BIN300- W2 Linkage between markers
1/14
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
15 Terms
Phase of the parents with genotype M1M2N1N2
M1N1/M2N2 (50%), M1N2/M2N1 (50%)
→ Phase either recombinant or non-recombinant gametes
Gametes transmitted from parent to offspring
All possible gametes (recombinant/non recombinant)
probabilities (as a function of Ɵ)
Identify gametes in offspring
sometimes not possble from gentoype information → non-informative offspring
Estimate recombination fraction
phase of the parents with genotype M1M2N1N2
Gametes transmitted from parent to offspring
Identify gametes in offspring
Non-infomrative offspring
•All heterozygots:
M1N1/M2N2 * M1N1/M2N2
M1M2N1N2
•Some homozygots:
M1N1/M1N2 * M1N1/M2N1
any possible offspring
For heterozygots not sure which chromosome came from which, also for homozygots
maximum likelihood estimation of Ɵ
From M1N1/M2N2 * M1N1/M1N1
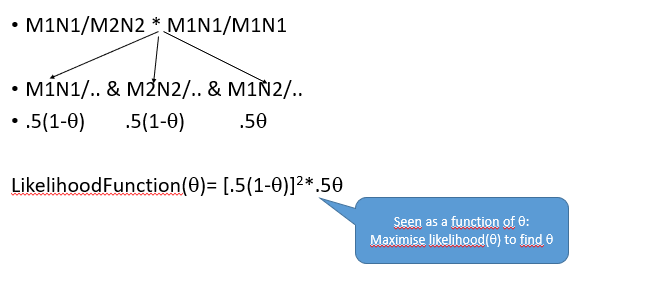
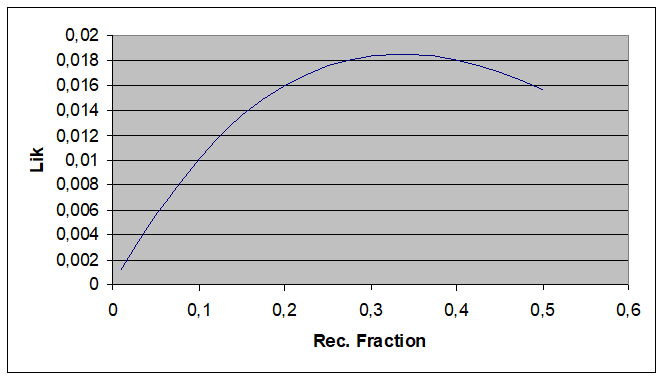
Plot of likelihood function
Also possible for family
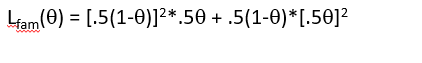
plot of Lfam (Ɵ)
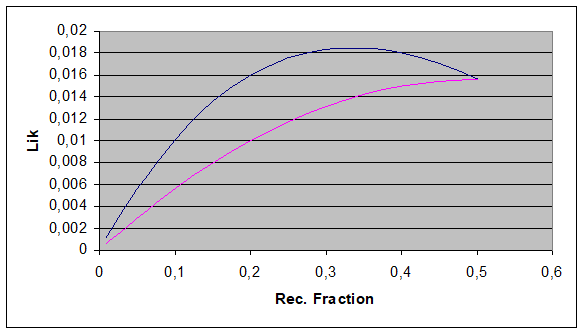
What happens if there are more families for the likelihood:
multiply L(Ɵ) of each family
D
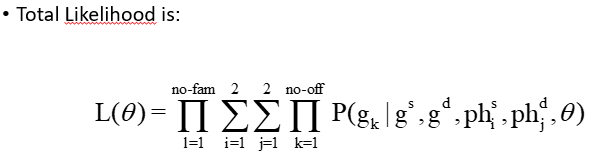
Detection of linkage
H0 = Ɵ = 0.5: unlinked, different chroms
H1: Ɵ = Ɵ1<0.5 (linked, same chrom)
LODscore
Z(Ɵ)>3: linkage (10³ = 1000 times more likely)
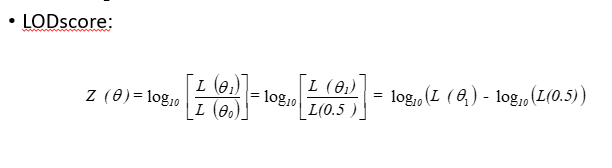
Design of mapping experiment
1 sire + 1 prog: 2 genotypings: 1 meiousis
1 sire + 10 prog: 11 typings: 11 meiosis
1 sire + 10 dams + 10 prog (HS fam): 21 typings: 20 meiosis
1 sire + 1 dam + 10 prog (FS fam): 12 typings: 20 meiosis
Large FS fams → most information
Homozygous parents are non-informative
need highly polymorphic markers
Design of mapping experiment - needs
Homozygous parents are non informative, need highly polymorphic marrkers, fullsib fams better than halfsib fams, larger families, small Ɵtrue → dense markers
linkage between SNPs
detected by linkage analysis
study inheritance of marker alleles from parent to offspring, how many recombinants/nonrecombinants
best desing: large full sib families, fish ideal species
uses maximum likelihood (ML)
write likelihood of data as function of parameters (Ɵ)
ask computer to maximise this function by varying Ɵ, gives best recombiantion rate Ɵ