FUNBIO 12: Genetic information flow: From DNA to RNA (Transcription, RNA polymerases, RNA processing)
1/15
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
16 Terms
Introduction question: what makes neutron cells different than muscle cells?
Key Concept: Different cells express different proteins, which is why muscle cells and neurons function differently, despite having the same DNA.
Muscle Cells: Produce motor proteins for movement.
Neurons: Produce proteins for conducting electrical impulses.
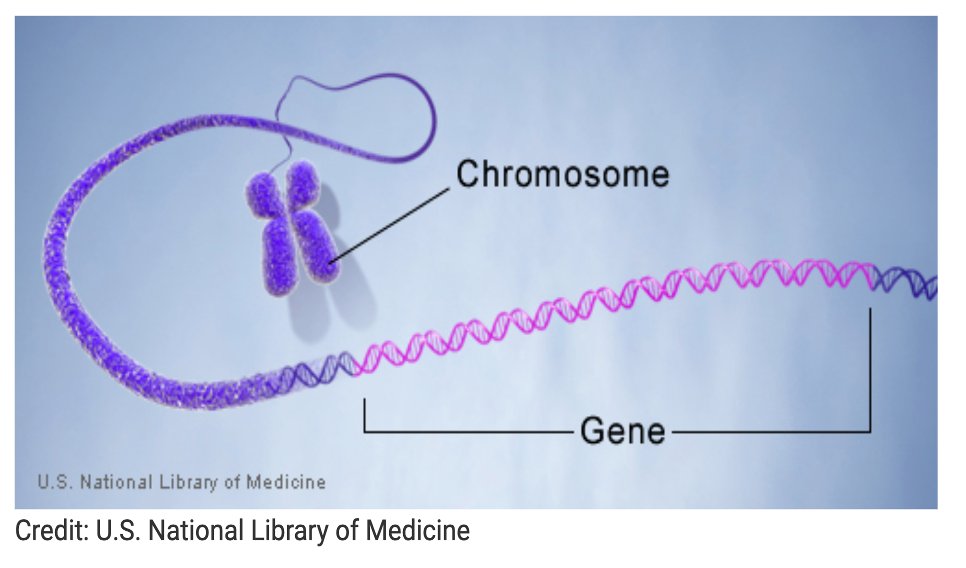
What is a gene?
Gene: The basic physical and functional unit of heredity, made of DNA sequences, and many genes code for proteins.
Human Genome: Estimated 20,000–25,000 genes (HUGO Project).
Genes vary in size from a few hundred to over 2 million DNA bases.
Define gene expression
Gene expression is the process by which the inheritable information in a gene, such as the DNA sequence, is made into a functional gene product, such as a protein
Gene expression can be detected by:
Gene expression can be detected by many molecular techniques
e.g.
-PCR &Real Time PCR
-Gel Electrophoresis and southern blotting
-Microarrays (as shown)
-FISH in situ hybridization (functional expression)
Gene expression can be promoted or impeded at….. (AO1/AO2)
1. DNA or Chromatin Modifications
DNA Methylation: Adding chemical groups (methyl) to DNA can silence genes by preventing transcription.
Histone Modifications: DNA wraps around proteins called histones. Changes to histones can either loosen or tighten this wrapping. Looser wrapping allows genes to be expressed, while tighter wrapping silences them.
Chromatin Remodeling: Enzymes can adjust how DNA is packed, making certain genes more or less accessible for transcription.
2. Transcription Regulation
Transcription Factors: These proteins bind to DNA to either help or block RNA polymerase from copying the gene into RNA.
Enhancers/Silencers: DNA regions that either boost or suppress transcription by interacting with transcription factors.
3. Post-Transcriptional Modifications
Splicing: Introns (non-coding sections) are removed, and exons (coding sections) are joined. Alternative splicing can produce different versions of proteins from the same gene.
5’ Capping and 3’ Polyadenylation: A protective cap and tail are added to the mRNA to prevent degradation and assist in translation.
4. RNA Transport and mRNA Degradation
mRNA Export: Only fully processed mRNA is exported from the nucleus to the cytoplasm for translation.
mRNA Stability: The lifespan of mRNA is regulated. Some mRNAs last longer and produce more protein, while others are degraded quickly to stop production.
5. Translation Regulation
Translation Control: Proteins control how often ribosomes start translating mRNA into proteins. Some RNAs (like microRNAs) can block translation or degrade the mRNA to stop it from being made into protein.
6. Post-Translational Modifications
Protein Folding: Proteins must fold into their correct shape to function.
Chemical Modifications: Proteins can be modified (e.g., adding phosphate groups) to change their activity, stability, or function.
Protein Degradation: Unneeded or damaged proteins are broken down to prevent buildup.
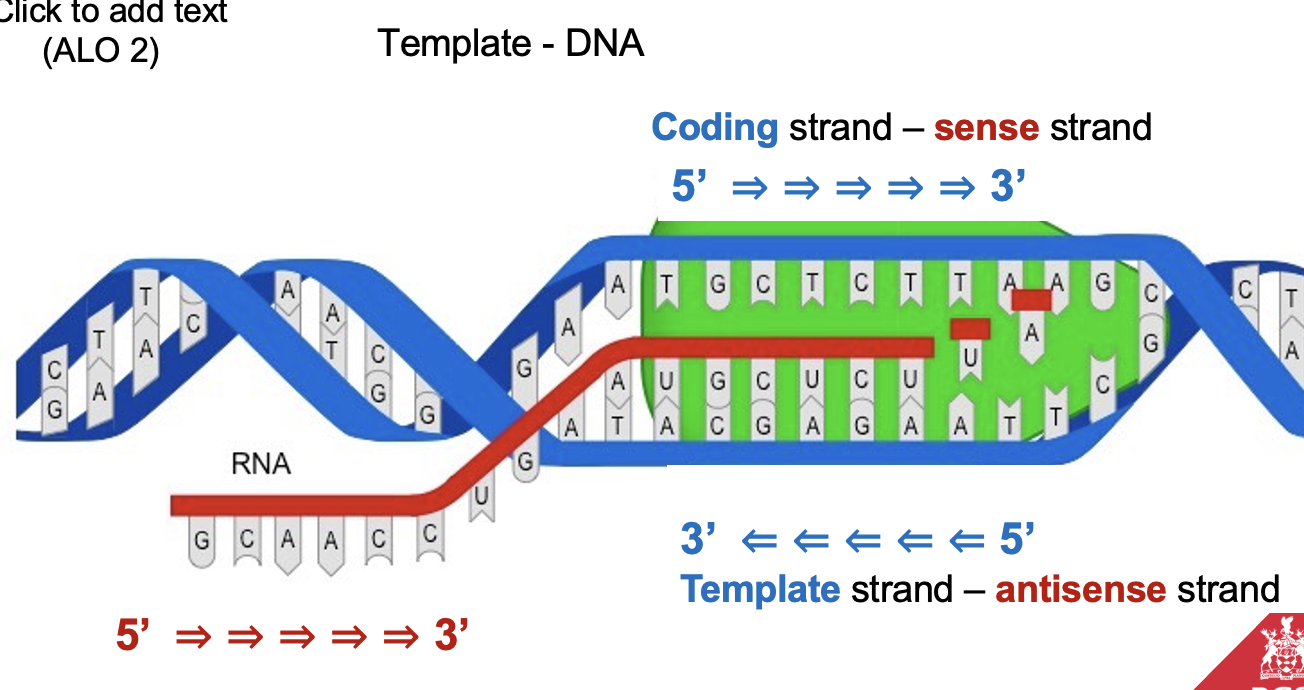
Discuss in detail how DNA is transcribed into RNA (ALO2)
Transcription process:
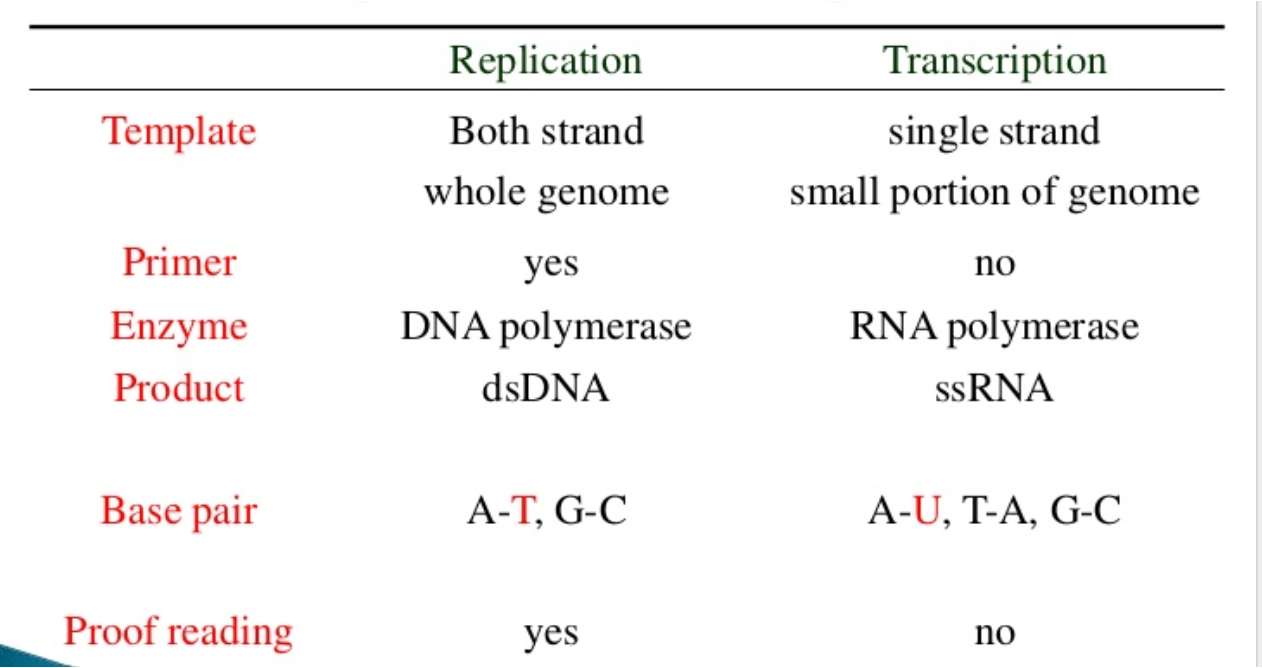
Primer: RNA polymerase doesn’t need a primer to start RNA synthesis.
Initiation
RNA polymerase (RNAP) binds to the promoter region in the 3’ to 5’ direction, unwinds DNA at a 17-18 base pair segment of the promoter, and initiates RNA synthesis.
Elongation
The RNAP moves along the template strand synthesizing RNA and adds nucleotides (in the 5’ to 3’ direction of the RNA chain) until it reaches the terminator region
Mg²⁺ Ions: They act as catalysts, helping RNA polymerase add ribonucleotides to form the RNA strand during transcription.
Termination
The terminator sequence is a specific sequence of nucleotides in the DNA that signals the end of transcription. When RNA polymerase (RNAP) reaches this sequence, it triggers the enzyme to stop or pause, and eventually dissociate (detach) from the DNA template.
What strands are involved in transcription? (ALO2)
Template Strand: The DNA strand used for transcription (antisense strand).
Coding Strand: The DNA strand with the same sequence as the mRNA (sense strand).
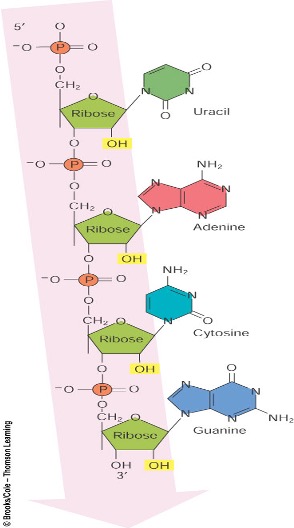
Describe the structure of RNA (ALO3)
RNA is a single-stranded molecule made of ribonucleotides joined by 3’ to 5’ phosphodiester bond
It has pentose sugar (ribose), a phosphate group, and a nitrogenous base (Adenine, Uracil, Cytosine, Guanine).
Conformation: Usually single-stranded (ssRNA)
What are the different types of RNA present in a cell (ALO3)
mRNA (messenger RNA): Carries the genetic code from DNA to ribosomes for protein synthesis.
tRNA (transfer RNA): Brings amino acids to the ribosome and matches them to the codons in mRNA.
rRNA (ribosomal RNA): Forms the core of the ribosome and catalyzes protein synthesis.
snRNA (small nuclear RNA): Involved in splicing and modifying pre-mRNA.
snoRNA (small nucleolar RNA): Involved in modifying rRNA and other RNA molecules.
ncRNA (non-coding RNA): RNA molecules that are not translated into proteins but have structural, regulatory, or catalytic roles.
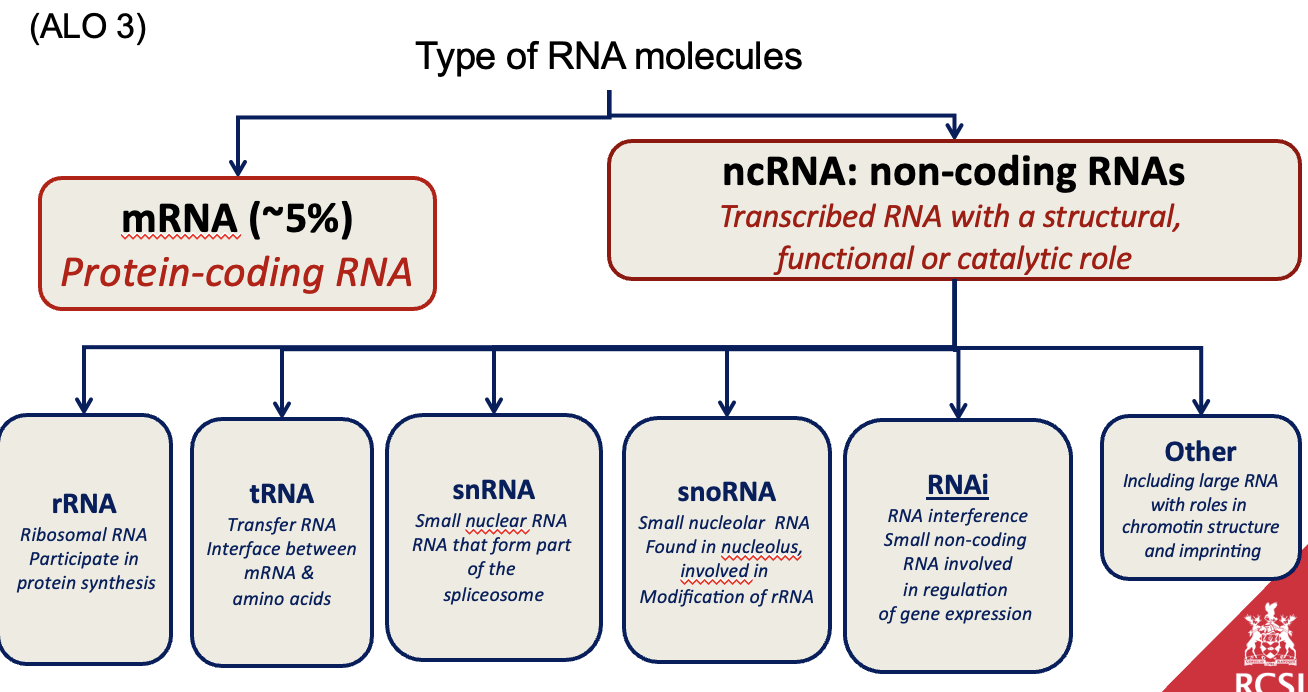
What are the RNA subtypes used in protein synthesis (ALO3)
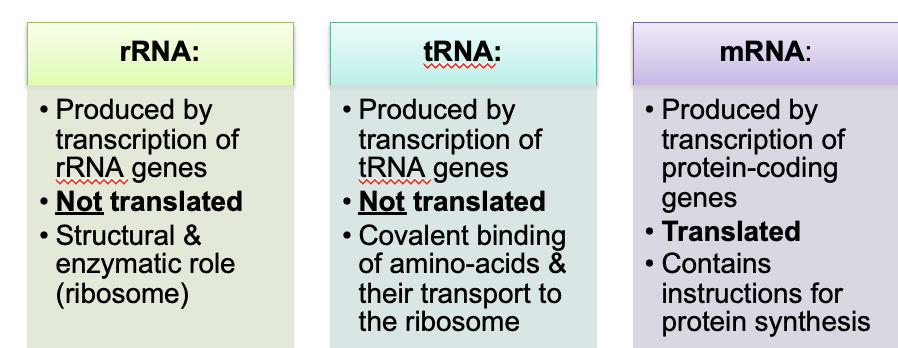
What is the difference between replication and transcription?
Explain the role of specialized enzymes in the transcription process. (ALO5)
RNA Polymerase: The main enzyme responsible for synthesizing RNA from the DNA template.
Although the structures of all three polymerase enzymes are homologous, they perform different functions.
RNA Polymerase I: Transcribes rRNA and is located in the nucleolus.
RNA Polymerase II: Transcribes mRNA and is located in the nucleus.
RNA Polymerase III: Transcribes tRNA and some rRNA, and is located in the nucleus.
Transcription Factors: Proteins that regulate gene expression by bind to specific base sequences in the promoter. They act as activators or repressers by facilitating or blocking the binding of RNA polymerase which will either initiate or inhibit transcription. (TF help turn on and off genes)
Helix-turn-helix (e.g. Oct-1)
helix-loop-helix (e.g. E2A),
zinc finger (e.g. glucocorticoid receptors, GATA proteins)
basic protein-leucine zipper [cyclic AMP response element binding factor (CREB), activator protein-1 (AP-1)
β-sheet motifs [e.g. nuclear factor-κB (NF-κB)
Helicase: Unwinds the DNA double helix to allow RNA polymerase to access the template strand.


Discuss post transcriptional RNA modifications: capping (ALO7)
Before the transcribed mRNA leaves the nucleus for translation in the cytoplasm, it undergoes three main processing steps:
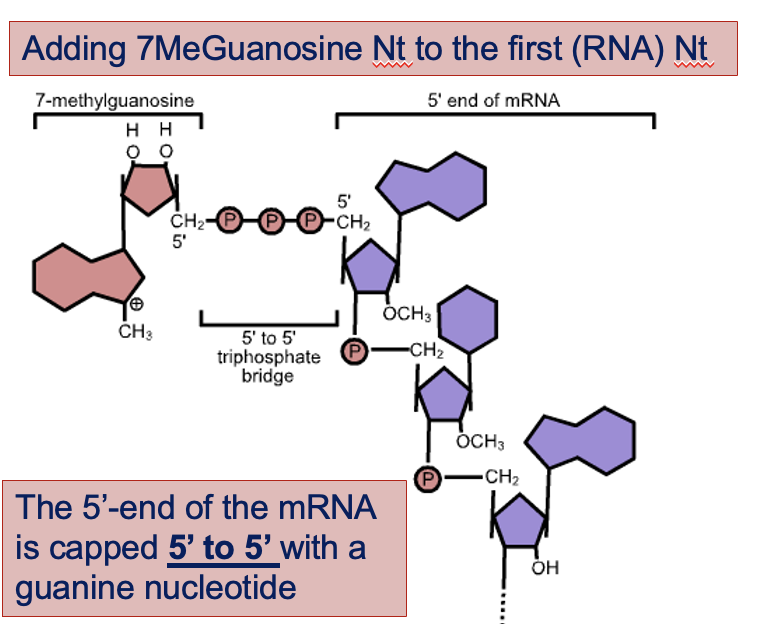
Capping
The 5' end of the mRNA is modified by the addition of a modified guanine nucleotide, known as the 5' cap.
This cap is crucial for:
Protecting the mRNA from degradation by exonucleases.
Essential for the ribosome to bind to the 5’ end of the mRNA
Enhanced translation
It helps ribosomes recognize and bind to the mRNA efficiently, ensuring that translation begins correctly at the start codon.
Enhanced transport from nucleus
Proteins that recognize the cap structure help transport the mRNA through the nuclear pore complexes, ensuring it reaches the cytoplasm where translation occurs. (Important for gene expression)
Enhances splicing of first intron for some mRNAs
Splicing factors recognize the cap and assist in the splicing process.
Promotes the recruitment of the spliceosome.
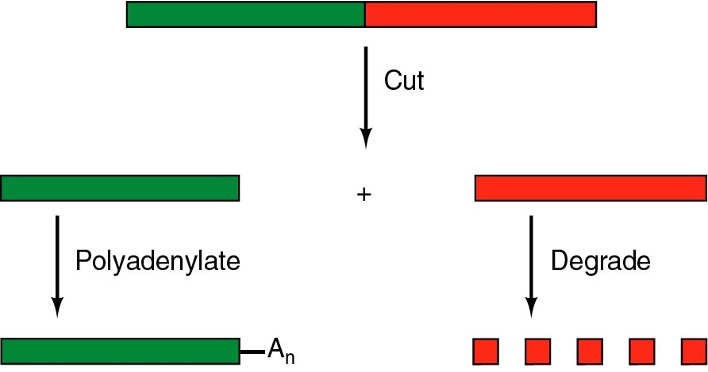
Discuss post transcriptional RNA modifications: Cleavage and Polyadenylation (ALO7)
The 3' end of the mRNA receives a poly-A tail, which is a long chain of adenine nucleotides (typically 50 to 250 adenines)
Most cytoplasmic mRNAs have a polyA tail (3’ end) of 50-250 Adenylates.
A notable exception is histone mRNAs
Added post-transcriptionally by an enzyme, polyA polymerase(s)
Turns over (recycles) in cytoplasm
This is crucial for:
Providing protection from protease degradation (degradation enzyme)
Promotes mRNA stability
Deadenylation (shortening of the polyA tail) can trigger rapid degradation of the mRNA
Enhances translation
Recruitment by Ribosomes: The poly-A tail helps ribosomes recognize and bind to the mRNA, facilitating the initiation of translation.
Poly-A Binding Protein (PAB1): In the cytoplasm, a protein called PAB1 binds to the poly-A tail, stabilizing the mRNA and helping recruit ribosomes for translation.
Synergistic Stimulation with the Cap: The combination of the 5' cap and the poly-A tail works together to increase the efficiency of translation. Both modifications are crucial for ribosome binding and effective protein synthesis.
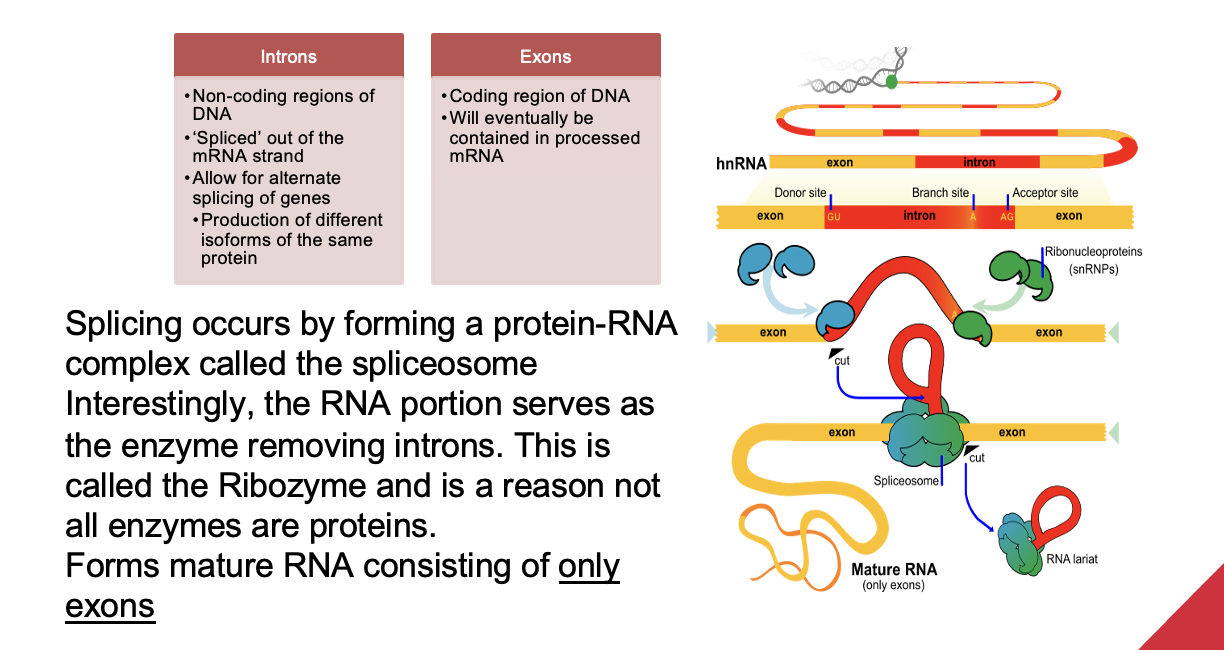
Discuss post transcriptional RNA modifications: RNA Splicing (ALO7)
Splicing occurs by forming a protein-RNA complex called the spliceosome
The RNA portion serves as the enzyme removing introns. This is called the Ribozyme and is a reason not all enzymes are proteins.
Forms mature RNA consisting of only exons
Heterogeneous nuclear RNA (hnRNA) contains both exons (coding regions) and introns (non-coding regions).
Spliceosomes, made up of small nuclear RNAs (snRNAs) and proteins, remove introns at 3’ end and join the exons together.
Introns are then degraded / broken down by the cell.
They play a role in allowing alternate splicing, which lets a single gene produce different protein versions (isoforms).
After splicing, the mature mRNA consists only of exons.