MBS 344: Final
1/125
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
126 Terms
natural selection vs evolution
evolution: change of characteristics over time
natural selection: individuals in a population have different levels of biological fitness due to variation in phenotypes
→ mechanism for evolutionary change
genotype vs phenotype
genotype: organism’s genetic information
phenotype: organisms observable physical traits
→ phenotype is associated with genotype bc of central dogma
Genotype→Phenotype
Define: homologous chromosomes, allele, homozygous, heterozygous
homologous chromosomes: have same genes in same locations
allele: alternative version of a gene
homozygous: alleles have identical DNA sequence
heterozygous: alleles have different DNA sequence
DNA, RNA, and Amino acids: need to know structure, how these molecules are linked together to form a macromolecule, function of each one in a cell, bonds that hold subunits and structure together.
DNA: very stable, stores all genetic information
RNA: product of transcription using DNA, less stable but multifunctional
protein: product of translation using RNA, perform many cell functions
Purpose and basic steps for PCR
→ amplify specific regions of DNA from small amounts
denature: separate DNA strands by breaking Hbonds bt bases
anneal: allow primers to anneal to template DNA
extend: optimal temp for DNA polymerase to add nucleotides (synthesize 5’→3’)
Purpose and basic steps for DNA cloning
→ combine DNA sequences from two different sources to create recombinant DNA
gene and plasmid cut by the same RE (digested)
sticky ends bind w/ corresponding BP
DNA ligase closes gaps in phosphodiester backbone→ create recombinant plasmid
Purpose and basic steps for RT-PCR
→ measure expression of a specific gene w/ cDNA
isolate RNA
cDNA conversion
anneal, denature, extend
intensity of bands~ gene expression
different from PCR bc use cDNA template (only exons bc derived from RNA strand)
Purpose and basic steps for qPCR
→ amplification detected by measuring an increase in florescence— gel electrophoresis is not used
isolate RNA
cDNA conversion
anneal, denature, extend
lower Cq value = higher gene expression
Purpose and basic steps for western blot
→ antibodies used to detect the amount of a specific protein on a membrane
denature protein (SDS-PAGE)
transfer to membrane
incubate w/ primary antibody
add secondary antibody that binds primary— used for visualization of product
thicker band~more protein
FISH vs southern blot
same?
techniques used to analyze specific DNA sequences within cell
use probes and hybridization to analyze sequences
different?
Southern: DNA must be isolated, cut with RE, and run on agarose gel
FISH: fixed cells on glass slide and florescent microscope
no DNA isolation, RE, or gel electrophoresis
reporter gene
→ used in fusion protein to study the function of regulatory DNA elements like promoter and enhancer
example: GFT, used in fusion protein to indicate protein location w/ in cell (highly fluor)
DAPI
fluorescent stain that binds strongly with all DNA→ used to localize nuclei or chromosomes under UV light
prokaryotic vs eukaryotic chromosomes
same?
bidirectional replication→ two rep forks, two leading and lagging strands
chromosomal DNA
different?
circular vs linear
one origin of replication vs thousands
naked vs protein containing
gene rich vs gene poor
presence of centromere and telomere
two stragetgies to compact prok chromosome
loop domain
supercoiling→ compaction and strand separation (rep or tranxn)
how? DNA underwinding→ remove turn on B DNA to induce strcutural stain and promote supercoiling
Euk DNA compaction levels
supercoiled DNA
10 nm→ histone octamer + DNA
30 nm→ histone octamer + DNA + H1
chromosome scaffold
further loops and coils
topoisomerases I and II
topo II: use ATP to break both DNA strands and promote DNA underwinding, then ligate w/ strains
topo I: break one DNA strand, relieve DNA underwinding, then ligate
histones vs nucleosomes
histone: proteins that DNA wraps around
one core histone=octamer of histones
octamer (2X): H2A, H2B, H3, H4
nucleosome: basic units of DNA packaging → octamer w/ DNA wrapped around it
chromatin remodeling complex
alter nucleosome arrangement on chromosome
use ATP to eject/replace nucleosome
histone variants
alternative substitutes for histones→ used to alter gene expression by CRC
variants→ H3 and H2A
no variants→ H2B and H4
histone modifications of H3K4, H3K9, H3K27
acetylation→ of all three residues causes increased transcription (euchromatin)
methylation→
H3K4me: increased transcription
H3K9 & H3K27: decrease transcription
X inactivation/Xist/histone modifications
x-inactivation: silencing one of two x chromosomes in female cells to allow dosage compensation between sexes
xist: lncRNA that coats one whole x chromosome and promotes histone modification (heterochromatin) by recruiting histone-modifying enzymes
Describe the functions of the following enzymes during DNA replication: DNA helicase, DNA ligase, topoisomerase, primase, DNA polymerase.
helicase breaks hydrogen bonds bt DNA strands to initiate DNA replication
topoisomerase removes supercoiling generated by helicase
primase synthesizes short RNA primers to give DNA pol a 3’ OH
DNA polymerase synthesize the new DNA strand in 5’ to 3’ direction
DNA ligase joins together fragments of DNA that are adjacent (okazaki fragments)
Leading vs lagging strands
leading: same direction as replication fork and continuous
lagging: opposite direction as replication fork and discontinuous (Okazaki fragments)
same: synthesized 5’ to 3’ direction, dNTDs
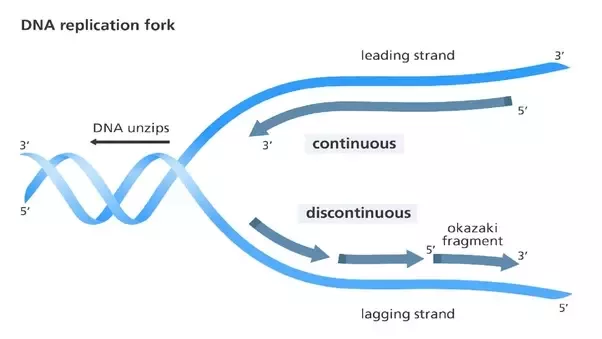
proofreading vs nick translation
proofreading: 3’ to 5’ proofreading is used by DNA pol I and III to identify and correct mistakes in DNA synthesis
nick translation: concurrent 5’ to 3’ excision of RNA nucleotide and DNA polymerization
Telomeres and Telomerase: telomeres, cell senescence, and end-replication problem; function of telomerase (TR vs TERT).
telomeres: repeat sequences at the end of linear chromosome for stability
end-replication problem: telomeres are shortened on both daughter strands after each round of replication
cell senescence: cells stop diving, even tho optimal condition, bc of excess telomere shortening
telomerase: enzyme that prevents telomere shortening by extending the 3’ overhang
composed of telomerase RNA and protein (TERT)
Need to know the differences between silent, missense, nonsense, and frameshift mutations. Which of these mutations is associated with disease? Why?
silent: base sub that changes DNA seq but does not alter aa sequence— protein function same
missense: base sub that changes DNA seq and alters aa sequence— protein function may be altered
nonsense: base sub that changes DNA seq and generate premature stop codon— protein function altered
frameshift: insertion or deletion that changes reading frame of gene seq— protein function is altered bc many aa changed
***disease? pt mutation must change protein function to cause disease, so all above apply except silent mutation
Describe the difference between loss-of-function (LOF) and gain-of-function (GOF) mutations. Describe some causes of LOF and GOF mutations.
LOF: change in DNA seq destroys protein function→ recessive
how? change protein shape or decrease gene expression
GOF: change DNA seq makes protein hyperactive (rare)→ dominant
how? change protein shape or increasing expression
What are spontaneous mutations? What are induced mutations? What causes them?Describe some similarities and differences between these two causes of mutations.
spontaneous: occur naturally, no mutagen involved
errors in DNA replication
oxidative damage (by ROS)
depurination
deamination
induced: caused by exposure to mutagen
DNA adduct (chemical attached to base)
UV light/exposure to radiation
same: lead to point mutations
Type of DNA damage fixed by BER, NER, MMR, NHEJ, and HRR. Which of these DNA repair mechanisms are associated with CRISPR/Cas9 gene editing system? What are their functions in the CRISPR/Cas9 gene editing system?
Base excision repair: repairs individual bases that are abnormal- repairs small distortions
one strand repair for oxidation, depurination, deamination
Nucleotide excision repair: corrects large distorts in double helix caused by exogenous sources
one strand repair for DNA adducts and thymine dimers
Mismatch repair: corrects mismatch bases— normal base, just incorrect position
one strand repair for mismatch
Nonhomologous end joining: corrects DSB w/ out a template strand
DSB repair from radiation
used in CRISPR/Cas9 when no donor DNA is present
Homologous recombination repair: correct DSB w/ a template strand
DSB from radiation
used in CRISPR/Cas9 when donor DNA is present
What are the roles of promoters, regulatory sequences, and terminator sequences in prokaryotic transcription?
promoter: binding site on DNA for RNA pol, always located next to gene seq
regulatory seq: binding site for TF that regulate rate of transcription, located close or far from gene seq
terminator: DNA seq at end of gene seq, ejects RNA pol from DNA
What are the three stages of transcription? What is the RNA polymerase doing in each stage?
stages of transcription:
initiation→ RNA pol binds to promoter, DNA strands are denatured to initiate transcription
elongation→ RNA pol leaves promoter and synthesizes RNA (5’ to 3’)
termination→ RNA pol reaches terminator seq and leaves template strand of DNA
What is the transcriptional start site (TSS)? What is the difference between the TSS and the promoter? What does upstream and downstream from the TSS mean?
TSS: base position on DNA where transcription starts (represented by bend arrow)
TSS is located downstream of promoter→ +1
downstream→ bases are in gene seq and have (+) values
upstream→ bases are in promotor/regulatory seq and have (-) values
What is the difference between the template and coding strands? What is the association between the template and coding strands with the RNA strand during transcription?
template: DNA strand transcribed by RNA→ complementary and antiparallel to RNA
coding: DNA strand not transcribed by RNA→ same seq and 5’ to 3’ orientation as RNA
What is the difference between the RNA polymerase core and RNA polymerase holoenzyme in prokaryotic transcription?
RNA pol core→ five subunits + RNA pol.
RNA pol holoenzyme→ six subunits (sigma factor)+ RNA pol.
What is the function of the sigma factor in prokaryotic transcription? What are functions of the -35 and -10 sequences in prokaryotic transcription?
sigma factor: binds to core and is responsible for finding promoter seq in DNA
**different sigma factors are used to recognize different promoters
-35 and -10 sequences on promoter are recognized by sigma factor→ after recognition RNA pol goes from loose to tight bind to DNA
**-10 seq is AT-rich so pol can separate strand
What is abortive initiation? What is the cause of abortive initiation?
abortive initiation: binding and release of short RNA strands from DNA at early stages of transcription
formation of ~10 phosphodiester bonds
elongation complex forms and continues until termination
why? instability of the Initial RNA-DNA Hybrid (weak H bonds)
Describe the similarities and differences between prokaryotic and eukaryotic transcription.
same?
RNA pol have similar structure and function
RNA pol binds to template strand of DNA- makes one RNA strand in 5’ to 3’ direction
different?
euk genes have more regulatory seq
euk regulatory seq are often far away from promoter
nucleosomes include transcription
euk cells have more RNA pols
euk RNA pol require more proteins for initiation
euk promoters are more complicated
How many RNA polymerases are used by eukaryotic cells? Describe the function of each eukaryotic RNA polymerase. Do they recognize the same promoter sequences or different promoter sequences?
Euk have 3 RNA polymerases
RNA pol I: produces most of rRNA in cell, produce large rRNA but small #
RNA pol III: produces all tRNA and some rRNA, produce small tRNA but high #
RNA pol II: produces all mRNA, microRNA, and some noncoding RNAs
recognition of promoter? transcription factors are used to initiate transcription ~sigma factor
Need to know the function of the following proteins during transcription: TFIID, TBP, TAFs, TFIIH, and mediator.
TFIID:binds TATA box in promoter using subunit called TATA-binding protein (TBP)
TBP→ binds TATA box near -30 position
binding bends DNA allowing other TF to bind promoter
TBP-associated factor (TAF): subunit of protein that recognized promoter and histone mods— recruit TFIID to promoters w/ out TATA box
TFIIH: final recruitment to promoter to form pre-initiation complex→ RNA pol II + 5 TF
Mediator: protein complex that mediates interactions b/t regulatory TF, GTF, and RNA pol II
recruits RTF to promoter for RNA pol II regulation
regulates final phosphorylation step to initiate elongation
What is a primary RNA transcript (aka pre-mRNA)? What type/order of modifications occur on primary RNA transcripts?
pre-mRNA: initial mRNA made from transcription, needs to be processed before used for translation (where it is a mature RNA)
modification?
5’ cap added
RNA splicing
3’ poly A tail attached
after transcription? RNA editing
What is the 5’ cap and what are its functions? Why is the 5’ cap often depicted as an upside-down nucleotide? steps?
5’ cap: modified guanine base (methylated) added to the 5’ end of RNA when transcript is ~25 nucleotides long
functions? protect mRNA from degradation, contribute to nuclear export, initiation of translation, increase efficiency for RNA splicing
upside-down bond? bc 5’,5’ triphosphate bond
steps:
RNA triphotophase (RTPase) hydrolyzes phosphate from base at 5’ end in pre-mRNA
GTase hydrolyses GTP and attaches GMP to base at 5’ end with diphosphate
forms 5’,5’ triphosphate bond
guanynyl-7-methyltransferase attaches a methyl gp to N7 on guanine base→ created 7-methyl guanosine
additional methyl groups often added to 2’ hydroxyl gp
GTase dissociates from mRNA, cap-binding complex (CBC) binds to 5’ cap
5’ cap is binding site for CBC
What is 3’ polyadenylation? What is the function of 3’ polyadenylation? steps?
3’ end modification of mRNA where repated adenines are added
function? protection from exoribonucleases
**polyA proteins are bound to c-terminus domain of RNA pol II
steps?
RNA pol II transcribes polyA signal (AAUUAA), and polyA proteins bind to signal
proteins recruit endonuclease to cleave RNA strand after polyA signal
polyA polymerase (PAP) adds many adenines to cleaved end (w/ out template)
polyA binding proteins (PABPs) bind to polyadenylation and protect 3’ end from degradation
What is the difference between exons and introns?
exons: DNA segments in euk gene used to make protein
intron: DNA segment in euk gene not used to make protein
What is RNA splicing? What is a spliceosome? What are small nuclear ribonucleoproteins (snRNPs) and snRNAs?
splicing: introns (and some exons) are removed from pre-mRNA using spliceosome
spliceosome: large RNA-protein complex responsible for carrying out RNA splicing
snRNP: RNA-protetin complexes that are critical for removing introns from pre-mRNA
they contain snRNA which is uses base-paring to identify seq (exon/intron) boundaries needed for splicing
Identify the consensus sequences that are important for splicing. Why are consensus sequences in an intron important for RNA splicing?
consensus seq indicate to spliceosome where to make two cuts:
5' → GU
branch pt→ A
3’ → AG
Describe the three steps and two reactions in RNA splicing.
snRNP bind to consensus sequences (5’ splicing site and branch point); form spliceosome position sites
rxn 1: 5’ splice site binds to branch site to form lariat and cuts away from exon 1 (branch point has 3 phosphate bonds: 2, 3, and 5)
rxn 2: 3’ OH on exon 1 creates phosphodiester bond to link exons and the lariat is free
each rxn needs OH and phosphate
What is alternative splicing? Is alternative splicing common for human genes?
alternative splicing: form of RNA splicing that allows a single gene to produce multiple proteins
70% of human genes undergo alt splicing
Describe the different types of alternative splicing discussed in class: exon skipping, intron retention, alternative 3’ splice site, and alternative 5’ splice site.
exon skipping: exon is cut out of pre-mRNA to make slightly different protein
intron retention: introns is left in mature mRNA to make slightly different protein
alternative 3’ splice sites: new 3’ splice site is used when cut
alternative 5’ splice sites: new 5’ splice site is used when cut
What is the difference between constitutive and alternative exons?
constitutive exons: exons necessary for protein function
alternative exons: exons unnecessary for protein function
What are splice site mutations? Why do splice site mutations cause disease? What are some examples of splice site mutations?
splice site mutations: change in DNA seq that alters function of spliceosome during RNA splicing
→ can promote intron retention and exon skipping
→ common cause of disease (~15% of point mutations causing disease are splice site mutations)
What is RNA editing? How does ADAR perform RNA editing? How does cytidine deaminase perform RNA editing?
RNA editing: changes to mature RNA seq without changing genomic DNA
function? genetic flexibility
some RNA undergo RNA editing co-transcriptionally → before or after RNA splicing
ADAR: base pair differs: A/T and I/C → change codon (I acts as G)
cytidine deaminase: converts cytosine to uracil through deamination→ change codon
What are some differences between constitutive and regulated genes? Provide some examples of both.
constitutive→ always expressed at same rate (aka housekeeping genes)
ex. rRNA or GTF genes
regulated→ genes expressed in response to change (depend on needs of the cell)
ex. metabolism genes like lactose genes
Describe the differences between general transcription factors and regulatory transcription factors.
GTF: binds promoter and recruit RNA pol to template strand (require all 5)
RTF (activator/repressor): bind regulatory sequences and regulate rate of transcription by influencing ability of RNA pol to transcribe specific gene→ allows differential gene expression
Describe three different strategies used by cells to regulate expression of genes.
transcriptional control: regulate activity of RNA polymerase (control amount of RNA)
translational control: regulate activity of ribosome and RNA stability (control amount of protein)
post-translational control: regulate protein activity using chemical mods on aa (control protein function)
Activators vs repressors. What are they? How do they regulate gene expression? How are they the same? How are they different?
both: RTF (bind reg sequences on DNA) and always expressed
activators bind enhancers→ increase rate of tranxn
promote RNA pol phosphorylation
accelerate TFIID binding DNA
repressors bind silencers→ decrease rate tranxn
masks sequence
hinder binding
block RNA pol phosphorylation
What are effectors? How are they used to regulate activators and repressors?
→ molecules that bind regulatory proteins to change function
effector=change in environment
activator (+) regulation:
gene usually on→ effector arrive→ impair ability of activator to bind DNA→ turn gene expression off
gene usually off→ effector arrive→ enhance ability of activator to bind DNA→ turn expression on
repressor (-) regulation:
gene usually off→ effector arrive→ impair ability of repressor to bind DNA→ turn gene expression on
gene usually on→ effector arrive→ enhance ability of repressor to bind DNA→ turn expression off
What is the difference between the major and minor grooves in the double helix? Which is used for transcription factor binding? Why is the groove preferred by transcription factors?
major vs minor groove? differ due to availability for non-covalent interactions
TF interact w/ major groove bc there is more non-covalent interactions
→ more hydrogen bonds available in the major groove compared to the minor groove; providing a much better discrimination between bases
What are regulatory sequences? Why are they often inverted repeats?
regulatory sequences: DNA sequences that RTF bind (enhancer or silencer)
inverted repeats signal binding of dimeric TF bc they bind more stably and specifically to DNA
What is an operon? What is a polycistronic RNA?
operon→ cluster of prok genes controlled by one promoter
polycistronic RNA→ product of tranxn of an operon, a mRNA that has more than one gene
**one polycistronic RNA can make multiple proteins
What is combinatorial control? Explain why it is used to regulate a high number of genes in different cell types.
→ genes use common regulatory proteins, but each gene requires a specific combination of reg proteins to activate or inhibit expression
same TF can be used in different complexes to regulate different genes→ multiprotein complexes formed
What is the transcriptional ground state of cells? Compare euk and prok TGS.
→ inherent activity of promoter without regulatory mechanism
prokaryotic TGS: “on”→ no proteins/chromatin in DNA→ genetic info is open
eukaryotic TGS: “off”→ due to chromatin structure
Why are many regulatory transcription factors homodimers? Why is it advantageous for eukaryotic cells to use transcription factors that are heterodimers?
homodimer→ one protein with two identical subunits: provide more protein-DNA interactions at regulatory sequences
heterodimers→ one protein with two different subunits: allow euk cells to create many functional tranxn factors from small number of individual proteins
What are epigenetic modifications? How are they similar to DNA mutations? How are they different?
epigenetic mods? chemical mods to DNA and histones that regulate tranxn
same? heritable + alter gene expression + cause disease
different? epigenetic mod do not alter DNA sequence while DNA mutations do. DNA mutations can be repaired but mods cannot be corrected.
Compare DNA methylation and histone modifications. How are they similar? How are they different?
same? influences gene expression
DNA methylation: chemical mod to DNA
Histone mod: chemical mod to histone **work with chromatin remodeling complexes
Are epigenetic modifications heritable? In other words, can they be maintained after DNA replication?
yes!
histone mods: some histone core subunits remain bound to DNA during replication, CRC used to recognize and maintain the modifications using bromodomain and HAT
methylation: DNMT localizes to replication fork during DNA replication and methylates CpG dinucleotides on daughter strand of DNA if CpG site on template strand is methylated
CpGs are symmetric
replication is semiconservative
Describe two ways DNA methylation can regulate expression of genes in eukaryotic cells.
prevents binding of GTF to a promoter
no GTF=no RNA pol binding= no tranxn initiation
prevents binding of RTF to regulatory sequence
no activator bound to enhancer= tranxn decrease
Explain what is meant by the phrase “a human body has one genome but many epigenomes”?
one genome bc different cell types in humans have the same DNA sequence
many epigenomes bc different cell types have different DNA methylation patterns
What is the function of ribosomes within cells? What are the functions of the small and large subunits of ribosomes?
ribosome: large protein complex that synthesizes proteins using genetic information within mRNA
small subunit: matches tRNAs to codons on mRNA
large subunit: generates peptide bonds between amino acids
What are ribosomes made of? Proteins, RNA, or both? If they have RNA, is it in the small subunit, large subunit, or both subunits?
ribosome complex is made of many proteins and at least one ribosomal RNA (rRNA) in each subunit→ for both euk and prok
prok: 40:60 (protein:RNA)
large (2): 23S and 5S
small (1): 16S
euk: 50:50 (protein:RNA)
large (3): 28S, 5.8S, 5S
small (1): 18S
Describe the process of ribosome biogenesis for the small and large subunits in prokaryotes. Start at transcription of rRNA then explain how ribosomes are made with mature rRNAs and ribosomal proteins.
ribosome assembly occurs while rRNA is being transcribed by RNA polymerase
after assembly, rRNA is process
pre-rRNA transcript has bases modified by enzymes— mods allow folding
pre-rRNA transcript is cut by ribonuclease to liberate 16S, 23S, 5S rRNA
mature 30S and 50S subunits combine to make functional 70S ribosomes
Describe the process of ribosome biogenesis for the small and large subunits in eukaryotes. Start at transcription of rRNA then explain how ribosomes are made with mature rRNAs, ribosomal proteins, snoRNPs, and assembly factors. Make sure you explain how all three polymerases contribute to this process.
45S pre-RNA is transcribed in nucleolus and processed/modified by snoRNP to mature, 5S rRNA is made in nucleus, ribosomal proteins made by translation of mRNA in cytoplasm
5S rRNA and proteins are imported into nucleolus
small and large ribosomal proteins bind to rRNAs
90S ribosome forms in nucleolus→ contains small and large subunits, rRNAs, snoRNPs, and assembly factors
inactive complex ^ imported to cytoplasm where 40S and 60S become functional subunits and can perform protein synthesis→ release assembly factors
**ribosomal biogenesis ends in the cytoplasm
Compare codons and anticodons.
codons: sequence of three nucleotides in DNA or RNA that correspond to a single amino acid during protein synthesis
**three letter= one amino acid
start→ AUG
stop→ UGA, UAA, UAG
anticodon: three base sequence in tRNA that is complementary to a codon in mRNA
What are wobble bases? Describe their location. How do cells use the wobble base to make the genetic code degenerate?
wobble bases→ base in codon and anticodon that do not always follow base-pair rules (more flexible bp)
examples?
tRNA w/ G or U in 5’ position can recognize two codons
tRNA with I (inosine) in wobble position can recognize three codons
contribute to degenerate? allow multiple base pairing to code for one amino acid
Describe at least one advantage for having a degenerate genetic code.
cells require fewer tRNAs→ greater efficiency and use of resources
silent mutation increase→ cells are able to absorb more mutations
Why do some amino acids require multiple tRNAs to recognize all codons?
amino acids with more then 3 different codons need more then one tRNA bc this maxes out the wobble base flexibility (with inosine)
ex. codon families like Val or Ser (6 codons)
What is the difference between the transcriptional start site, the start codon, and untranslated regions? Describe their locations.
TSS: first base that is transcribed @ DNA
start codon: first amino acid translated @ mRNA
UTR: space between TSS and start codon that does not code for protein @ mRNA
What is an open reading frame?
→ long segment of DNA nucleotides in a gene sequence with no stop codons signaling a region that is translated
**statistical anomaly→ if mRNA is translated with the wrong reading frame, a stop codon is statically likely to arise
What is the function of aminoacyl tRNA synthetases? Describe how these proteins perform their function in two steps.
ARS→ protein that attaches amino acids to 3’ end of mature tRNA in cytosol
how?
ATP and aa bind to ARS, ATP is used to link aa to AMP forming aminoacyl-AMP
tRNA binds ARS, aa is transferred from AMP to tRNA to form aminoacyl-tRNA→ tRNA is now charged
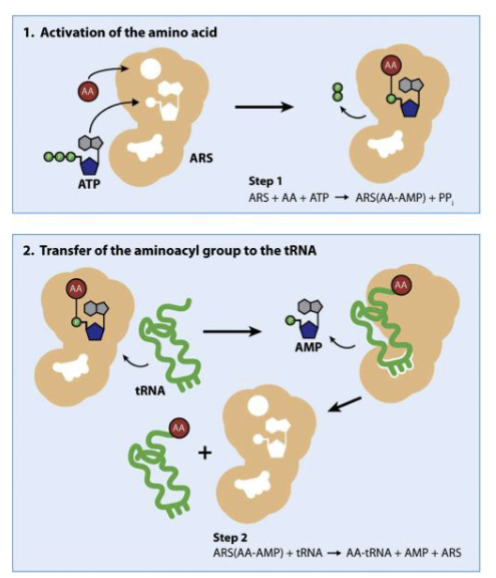
What is the initiation complex in translation? Describe the sequence of events that leads to the formation of the initiation complex in prok.
initiation complex: ribosome + mRNA + initiator tRNA form a complex around start codon to initiate translation
recognition of mRNA
binding of small subunit to mRNA
initiator tRNA recruited to P site
large subunit joins
Identify the three sites in a ribosome that are used for translation and describe the function of each site.
Aminoacyl site: binding site for charged aminoacyl tRNA
Peptidyl site: site where polypeptide chain is attached to peptidyl tRNA
Exit site: exit site for uncharged tRNA
Identify and describe the three stages of translation used by prokaryotes and eukaryotes.
Initiation: ribosomal subunits, mRNA, and initiator tRNA form initiation complex at start codon— reading frame established, initiator tRNA @ P site,
Elongation: ribosome moves down mRNA and adds amino acids to growing chain by forming peptide bonds— charged tRNAs enter A site and add amino acid to chain
Termination: ribosome reaches stop
What is a Shine-Dalgarno sequence? What is its function? Where is it located?
shine-dalgarno sequence→ consensus sequence on 5’ end of prok mRNA that binds to 16S rRNA in small subunit of ribosome
function? this is how the small subunit binds mRNA in prok cells
What is the initiator tRNA? Why does methionine have two tRNAS bur only one codon?
initiator tRNA: binds start codon at initiation @ P site
elongator tRNA: codes for methionine @ A site
Describe how three different initiation factors promote initiation of translation in prokaryotic cells. Why is GTP hydrolysis important for the initiation stage?
IF-1 binds small subunit and blocks tRNA binding
IF-3 binds small subunit and prevents premature activation of ribosome
**rRNA binds Shine-Dalgarno
IF-2 (GTP-binding protein) w/ GTP attached recruits initiator tRNA to start codon in P site of small subunit
**GTP hydrolyzed by ribosome→ IF released, large subunit binds, initiation complex formed
Describe the three steps of elongation. Why is GTP hydrolysis important for the elongation stage?
aminoacyl-tRNA binds to A site in ribosome using base-pairing between anticodon and codon
aminoacyl-tRNA binds EF-Tu w/ GTP attached→ delivers tRNA to A site and only hydrolyzes if correct bp match
EF-T is a GEF for EF-Tu, attaches GTP so it can bind another tRNA
peptide bond formed bt amino acid in P and A sites→ growing polypeptide transferred to A site
peptidyl transferase rxn @ large subunit
translocation shifts tRNA without amino acid to E site, tRNA w/ polypeptide to P site, and new codon in A site
EF-G-GTP (mimics EF-Tu-aminoacyl tRNA) binds A site and hydrolyzes to EF-G-GDP which promotes translocation
cycle goes on and on!
What are the two goals of termination? How do prokaryotic cells accomplish each goal? Why is GTP hydrolysis important for the termination stage?
release of polypeptide
release factors (RF) work to promotes release of polypeptide chain→ 1st RF release polypeptide and 2nd RF uses GTP hydrolysis to removed 1st RF from A site
dissociation (and recycling) of subunits, mRNA, and tRNA
ribosome recycling factors (RRF) has same shape as tRNA and fit A site with stop codon→ recruit EF-G-GTP to A site
translocation through hydrolysis→ RRF @ P site and IF-3 binds small subunit to break ribosome
Describe how the beginning of initiation in eukaryotes is similar to prokaryotic cells. How is it different?
same?
initiation factors separated and prevent tRNA binding to A site
establish reading frame with start codon
different?
euk 43S pre-initiation complex forms prior to mRNA bindings
recruitment of mRNA: Shine-Dalgarno sequence vs recognition of the 5' cap
IF vs eIF
initiator tRNA w/ formyl gp vs without
Compare non-coding RNA (ncRNA) and coding RNA.
ncRNA: transcribed from the genomic DNA (like mRNA) but is not translated into protein
interact with DNA/RNA to regulate gene expression in euk cells
detect and degrade viral DNA in prok cells
~80% of expressed genes
coding RNA: transcribed and translated genomic DNA
Compare and contrast long non-coding and small non-coding RNAs. How are they similar? How are they different?
lncRNA: longer than 200 nucleotides and are often used to localize proteins to specific locations on DNA→ bind DNA/RNA using protein & bp interaction
microRNA: shorter than 200 nucleotides and typically bind to mRNA to influence gene expression→ bind DNA/RNA using complementary base pairing
What is HOTAIR? How does HOTAIR regulate gene expression? How many genes does HOTAIR regulate?
lncRNA that recruits histone-modifying enzymes to their target genes to inhibit gene expression→ secondary structure + bp abilities
**regulates ~10 genes on chromosome 2
H3K27me3 (repressive mark addition)
H3K4 demethylation (active mark removal)
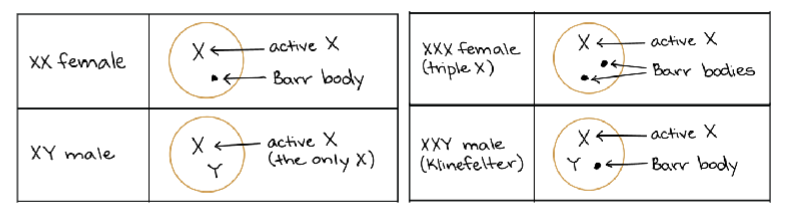
What is X-inactivation? What is a Barr body? What is Xist?
x-inactivation ia s dosage compensation mechanism in females to only express one x chromosome
express Xist gene and produce X-ist lncRNA to condense one x chromosome→ form a barr body via histone mods (epigenetic effect)
heterozygosity at x chromosome can lead to somatic mosaicism
Describe why individuals with Turner, Klinefelter, and Triple X syndromes have altered expression from the X chromosome.
deviation from XX or XY causes several health issues
Turner (XO): missing X chromosome (reduced expression of X-linked genes—no escapies
Klinefelter (XXY): x-inactivation shuts doesn’t extra chromosome—incorrect expression bc escapies
Triple X (XXX): x-inactivation shuts down two x chromosome—incorrect expression bc too many escapies
What is DNA fluorescence in situ hybridization (DNA FISH)? What is the purpose of performing DNA FISH?
DNA FISH: lab technique to visualize specific DNA sequence or entire chromosome within a cell
purpose? allows scientist to detect and diagnose genetic diseases
collect cells arrested (usually @ metaphase) and fixed to side
denature DNA and add probe w/ fluorescence to hybridize
stain and analyze with microscope
What is RNA FISH?
RNA FISH: lab technique allows visualization of specific mRNAs within a cells
collect cells arrested (usually @ interphase) and fixed to side
add many probe w/ fluorescence to hybridize
stain and analyze with microscope
What is RNA interference (RNAi)? Describe how RNAi regulates gene expression.
RNAi: dsRNA processed by cell to induce sequence-specific gene silencing
→ translational control
miRNA: natural
siRNA: experimentally
function^? use base pair to identify mRNA and either degrade mRNA (w/ nuclease) or halt translation
What are sRNAs? What is a riboswitch?
sRNA: regulatory RNA (50-500 nucl) that bind mRNA to regulate gene expression in prok
riboswitch: structure in 5’UTR in mRNA that binds to small molecules allowing regulation of gene expression
**sRNA bind to riboswitches to promote (positive regulation) or inhibit (negative regulation) translation
The CRISPR Cas system uses two non-coding RNAs and different Cas proteins. Describe how the CRISPR-Cas system works in prokaryotic cells and explain the role of non-coding RNAs and Cas proteins in this process.
adaptation phase: bacteria integrates viral DNA into chromosome
Cas1/Cas2 cut up viral DNA
expression phase: bacteria express crRNA (integrated viral DNA), tracrRNA, and Cas9 proteim
tracrRNA (scaffold) bind crRNAs (guide) using complementary base pairing
interference phase: crRNA detects second viral infection
tracrRNA-crRNA complex binds Cas9 (a nuclease)
crRNA recognized viral DNA and Cas9 destroys it
What is DNA sequencing (aka Sanger sequencing)?
→ determining the sequence of bases for a fragment of DNA
sequencing=replication in a tube
What are the critical components of a DNA sequencing reaction? What is the function of each component?
components:
template strand of DNA: desired sequence
one sequencing primer: short DNA strand that is complementary to template and primes for SNA synthesis (provides 3’OH)
nucleotides (dNTP and ddNTP): add to new DNA strand
DNA polymerase: connect nucleotides to make a new DNA strand
What is the human genome project? How long did it take? How much did it cost?
human genome project: international collaboration to sequence entire human genome using DNA sequencing technology
→ product was the reference sequence: high quality genome sequence
13 years
$3 billion dollars