BIOL 3000 DNA Barcoding
1/25
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
26 Terms
Linnaean Taxonomy System
Based on morphological characteristics of individuals. Classical taxonomy falls short in cataloging biodiversity.
Kingdom Phylum Class Order Family Genus Species
DNA Barcode
Unique pattern of DNA sequence that identifies each living thing
Dr. Paul Hebert
“Father of DNA Barcode”
Help with “Hard-to-Identify” specimens
A new tool allowing non-experts to identify specimens
Limitations of Traditional Taxonomy Approach
Phenotypic plasticity employed for species level identification can lead to incorrect IDS
Morphologically cryptic taxa can be overlooked
Morphological keys are often effective only in certain life stages
Expertise is required for sample analysis and identification
What are we looking for in a DNA Barcode?
Universal, differentiating, and size
Universal
It has to be easily identifiable in all species
Differentiating
Variable enough to provide a unique sequence for each species
Size
Long enough to provide differential sequence but short enough to sequence quickly, efficiently and cheaply
How many base positions?
600-700 base pairs, so 15 positions and there are 4 alternate characters for each position
Which gene?
Mitochondrial genome
Mitochondrial genome
Plentiful mitochondria in the cell
Lacks introns
Limited recombination because haploid
Mix of highly variable and conserved regions
Mitochondrial genes
Only codes for 13 individual genes, energy production (ETC). Can only use any gene of the ETC, but settled on the Cytochrome C Oxidase (COX)
Cytochrome C Oxidase (COX)
Robust primers
Increased phylogenetic signal
How does DNA Barcoding work?
Sample collection
DNA amplification
Sequencing and Analysis
Sample collection
Effective DNA barcoding depends on the quality of the biological material
Animals: freeze whole specimens at -20C, fixation in 95% pure Ethanol
Plants: Silica gel
Ancient DNA: Depends on age of samples and source used for DNA extraction
DNA Amplification
Polymerase Chain Reaction (PCR)
Polymerase Chain Reaction (PCR)
Molecular biology technique used to amplify a small amount of DNA several orders of magnitude
What is used in PCR?
Template DNA, specific primers (sense and antisense), DNA polymerase (Taq polymerase), Di-nucleotides (dNTPs of A, T, C, and G), buffer system, and cations (Mg+2, K+2)
Taq polymerase
Isolated from bacteria that lives in hot environments
Steps of PCR
Denaturation (94-96°C): Hydrogen bonds tend to break first
Annealing (65-68°C)
Elongation (72-74°C): Turns one strand into two. Exponential amplification of DNA fragment of interest
Sequencing and Analysis
Sanger Sequencing
Sanger Sequencing
Using purified DNA template, specific primer, Taq polymerase, Di-nucleotides (dNTPs of A, T, C, and G), labeled di-deoxynucleotides (ddNTPs of A, T, C, and G), buffer system, and cations (Mg+2, K+2)
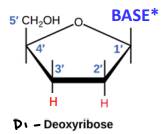
Di-deoxyribonucleotides
Cannot add anything to the 3’ carbon
Steps of Sanger Sequencing
Denaturation (94-96°C)
Annealing (65-68°C)
Elongation (72-74°C): Adds every base at line until at random it will grab a di-deoxyribose which will stop elongation.
Why barcoding?
Works with DNA fragments
Works with all stages of life
Allows look-a-likes and mimics to be identified
Reduces ambiguities in identification
Makes expertise go further, doesn’t require much
Speeds up determination of the Encyclopedia of Life
How is DNA Barcoding used?
Species Identification
Consumer Protection
Health related issues
Identify ancient specimen