops reaction
Denaturation
Can occur when changes in enzyme structure are too significant to be reversed
What can affect enzyme reaction rates?
Temperature, pH, Substrate Concentration
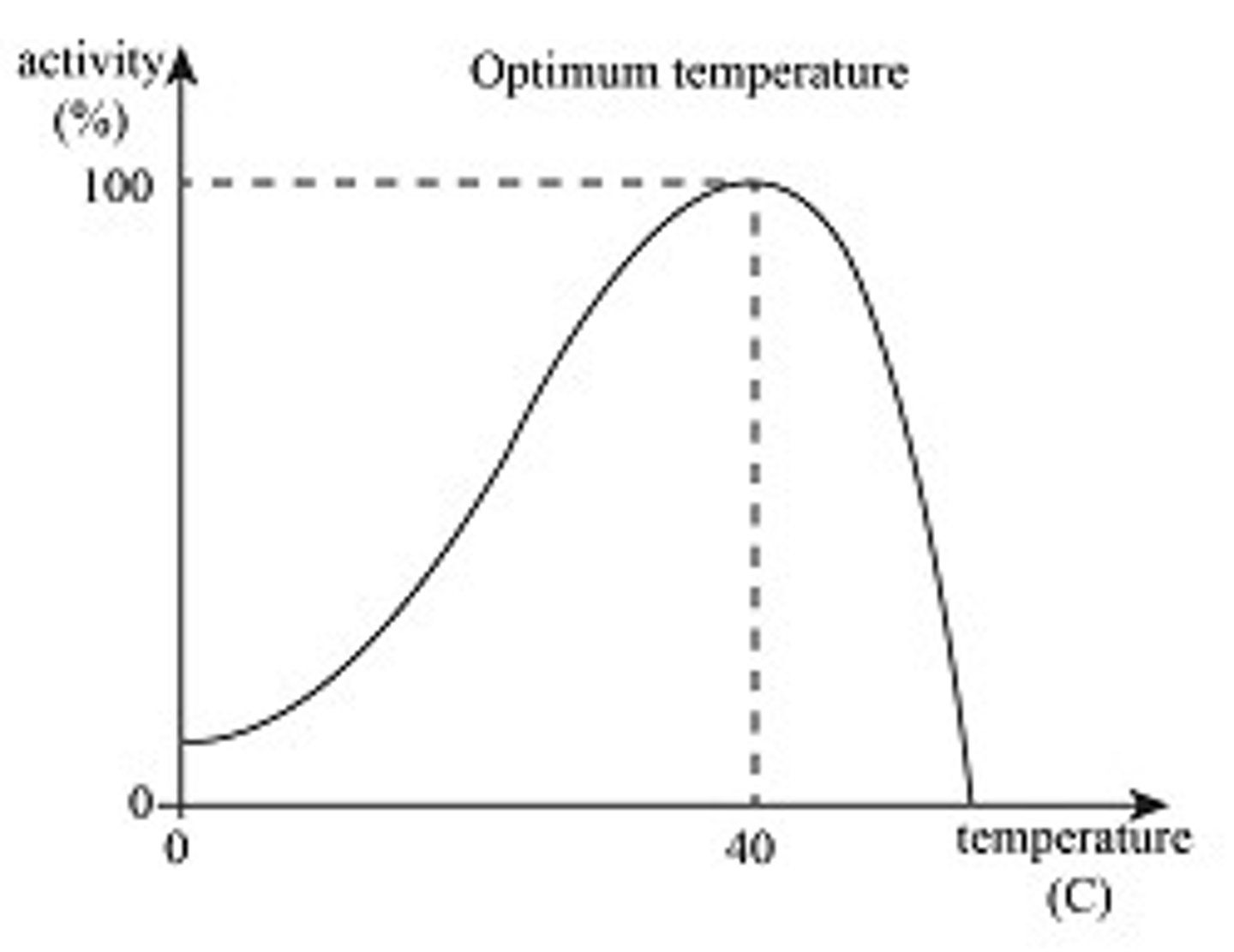
Temperature
Increase thermal energy = increase kinetic energy = increase particle motion = increase #collisions (and bond vibration rate) = increase reaction rate
-Optimal temperature about 37 C, body temp
Peak enzyme activity = greatest # of molecular collisions
-higher temp (too high) disrupts H-bonds, causing enzyme to denature, active site loses shape and enzyme loses function
*lower temp decreases rxn rate*
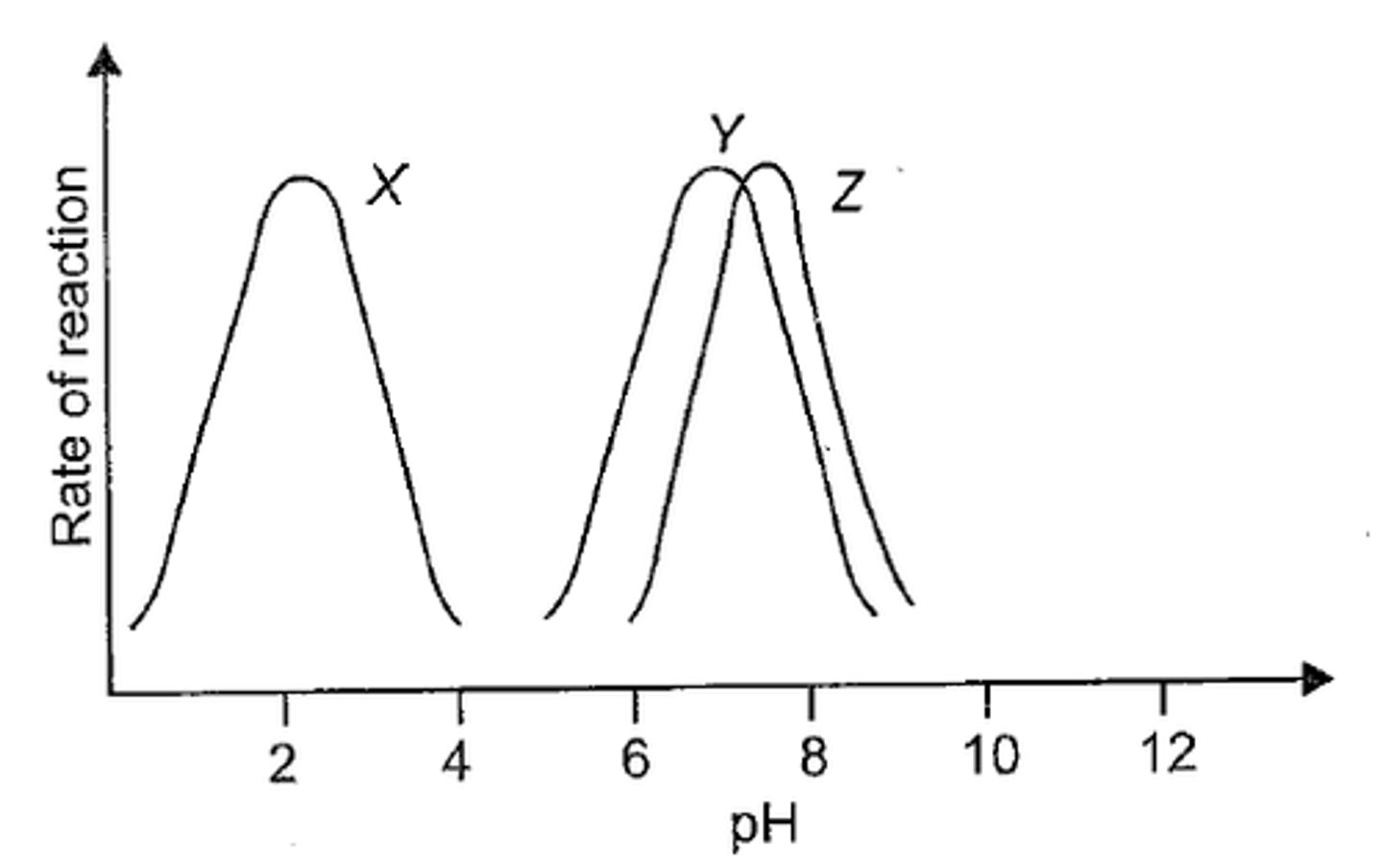
pH
adding or removing H+ changes pH
-pH measure {H+}, logarithmic scale, changing pH by 1 alters soln by 10x
Optimal pH
-moving away from this range decreases enzyme activity
-affects enzymes charge and disrupts ionic bonds
-denaturation changes overall shape and decreases enzyme function
-most human enzymes; pH 6-8
-pepsin, pH 2-3
Substrate Concentration
Increase substrate [] = increase enzyme activity = increase rxn rate
-higher substrate [] allows more frequent collisions w/ enzymes, although at some point rxn rate eventually plateaus when all active sites become occupied (saturated) and are unavailable until product is released
**rxn has reached its maximum rate or Vmax (velocity maximum)
![<p>Increase substrate [] = increase enzyme activity = increase rxn rate</p><p>-higher substrate [] allows more frequent collisions w/ enzymes, although at some point rxn rate eventually plateaus when all active sites become occupied (saturated) and are unavailable until product is released</p><p>**rxn has reached its maximum rate or Vmax (velocity maximum)</p>](https://knowt-user-attachments.s3.amazonaws.com/19e1a444-1207-4372-8fe9-6d4a78cc9f2c.png)
Activation Energy
Amount of energy required to begin a chemical reaction, enzymes increase reaction rate by decreasing activation energy
-enzyme binding stresses and destabilizes bonds of substrate, reducing its energy level so less energy required to become a product
*net amount of energy in rxn the same*
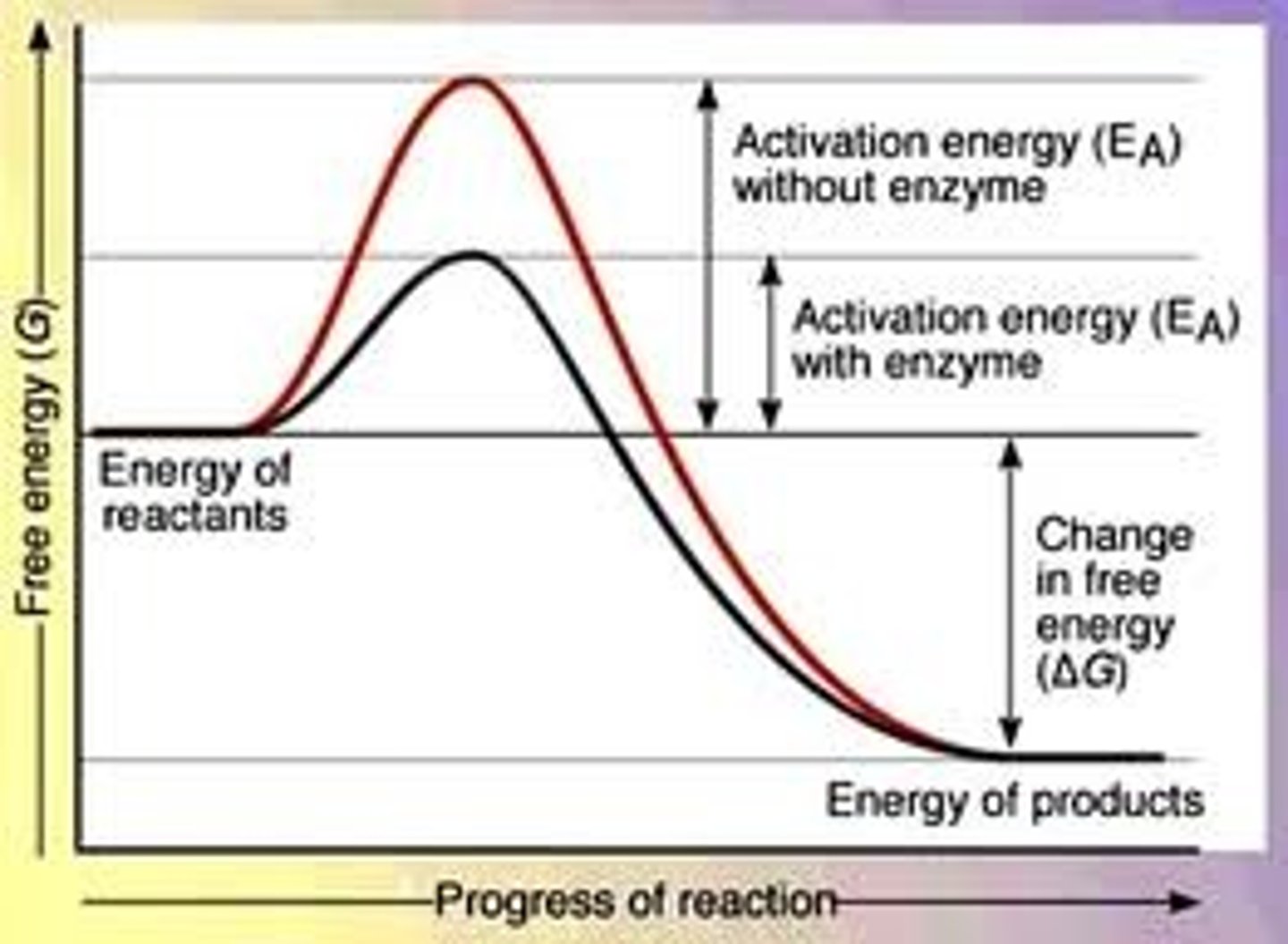
How do enzymes affect activation energy?
By acting as templates to allow substrates to reach optimal orientation and place stress on necessary bonds
By acting as suitable microenvironments to maintain certain pH
Types of Inhibition
Non-competitive, Competitive, End-Product Inhibition, Mechanism-Based Inhibition
Non-Competitive Inhibition
Inhibitor binds to allosteric site and changes overall shape of enzyme and therefore its active site
-sometimes the changes activates the enzyme (activator), but typically inhibits
Substrate cannot bind since two are no longer complementary
-because inhibitor does not "compete' with substrate, increase in [substrate] doesn't overcome inhibition
Competitive Inhibition
Inhibitor is structurally and chemically similar to substrate and binds to active site, prevents substrate from binding and remains bound longer than substrate
-increase in [substrate] usually overcomes inhibition
*covalently bonded inhibitors irreversible tho*
Competitive vs Non-Competitive Inhibitors
Competitive: Increasing [substrate] will alter the effect of competitive inhibition bc attaches to same active site, therefore Vmax is eventually the same but Km ([substrate] when rxn rate is 50% of Vmax) is lower
Non-Competitive: Increasing [substrate] will not alter the effect of non-competitive inhibition bc different site, therefore the Vmax is lower in the end
![<p>Competitive: Increasing [substrate] will alter the effect of competitive inhibition bc attaches to same active site, therefore Vmax is eventually the same but Km ([substrate] when rxn rate is 50% of Vmax) is lower</p><p>Non-Competitive: Increasing [substrate] will not alter the effect of non-competitive inhibition bc different site, therefore the Vmax is lower in the end</p>](https://knowt-user-attachments.s3.amazonaws.com/f59a6212-4f9e-4587-98ef-de7a392ad23c.png)
End-Product Inhibition
In chain/cycle pathway, product of one enzyme is used by next enzyme in metabolic pathway until final product made
-if too much final product, the final product acts as a noncompetitive inhibitor by binding to 1st enzyme's active site, stops production
-Once product levels decrease, the increase in [substrate] dislodges the inhibitor and pathway resumes to make product, or substrate attaches to second allosteric site and acts as an activator to cause inhibitor to release
Feedback inhibition regulates amount of final product to conserve energy, intermediate products are not made and do not accumulate
Mechanisms-Based Inhibition
Irreversible Inhibitors, inhibitor permanently binds to active site via covalent bond, results in stable enzyme-inhibitor complex that never forms a product
-may kill organism if the inhibited enzyme is vital
ex. penicillin to kill most gram-positive bacteria (thick cell wall)