IMED1002 - RNA and Genes (L14)
1/24
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
25 Terms
RNA
- second major form of nucleic acid in cells
- found in nucleus and cytoplasm
- some RNA has roles as intermediary in conveying and converting genetic info into functional proteins
RNA components
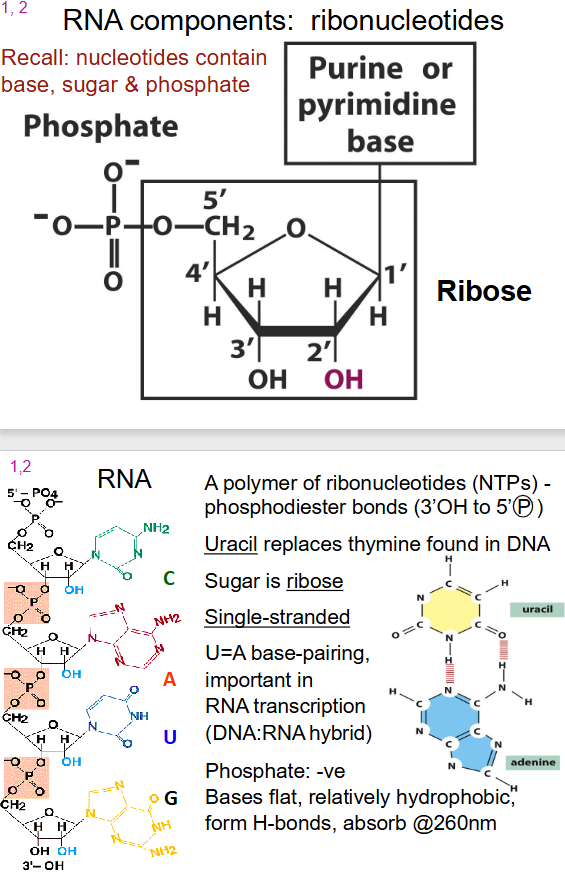
- ribonucleotides: contain base, sugar and phosphate
- purine or pyrimidine base
- phosphate
- ribose sugar
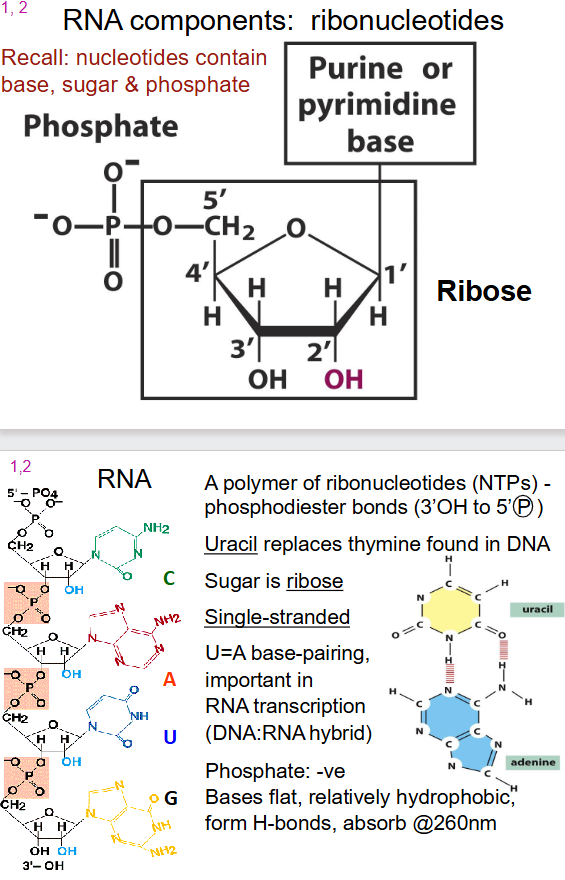
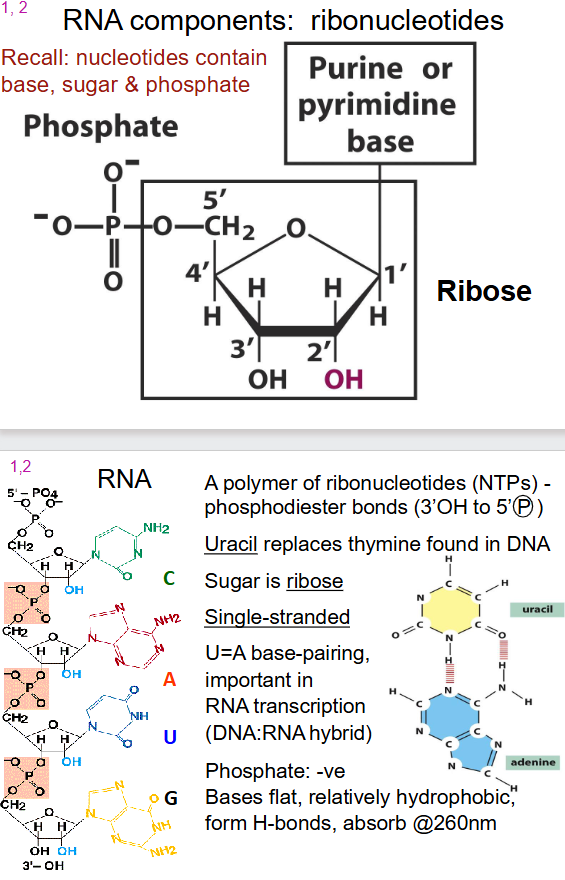
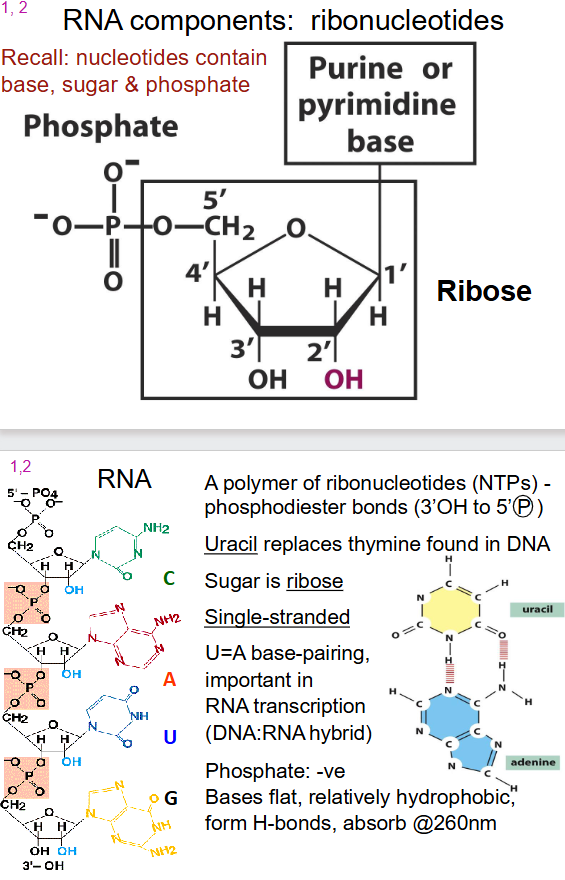
General Structure of RNA
- a polymer of ribonucleotides (NTPs) - phosphodiester bonds (3' OH to 5' (P))
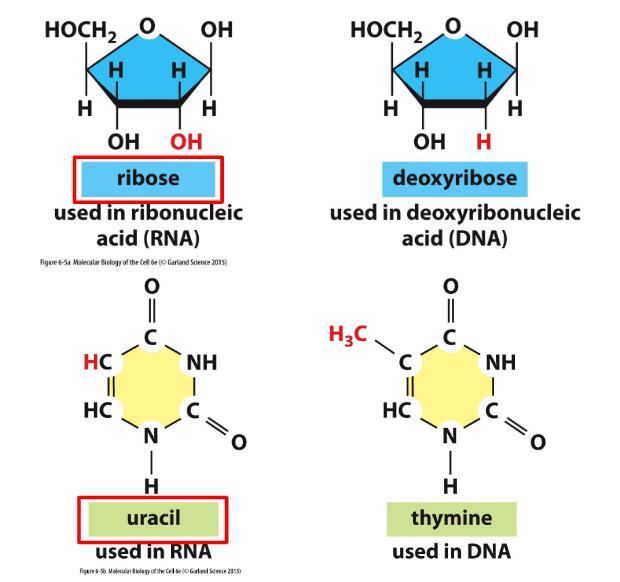
- Uracil instead of thymine
- Sugar is ribose
- single stranded
- U=A base pairing
- Phosphate: -ve
- Bases flat, relatively hydrophobic, form H-bonds, absorb at 260nm
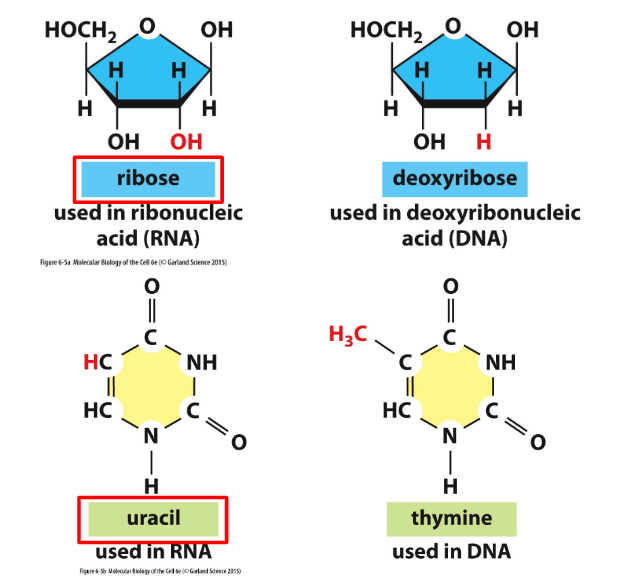
Key Differences between DNA and RNA
- ribose vs deoxyribose: extra hydroxyl group makes it a little more unstable.
- uracil vs thymine: uracil is demethylised version of thymine
RNA Primary Structure
- Sequence of Ribonucleotides from 5' to 3'
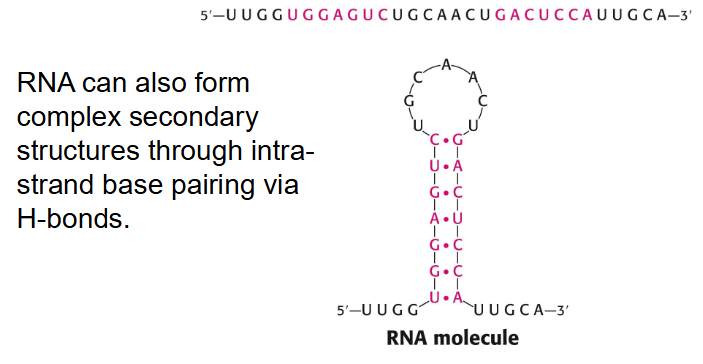
- RNA can also form complex secondary structures through intrastrand base pairing via H-bonds
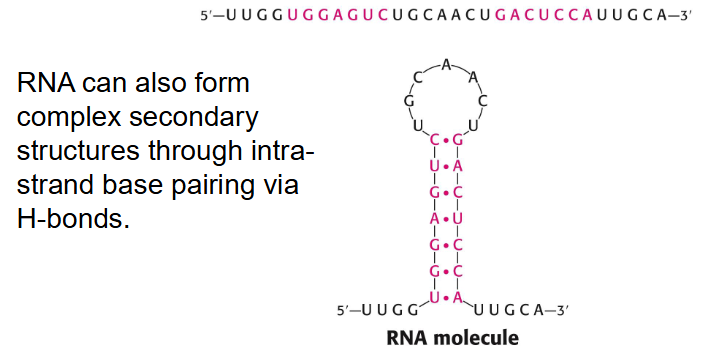
RNA Secondary Structure
Self-complementary sequences can lead to more complex and specific structures. RNA can base pair with complementary strands of DNA or RNA.
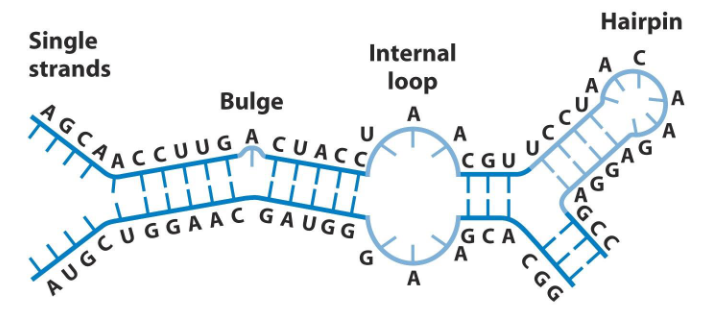
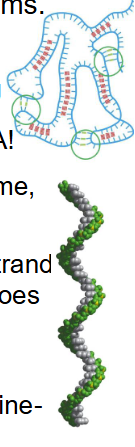
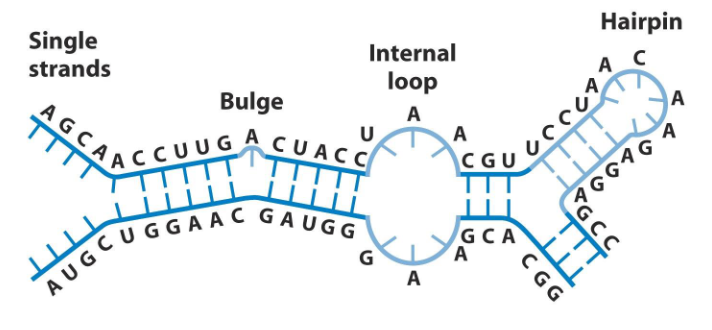
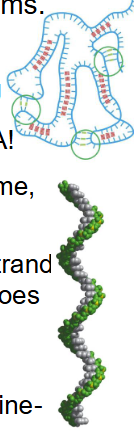
RNA Complex Secondary Structures
- Can base-pair conventionally: A:U and G:C
- Can also form unusual base pairs like U-U and G=U
- 2 H-bonds in U=G are weaker then 2 in U=A
- Folded RNA can have enzymatic activity = ribozyme, or it can have structural roles
- All RNA is synthesised (transcribed) as a single strand within the nucleus (rRNA in nucleolus) but this does not mean RNA has random structure
- single strands tend to form a right-handed helix dominated by base-stacking interactions, with purine-purine stacks being the strongest.
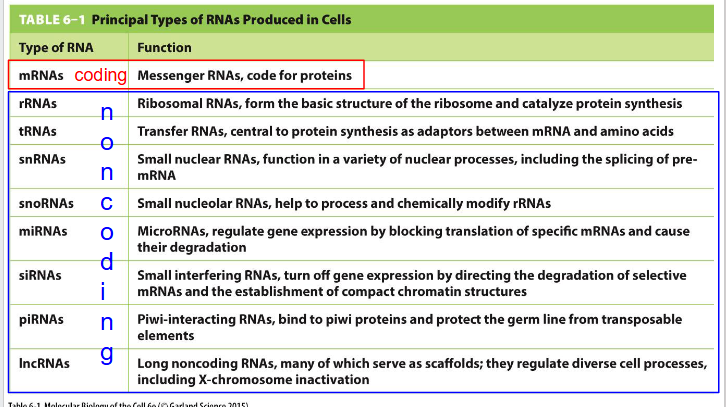
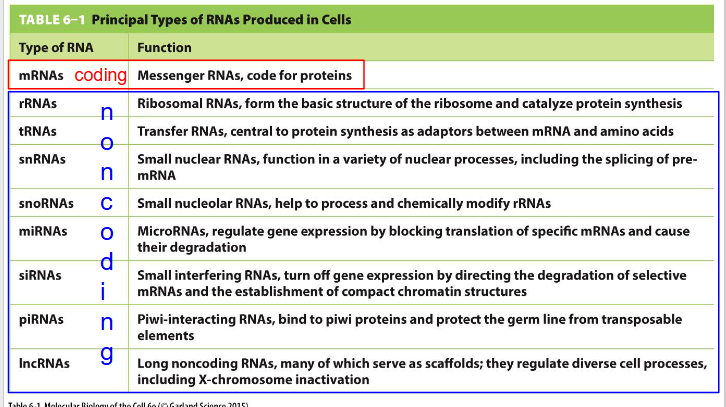
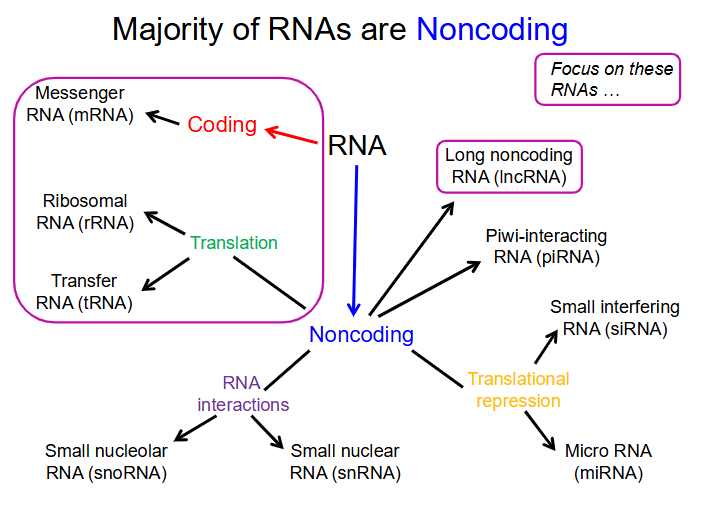
How is RNA grouped
Coding and Non-coding
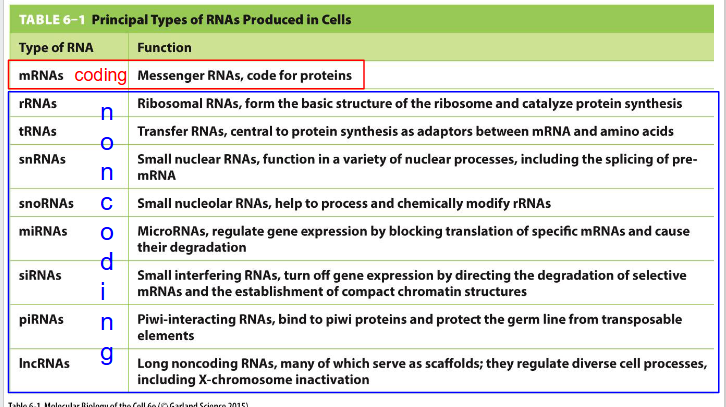
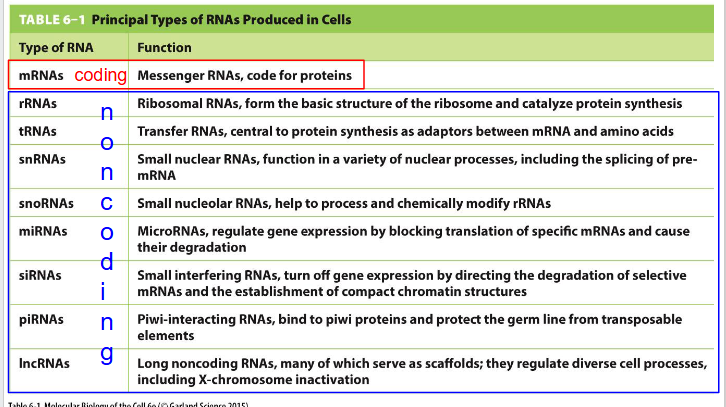
Functions and where they are made in cell of Coding and Non-coding RNA: need to know: mRNA, rRNA, tRNA, lncRNA
- mRNA (Messenger RNA): Coding. code for proteins. made in nucleus (not nucleolus)
- rRNA (Ribosomal RNA): Non-coding. form the basic structure of the ribosome and catalyse protein synthesis. Made in nucleolus
- tRNA (Transfer RNA): Non-coding. central to protein synthesis as adaptors between mRNA and amino acids. Made in nucleolus
- lncRNA (long noncoding RNAs): Non-coding. many serve as scaffolds; they regulate diverse cell processes including X-chromosome inactivation. Made in nucleolus
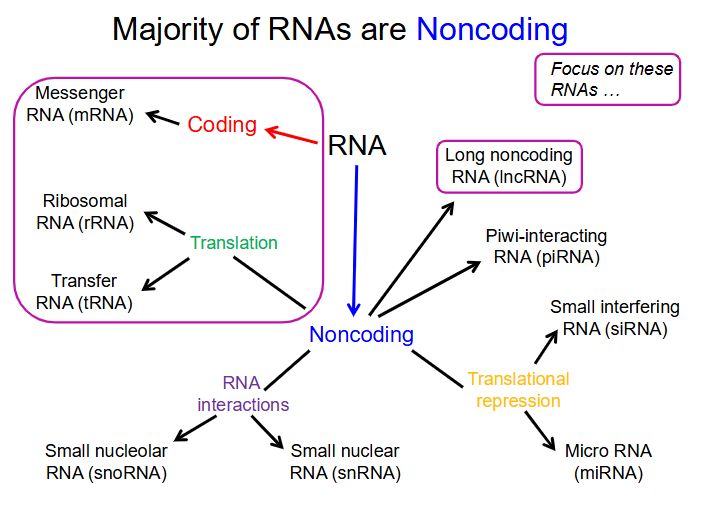
Majority of RNA is
non-coding
3 essential RNAs for Protein Synthesis
- mRNA: AA sequence code
- rRNA: structural and functional part of ribosome = site of protein synthesis
- tRNA: delivers AA to ribosome for incorporation into protein
Ribosomes
- protein synthesis takes place when tRNA and mRNA associates with ribosomes
- tRNA and ribosomes translate mRNA language of nt sequence into AA sequence in protein
- consists of a small and large subunit with different rRNAs and proteins in each
- 50% of ribosome mass is RNA and 50% is protein: RNA is catalytic in protein synthesis
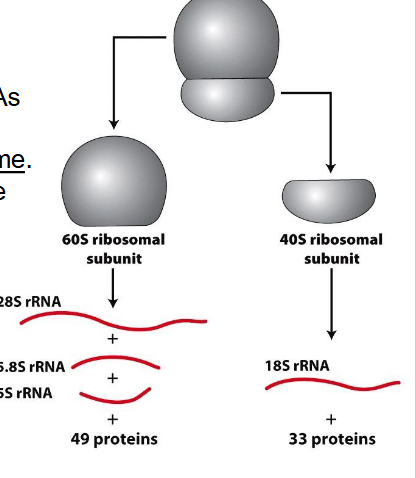
Ribosome naming nomenclature
- S = Svedburg unit, an indication of size due to sedimentation in centrifuge
- Eukaryotic Ribosomes = 80S
- since RNAs fold into specific secondary structures, it enables rRNAs to have function (catalytic activity = ribozyme)
- Peptide bonds form in the large ribosomal subunit through the action of 28S rRNA
- dont add up because the unit indicates density, doesnt need to add up
- DONT NEED TO REMEBER NUMBERS
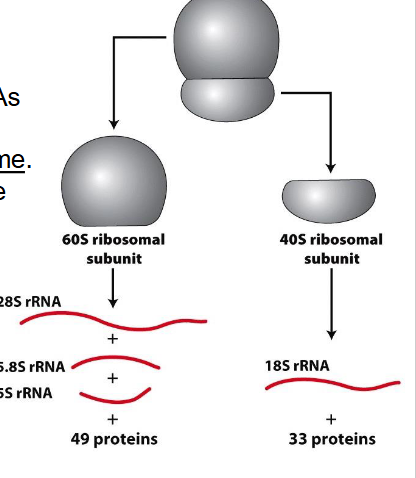
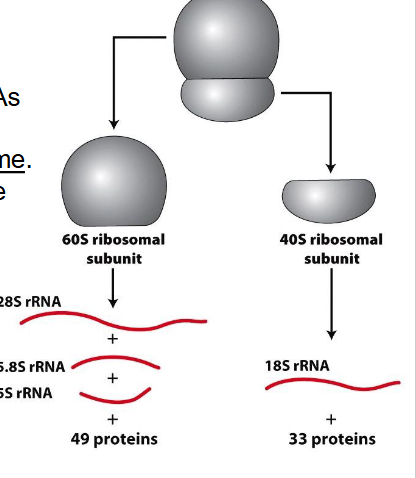
Peptide bonds (protein formation) are formed in the
- large ribosomal subunit (60S) through action of 28S rRNA, 28S rRNA is the catalytic unit
- NOT NEEDED TO KNOW
DIAGRAM ON SLIDE 14
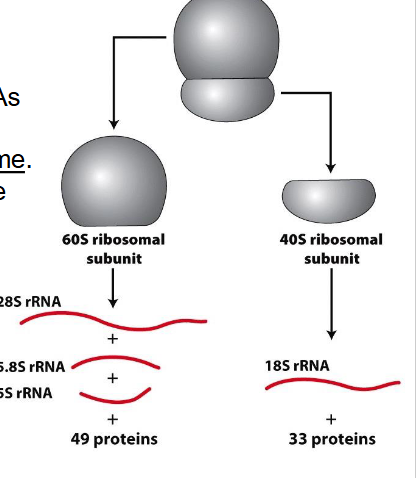
rRNA is synthesised in the nucleolus from which two genes
- 18S, 5.8S and 28S made as large (45S) precursor rRNA, which is extensively modified prior to use
- 5S expressed from a separate gene
- rRNAs and ribosomal proteins start to assemble into ribosomes within the nucleolus and continue to be processed in the cytoplasm before the two subunits combine
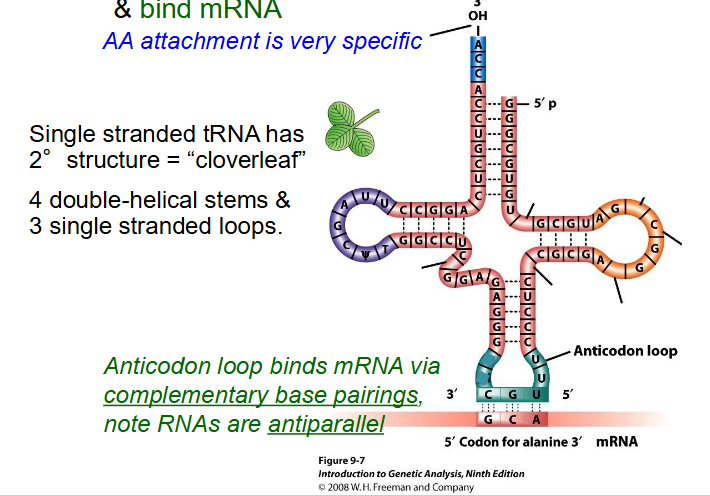
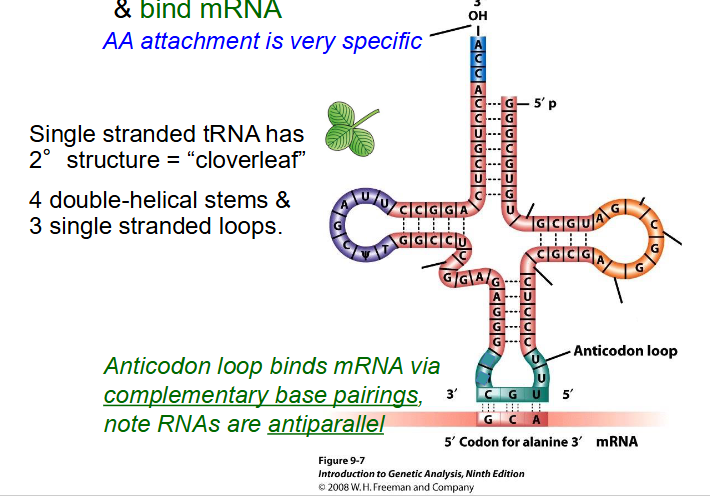
Function of tRNA
- transfer amino acids to ribosome and bind mRNA
- AA attachment is very specific
- single stranded tRNA has secondary structure = "cloverleaf"
- 4 double-helical stems and 3 single stranded loops
- Anticodon loop binds mRNA via complementary base pairings. RNAs are antiparallel.
- if u trace it with ur finger u would notice its still single stranded
lncRNA and X Chromosome Inactivation
- only one active X per cell. Caused by Xist gene which encodes a lncRNA that coats the DNA and turns off transcription of that X Chr
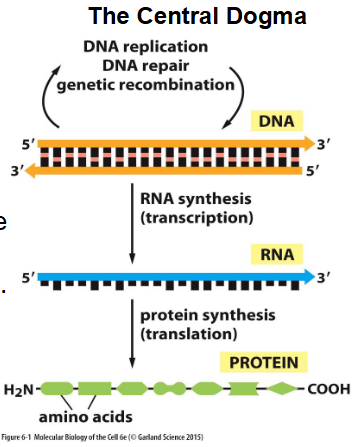
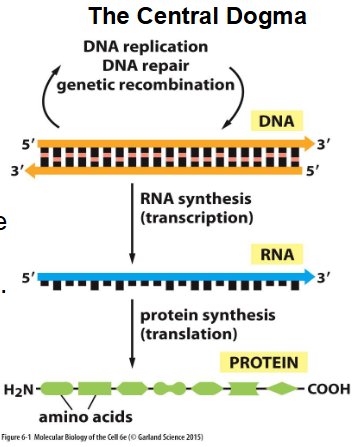
Central Dogma
theory that states that, in cells, information only flows from DNA to RNA to proteins
Why we have both DNA and RNA
- DNA is more stable and less reactive then RNA. DNA is better for long term genetic store
- Deoxyribose in sugar-phosphate backbone is stable over greater lengths
- Double helix and thymine is less prone to chemical damage then uracil. Also, errors in DNA replication repaired by DNA pol 3' to 5' proofreading.
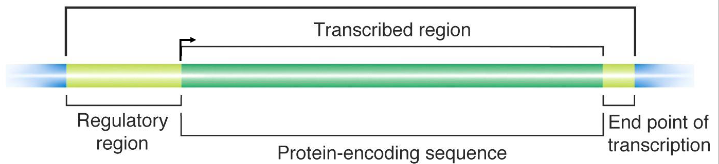
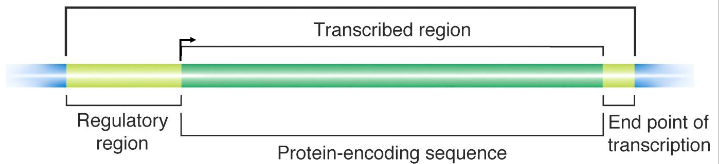
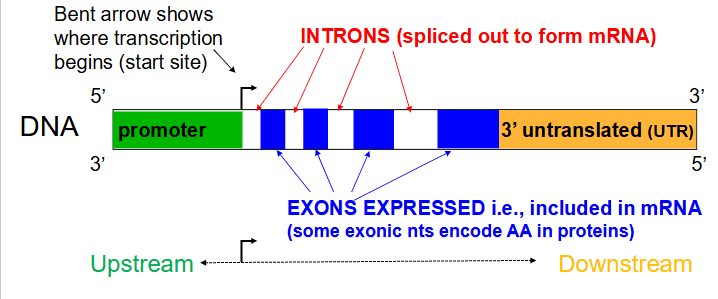
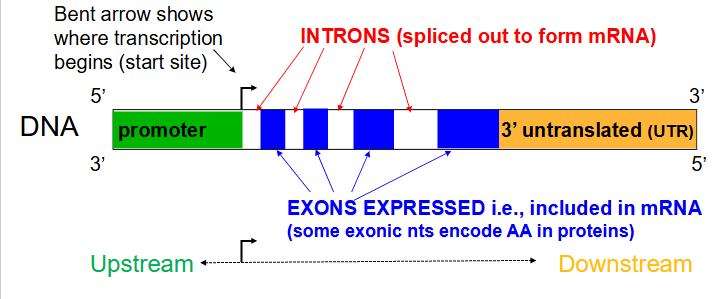
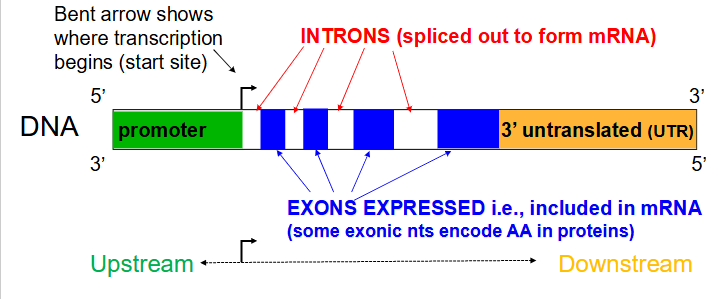
Gene structure (for a protein-encoding gene)
- Transcribed regions contain information for the nt sequence of the gene product. Regulatory regions contain sequences that are recognised and bound by proteins that transcribe the DNA into RNA
- Proteins also influence the amount of RNA made
- Regions close to the transcriptional start site (bent arrow) form the promoter, while sequences far away are enhancers (further away from transcribed region in the same direction). Many proteins associate with a gene prior to starting transcription.
- arrow in diagram always faces away from regulatory region and towards the transcribed region
Not all DNA for a gene codes for an RNA
- a region immediately upstream (i.e 5' to the coding area) called the promoter, is involved in initiation of transcription. An area downstream (3' to the coding region) is involved in termination of transcription.
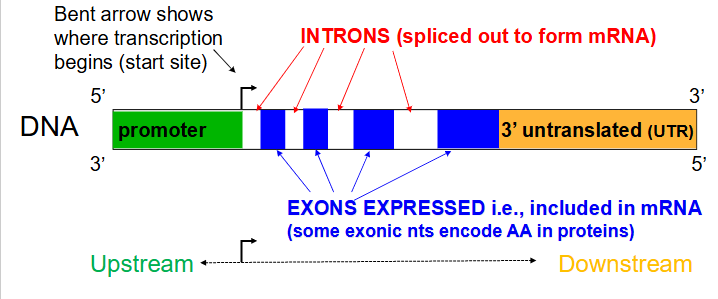
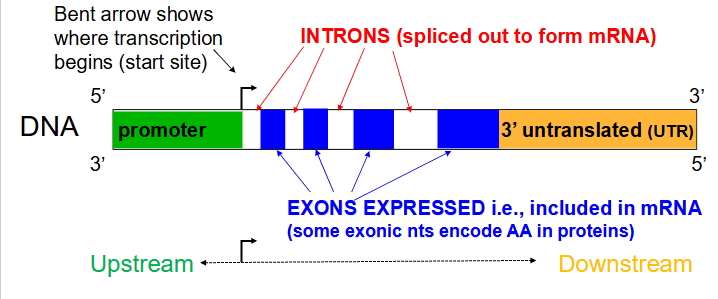
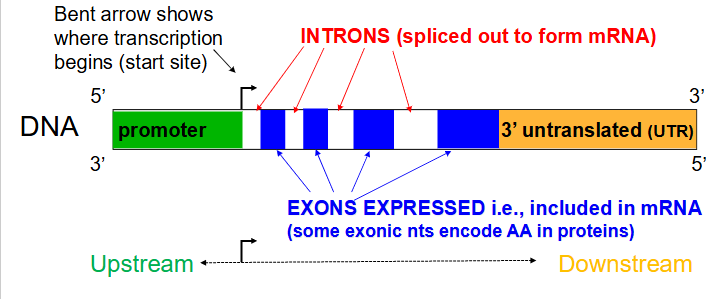
Introns
Noncoding segments of nucleic acid that lie between coding sequences. (spliced out to form mRNA)
Exons
Coding segments of eukaryotic DNA. (rmbr that Exons are expressed)
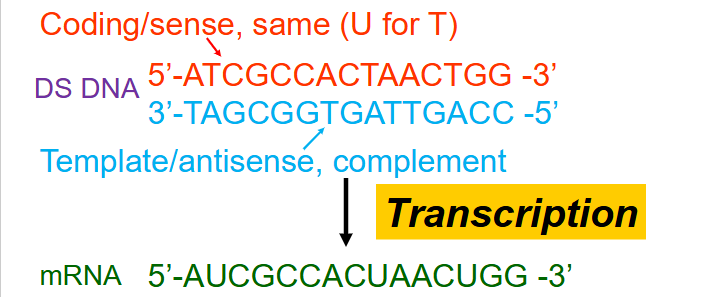
Naming scheme for strand produced during transcription
- Coding/sense = same (U for T)
- template/antisense, complement
- by convention, always state sequence of DNA and RNA 5' to 3'. AND Always refer to the coding strand (since it has the same sequence and direction as the mRNA)
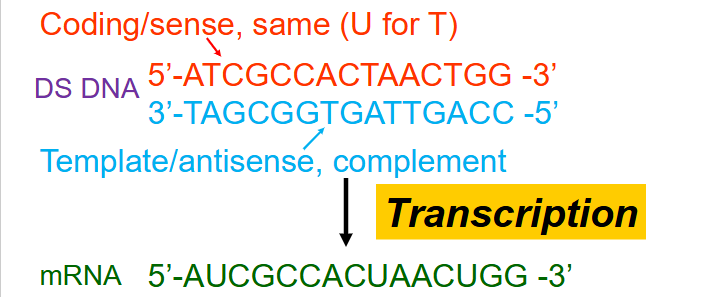
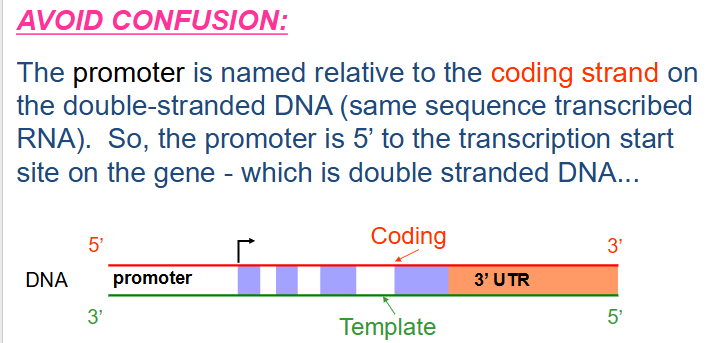
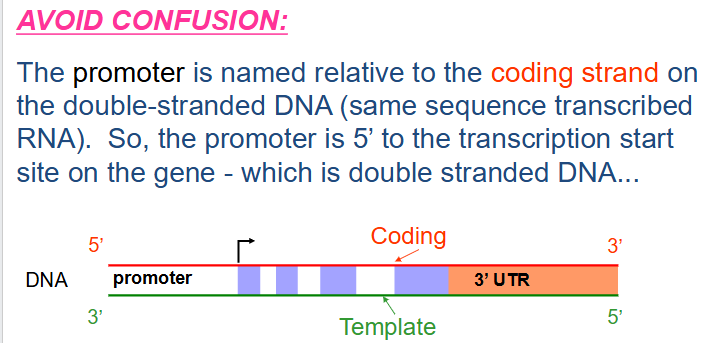
How to avoid confusion in transcription
- the promoter is named relative to the coding strand on the double stranded DNA (same sequence transcribed RNA). So the promoter is 5' to the transcription start site on the gene - which is double stranded DNA.