MICR5831 L8: Protein Transport and Folding 8/6/25
1/94
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
95 Terms
How do proteins self-fold without requiring extra energy from the cell? (slide 4)
1) Hydrophobic collapse
2) Secondary structures create foci for more flexible sections
3) Molten globule forms, side chains create final “native conformation”
Which chaperone is required to associate with a ribosome to initiate self-folding? How does it work? (slide 6)
Trigger Factor (TF)
-Binds to the ribosome's exit tunnel
-Interacts with the nascent polypeptide chain as it emerges
-Helps to prevent aggregation and promote proper folding.
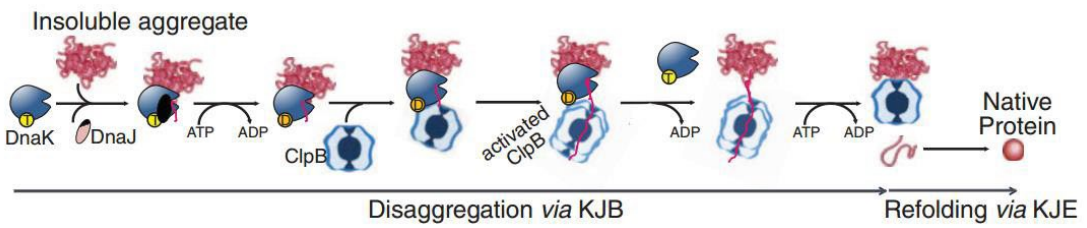
What are the chaperones and steps in the protein Disaggregation Pathway? (slide 7)
Include:
1) DnaK (Hsp70) and DnaJ (Hsp40) complex
3) ClpB foldase (Hsp100)
1) DnaK (Hsp70) clamps onto insoluble aggregates, recruits DnaJ (Hsp40)
2) DnaK/J use ATP to refold a short nascent peptide
3) ClpB foldase (aka Hsp100) binds/recognizes peptide
4) DnaK/J complex is released, ClpB uses ATP to continue with re-folding
5) ClpB releases native protein
What macromolecular machine is required for protein refolding?
What are the components of this machine? (slide 8)
GroEL/GroES Chaperonin Machine:
1) GroEL Ring: Cylindrical structure, 7 subunits stacked on top of each other
2) GroES Cap: Also has 7 subunits
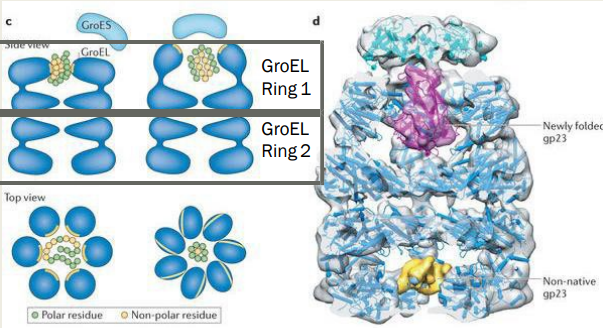
How does the GroES/EL chaperone result in protein refolding? (slide 9)
1) Misfolded proteins bind hydrophobic face of GroEL Ring
2) GroES cap on top changes conformation of cylinder
3) Hydrophilic face presents towards the center
4) 7 ATP molecules bind to the GroEL ring and triggers conformational change, expose hydrophilic patches
5) Hydrophobic patches interact with/unfold misfolded protein
6) 14 ADP + GroES is released from GroEl Ring 2
7) ATP + GroES bind to GroEl Ring 1, shifts are repeated
True or False: During the GroES/GroEL refolding pathway, at any given time one of the rings has ATP + GroES bound and the other ring is empty
True
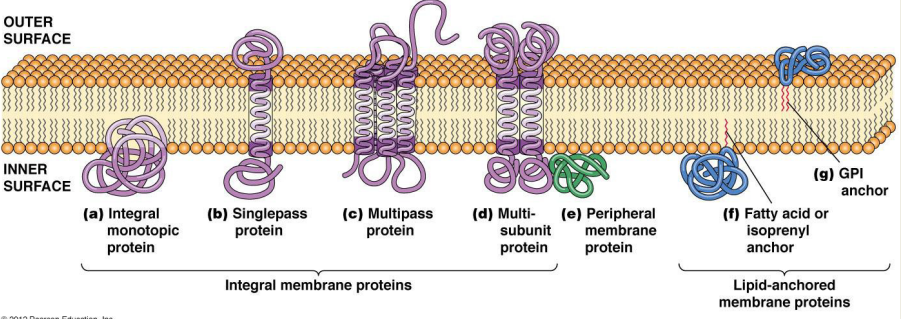
What are the common characteristics of integral membrane proteins? (slide 11)
Include:
1) Single/Multipass
2) Integral Monotropic
3) Peripheral Membrane
4) Lipid Anchored
1) Single/Multipass Proteins: Hydrophobic domains that sit inside the phospholipid bilayer (alpha helices)
2) Integral Monotropic Proteins: Hydrophobic domain that strongly associates with lipids, does not cross the membrane
3) Peripheral Membrane Proteins: Hydrophilic surface binds → hydrophilic headgroups of phospholipids
4) Lipid Anchored Proteins: Lipid anchor at one end (isoprenyl in bacteria)
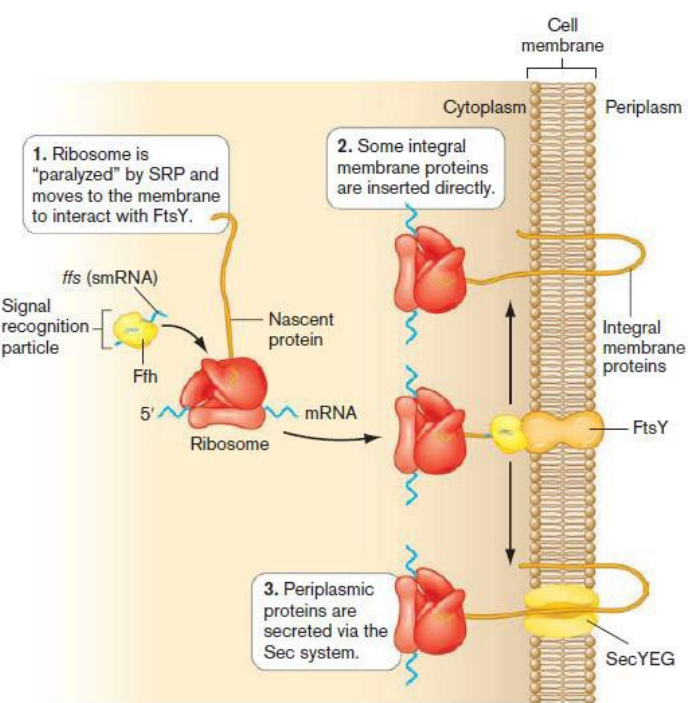
Describe the steps in the signal recognition particle (SRP) pathway for protein transport
Include:
1) Ffh protein + sRNA complex (binds signal)
2) FtsY protein (targets proteins)
3) Sec System (secretes)
1) Proteins have a hydrophobic signal sequence at the Nterminus → destined for inner membrane
2) Ffh protein + sRNA complex bind to signal sequences emerging from ribosome
3) Ffh protein + sRNA complex binds to the FtsY protein in the cytoplasmic membrane
4) Integral membrane protein is directly inserted into the membrane as it is translated
5) Periplasmic protein is completely synthesized and FtsY carries it to the Sec system for secretion
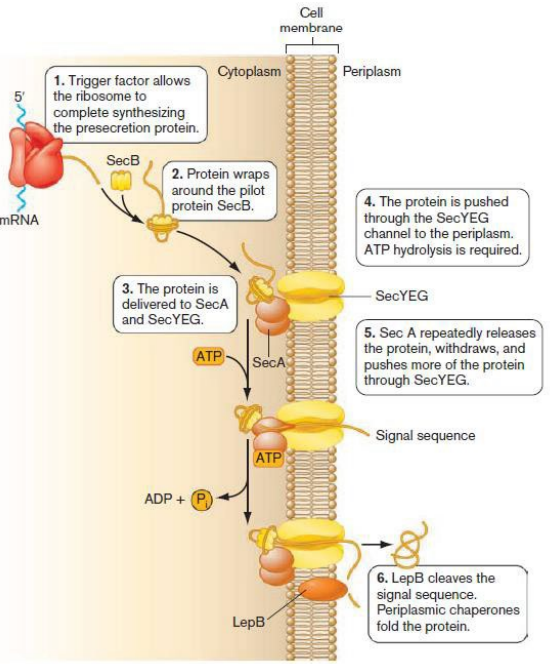
Describe the steps in the Sec-dependent protein transport pathway for perisplasmic bacterial proteins. (slide 13)
Include:
1) SecB piloting protein
2) SecA ATPase
3) SecYEG channel/pore protein in inner membrane
4) LepB signal peptidase
1) Protein is completely translated in the cytoplasm
2) SecB captures unfolded protein and delivers it to to SecA
3) SecA forms complex + SecYEG channel protein
4) SecA breaks down ATP and pushes protein → SECYEG pore → periplasm
4) LepB signal peptidase removes protein signal sequence
5) Proteins released into the periplasm, folded by chaperones
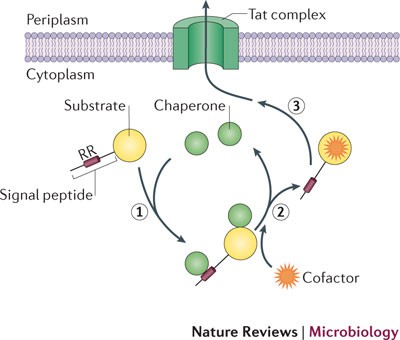
Describe the steps in the Twin Arginine Transport (TAT) pathway for bacterial proteins (slide 14)
Include:
1) Tat A/B/C (Tat Complex)
2) Signal peptidase
4) PMF
1) Protein is fully translated and folded in the cytoplasm and is tagged for association with TatA/B/C
2) First loop of the protein is transported into the periplasm
3) Signal peptidase (SPase) cleaves the signal sequence
4) TatA/B/C uses the proton motive force (PMF) to flip the folded protein into the periplasm
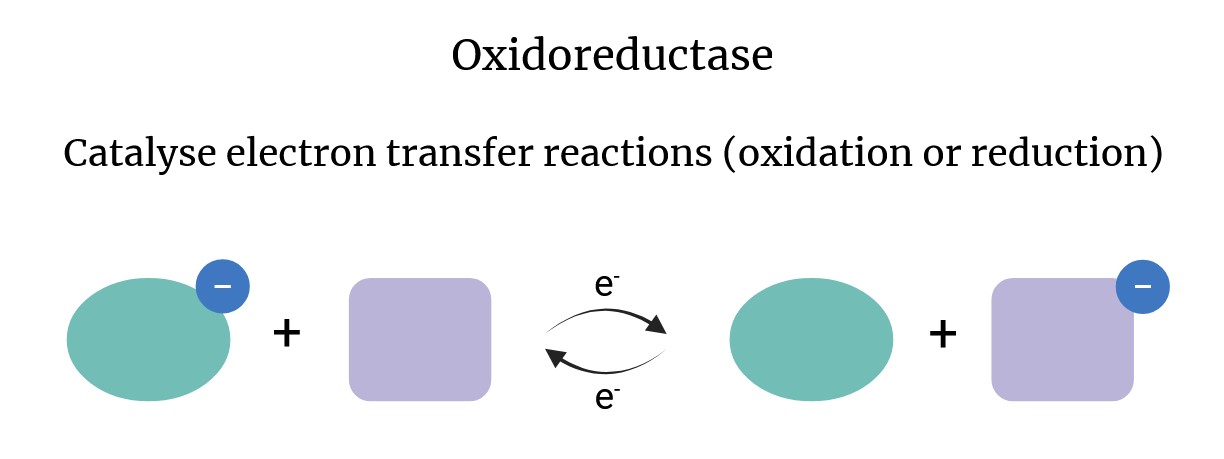
What is the role of Oxidoreductases such as DsbA in bacterial protein folding?
How do they operate? (slide 15)
-Introduce Disulphide bonds into the correct position
– DsbA is oxidized + gets a disulphide bond from DsbB
– DsbA donates the disulphide bond to a protein
– DsbA is reduced and is re-oxidsed by DsbB
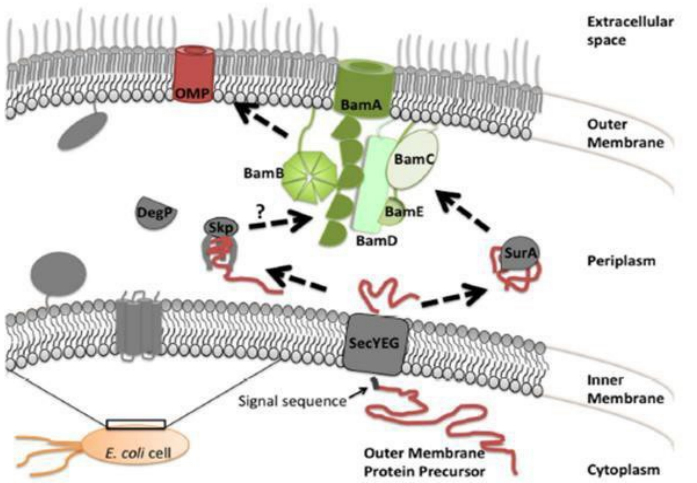
How are Outer Membrane Proteins exported to the outer membrane of Gram Negative bacteria? (slide 16)
Include:
1) Sec translocon (transporter)
2) SurA (isomerase)
3) Skp (isomerase
4) Bam complex (channel)
1) OMPs are translated in unfolded state in cytoplasm
2) Signal sequence is guided to the Sec transporter
3) SurA and Skp isomerases bind/fold the protein
4) SurA and Skp guide OMP across the periplasm to Bam complex
4) Bam complex flips OMP into the Outer Membrane
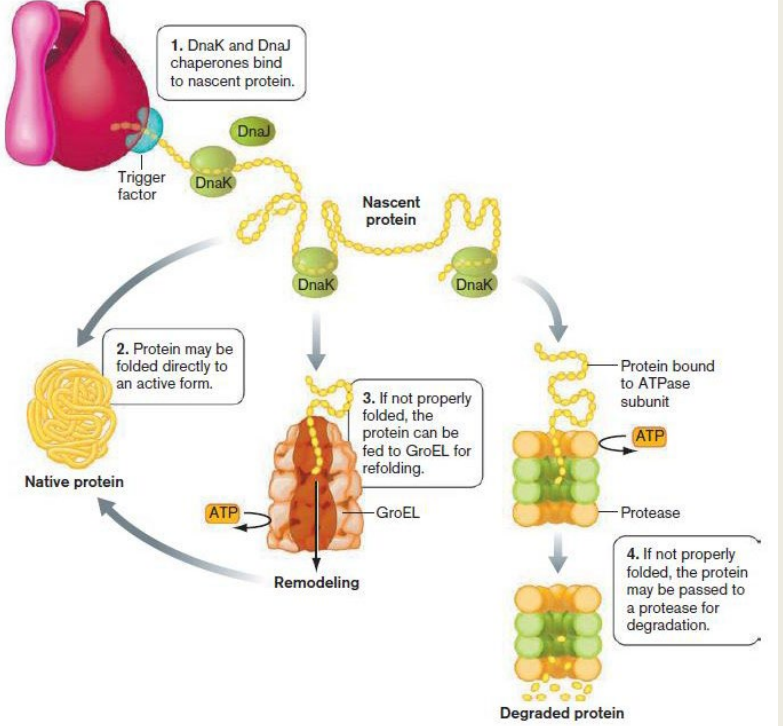
Describe the components of the protein Degradation Pathway and how the pathway works (slide 20)
Include:
1) ClpP (ATP-dependent endoprotease)
1) Clp contains a proteolytic core made of two homoheptameric rings of the protein ClpP
2) Associates with a hexameric ATPase cap
3) Uses ATP to unfold and digest the protein
True or False: Many proteins do not require extra energy input to fold by themselves
True
True or False: The intracellular environment of the cytoplasm is mostly empty space with a few proteins
False, it is very crowded (300-400 mg/mL protein)
True or False: When they are synthesized, most proteins will be unfolded/inactive and made incorrect contacts with other proteins
True
What determines the 3D structure of a protein?
-Linear sequence of amino acid residues
-Hierarchical folding pattern
What is the first step of a hierarchical protein folding pattern?
Hydrophobic Collapse:
-Snapping into a compact globular structure
-Get hydrophobic residues away from water
-Formation of secondary structural elements
What happens after this step during protein folding?
1) Hydrophobic collapse to get away from water
-Secondary structures form in milliseconds
-Foci for flexible sections to naturally fold around
-Molten globule forms
What happens after this step during protein folding?
2) Secondary structures form foci for flexible sections to fold around and create molten globules
Side chains undergo movement to attain the final "native conformation"
True or False: Protein folding does not require any extra energy in the form of ATP from the cell
True
True or False: Native conformation of most proteins requires the most energy to maintain and therefore is unstable
False
True or False: Being in a misfolded conformation is a high energy state yet proteins can become stuck in it
True
What do you use to rescue misfolded proteins?
Chaperones
What is this?
-Chaperones
-Associate with target proteins then dissociate after target protein is completely folded
-Originally identified as proteins produced in response to cellular stress
What are some examples of cellular stress that might cause proteins to unfold?
-Temperature/pH changes
-Exposes hydrophobic amino acids which clump together and form aggregates
What happens to 70% of proteins after they are released from a ribosome?
Fold themselves into low energy native conformation state
What is this?
-Trigger Factor (TF)
-Peptidyl-prolyl cis-trans isomerase (PPlase)
-Binds to proteins emerging from the ribosome
-Speeds up proline isomerization from trans to the cis
-Does not require ATP
True or False: Using Trigger Factor (TF) to speed up proline isomerization (trans to cis) requires ATP for energy
False
True or False: If a protein unfolds due to heat stress, a chaperone is required to refold it
True
What are some chaperones that exist in the cytoplasm and are used by 30% of proteins to reach native conformation?
1) DnaK/DnaJ complex
2) GroES/EL complex
What are some chaperones that exist in the periplasm and are used by 30% of proteins to reach native conformation?
1) PPlases
2) Oxidoreductases
Describe the Disaggregation pathway
1) DnaK (Hsp70) clamps onto insoluble aggregates, recruits DnaJ
2) DnaK + DnaJ use ATP to refold short nascent peptide
3) Nascent peptide is recognized by ClpB foldase
4) ClpB binds the protein and DnaK/DnaJ complex is released
5) ClpB uses ATP to continue re-folding
6) Native protein is released from ClpB
7) Medium proteins with a complex structure
What are the first 3 steps of the Disaggregation pathway?
1) DnaK (Hsp70) clamps onto insoluble aggregates, recruits DnaJ
2) DnaK and DnaJ use ATP to refold short nascent peptide
3) Nascent peptide is recognised by ClpB foldase
What are the last 4 steps of the Disaggregation pathway?
4) ClpB binds the protein and DnaK/DnaJ complex is released
5) ClpB uses ATP to continue re-folding
6) Native protein is released from ClpB
7) Medium proteins with a complex structure
What is this? (Disaggregation pathway)
-ClpB (Hsp100)
-Foldase that uses ATP to refold proteins
-Recognizes nascent peptide and binds to it
-Releases DNaK/DnaJ complex before refolding
What is this? (Disaggregation pathway)
-DnaK (Hsp70)
-Found in cytoplasm
-Clamps onto insoluble aggregates
-Recruits DnaJ (Hsp40)
-Uses ATP to refold short nascent peptide
-Released when ClpB binds to complex
What is this? (Disaggregation pathway)
-DnaJ
-Found in cytoplasm
-Recruited by DnaK (Hsp70)
-Uses ATP to refold short nascent peptide
-Released when ClpB binds to complex
What type of proteins can use the DnaK, DnaJ, and ClpB Disaggregation pathway?
Medium length proteins w/ complex structure
What is different about the Disaggregation pathway with ClpB for very long proteins with complex tertiary structures?
-ClpB releases partially folded protein
-Protein enters GroES/EL complex to complete folding
Describe the Refolding pathway
1) Misfolded proteins bind to the exposed hydrophobic face of the GroEL ring
2) GroES cap is attached to the top
3) Cylinder conformational change with hydrophilic face towards the center
4) 7 ATP molecules bind to the GroEL ring, triggers conformational change, exposes hydrophobic patches that unfold misfolded protein.
5) 14 ADP + GroES is released from Ring 2 and ATP + GroES bind to Ring 1, the shifts are repeated
6) At any given time one ring has ATP + GroES bound and the other is empty
What are the first 3 steps of the Refolding pathway?
1) Misfolded proteins bind to the exposed hydrophobic face of the GroEL ring
2) GroES cap is attached to the top
3) Cylinder undergoes conformational change with hydrophilic face towards the center
What are the last 3 steps of the Refolding pathway?
4) 7 ATP bind to the GroEL ring and trigger conformational change, exposes hydrophobic patches that unfold misfolded protein.
5) 14 ADP + GroES is released from Ring 2 and ATP + GroES bind to Ring 1, shifts are repeated
6) At any given time one ring has ATP + GroES bound and the other ring is empty
What is this?
-GroEL-GroES Chaperonin Machine
-Nanocage for protein folding
-Partially folded proteins will leave ClpB to enter it if they are too long and complex for the disaggregation pathway
What is this?
-GroEL
-Hydrophobic cylindrical structure
-7 subunits in 2 stacked rings
-Misfolded proteins attach to exposed face
-7 ATP bind to it after conformational change
-Exposes more hydrophobic patches to fix misfolded protein
What is this?
-GroES
-7 subunits that form a cap attached to top
-Binds to ring, causing conformational change
-Released with 14 ADP from Ring 2
-Binds with ATP to Ring 1
How does the GroEL Ring 1 help unfold a misfolded protein?
-7 ATP bind and trigger conformational change
-Subunits twist and tilt
-Expose hydrophobic patches that can unfold the misfolded protein
True or False: At any given time during the refolding pathway, one of the GroEL rings will have ATP + GroES bound to it but the other ring will be completely empty
True
What are the 3 pathways for protein selection and transport to the inner membrane/periplasm?
1) Signal Recognition Particle (SRP) co-translational pathway
2) Sec-dependent general secretion pathway
3) Twin arginine transport (TAT) pathway
What are these selection/transportation pathways best used for?
1) Signal Recognition Particle (SRP)
2) Sec-dependent general secretion
3) Twin arginine transport (TAT) pathway
1) Integral membrane proteins
2) Unfolded proteins into periplasm
3) Folded proteins into the periplasm
What are these selection/transportation pathway do these proteins use?
1) Integral membrane proteins
2) Unfolded proteins → periplasm
3) Folded proteins → periplasm
1) Signal Recognition Particle (SRP)
2) Sec-dependent general secretion
3) Twin arginine transport (TAT) pathway
What enzymes are responsible for folding incomplete proteins that use the Sec-dependent general secretion to enter the periplasm?
1) PPlases
2) Oxidoreductases
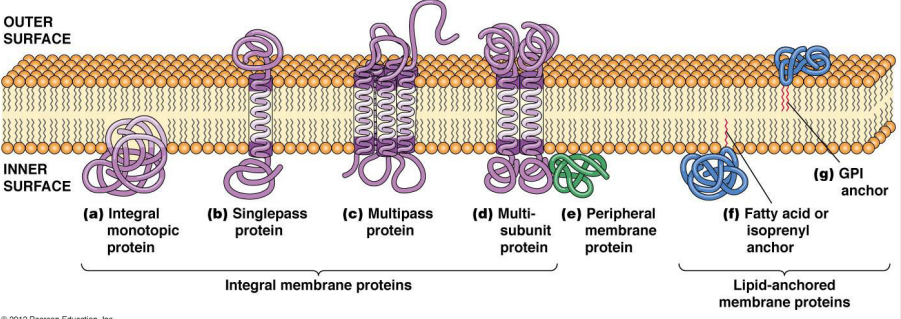
What special structural features could these integral membrane proteins have?
1) Single/Multipass Integral Membrane proteins
2) Integral Monotropic proteins
1) Hydrophobic domains that sit inside the phospholipid bilayer (alpha helices)
2) Hydrophobic domain that strongly associates with the lipids but does not cross the membrane
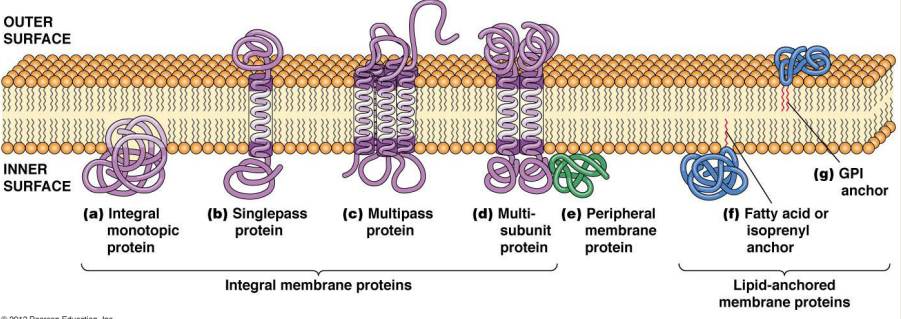
What special structural features could these integral membrane proteins have?
1) Peripheral Membrane proteins
2) Lipid Anchored proteins
1) Hydrophilic surface that binds to the hydrophilic headgroups of phospholipids
2) Tagged w/ lipid anchor (isoprenol) at one end
What happens during SRP Co-translational pathway?
1) Inner membrane proteins have a 15-30 amino acid hydrophobic sequence at the Nterminus
2) Signal sequences emerge from the Ribosome
3) Ffh protein + sRNA binds to signal sequence
4) FtsY protein binds to complex in the cytoplasmic membrane
5) Protein is either directly inserted/translated into membrane or sent to Sec system via FtsY protein
How do you tell if proteins are destined for the Inner Membrane?
-Signal sequence of 15-30 amino acids
-Hydrophobic sequence at the N-terminus
What binds to signal sequences of inner membrane proteins as they emerge from the Ribosome?
-Ffh protein
-Complexed with sRNA
What is this? (SRP Pathway)
-Ffh protein
-Forms complex with sRNA
-Binds to hydrophobic signal sequences for inner membrane proteins emerging from Ribosome
What happens to the Ffh + sRNA protein complex that binds to a inner membrane protein signal sequence emerging from a ribosome?
Binds to FtsY protein in cytoplasmic membrane
What is this? (SRP Pathway)
-Protein found in cytoplasmic membrane
-Binds to the Ffh/sRNA protein complex for signal sequences
-Can carry protein to Sec system if protein is completely synthesized
FtsY
What happens after a SRP signal sequence complex binds to FtsY in the cytoplasmic membrane?
1) Protein is directly inserted into the membrane as it is translated
2) Protein is completely synthesized and FtsY carries it to the Sec system for secretion
True or False: Protein translation occurs completely in the cytoplasm
True
What happens during Sec-dependent export to the periplasm?
1) Trigger Factor (TF) allows ribosome to complete synthesizing the presecretion protein.
2) Protein wraps around the pilot protein SecB.
3) The protein is delivered to SecA and SecYEG.
4) The protein is pushed through the SecYEG channel to the periplasm using ATP hydrolysis
5) Sec A repeatedly releases the protein, withdraws, and pushes more of the protein through SecYEG
6) LepB cleaves the signal sequence. Periplasmic chaperones fold the protein.
What does Trigger Factor (TF) do in the Sec-dependent export to the periplasm?
Allows ribosome to complete presecretion protein synthesis
What is this? (Sec-dependent pathway)
-Piloting protein
-Captures unfolded protein after it’s completely translated
-Delivers unfolded protein to SecA
SecB
What is this? (Sec-dependent pathway)
-ATPase that uses ATP to push unfolded protein through SecYEG pore
-Receives unfolded protein from SecB
-Forms inner membrane channel complex with SecYEG
-Pushes protein into the periplasm
SecA
What is this? (Sec-dependent pathway)
-Forms channel complex with SecA across inner membrane
-Allows SecA to push unfolded proteins into periplasm
SecYEG
Where does SecA get the energy to push an unfolded protein through SecYEG pore into the periplasm?
ATP
How can proteins that are destined for the periplasm be distinguished?
-Cleavable signal sequence
-LepB signal peptidase removes it before release
What is this?
-Signal peptidase that removes the signal sequence from proteins destined for the periplasm (NOT the inner membrane)
LepB
What happens during Sec-dependent export after the signal sequence for a protein destined for the periplasm is removed by signal peptidase LepB?
Proteins are released into periplasm to be folded by periplasmic chapterones
What Sec-dependent chaperones are responsible for completion of protein folding in the periplasm?
1) Peptidyl prolyl cis trans isomerases
2) Oxidoreductases
What is this? (Periplasm pathway)
-Responsible for folding OMPs in periplasm
-Examples include SurA and Skp
-Similar function to cytoplasmic Trigger Factor
Peptidyl prolyl cis trans isomerases
What is this? (Periplasm pathway)
-Chaperone responsible for folding proteins in periplasm
-Normally has a disulfide bond with DsbB
-Introduces disulfide bonds between cysteines, is reduced
-Re-oxidized by DsbB
Oxidoreductases (DsbA)
What is this? (Periplasm pathway)
-Oxidizes DsBA in the inner membrane
-Re-oxidizes DsbB every time it becomes reduced from donating a disulfide bond
DsbB
What happens after an Oxidoreductase like DsbA introduces a disulfide bond between cysteines into the correct position?
-DsbA is reduced after donating the disulfide bond
-DsbA is reoxidized by DsbB
True or False: The periplasm is a strongly reductive environment
False
True or False: The periplasm is a strongly oxidizing environment, therefore cysteines will become easily form covalent disulfide bonds if close together
True
True or False: Disulfide bonds are not very stable, therefore if they form in the wrong place they can inactivate the protein
False, disulfide bonds can inactivate the protein in the wrong spot but are very stable
What feature do Outer Membrane proteins tend to have?
-Large beta barrel structures
-Huge protein that forms a pore
-Hydrophobic outside, hydrophilic core
What is the advantage of the OM beta barrel structure having a hydrophobic outside and hydrophilic core?
Hydrophobic Outside:
-Can sit in the phospholipid membrane
Hydrophilic Core:
-Can transport molecules in the aqueous phase
True or False: Outer Membrane Proteins (OMPs) are translated in a folded state in the cytoplasm
False
What happens after unfolded Outer Membrane proteins are translated in the cytoplasm?
Signal sequence is guided to Sec transporter
What happens after unfolded Outer Membrane proteins leave the Sec translocon?
-SurA and Skp bind to the protein and fold it
-Guide protein across periplasm to Bam complex
-Flip protein into Outer Membrane
What do these macromolecular secretion systems do?
1) RND pumps and ABC transporters
2) Type III and Type IV systems
1) Remove toxins
2) Inject proteins into plant/animal cells
How do you keep a low level of certain proteins?
-Degradation signals (degrons) dictate protein stability
-Each protein has a given half-life
What is this?
-Degron
Degradation signal that dictates protein stability
What is this?
-N terminal rule
Beginning amino acid of protein (N-terminus) is directly correlated → stability
-Arg, Lys, Phe: Short half-life (2 min)
-Asp, Cys, Gly: Long half-life (10 hours)
What half life would a protein beginning with these N-terminal amino acids have?
1) Arginine (Arg)
2) Lysine (Lys)
3) Phenylalanine (Phe)
Short (Two minutes)
What half life would a protein beginning with these N-terminal amino acids have?
1) Aspartic Acid (Asp)
2) Glutamic Acid (Glu)
3) Cysteine (Cys)
Long (10 hours)
What is this?
-Degrades abnormal proteins into smaller and smaller pieces
Proteases
What are the steps in the Proteasome degradation pathway?
1) DnaK and DnaJ chaperones bind to nascent protein.
2) Protein is folded directly to an active form.
3) If not folded, feed to GroEL for refolding.
4) If not folded, protein may be passed on to a protease for degradation.
What is this? (Proteasome degradation pathway)
-Contains proteolytic core
-Made of two homoheptameric rings of protein ClpP
Clp
What is this? (Proteasome degradation pathway)
-ATP dependent endoprotease
-Associates with hexameric ATPase cap
-Uses ATP to unfold and digest the protein
ClpP
What determines the optimal 3D structure of a protein?
Linear sequence of amino acid residues