2. RNA Synthesis- Transcription
1/17
Earn XP
Description and Tags
IN THIS LECTURE HE SAID HE IS TESTING THE MOST ON REGULATION!!!!! NOT THE ACTUAL STEPS
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
18 Terms
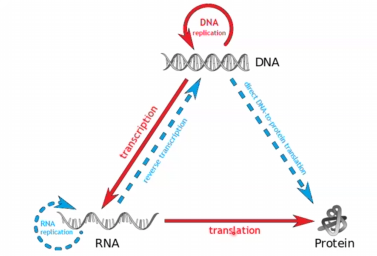
Central Dogma of Molecular Biology
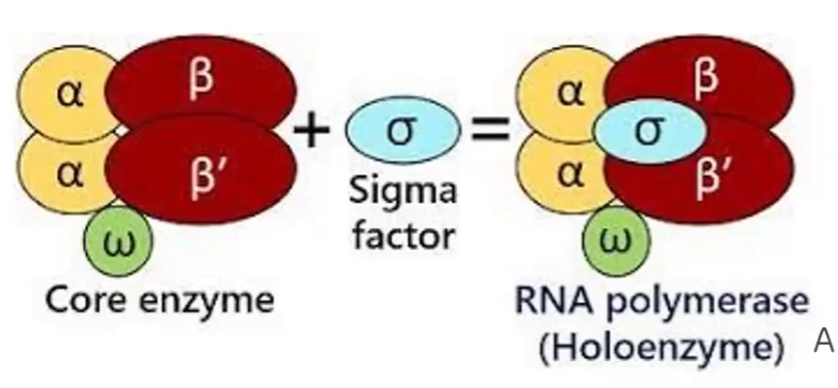
RNA Polymerase in Prokaryotes:
Core enzyme/Apoenzyme
Holoenzyme
Sigma Factor
Core enzyme: 2 alpha subunits, beta and beta’ unit and omega unit
Holoenzyme: core enzyme + sigma factor
Sigma Factor: Recognizes promoter
RNA Polymerases I, II and III
RNA pol I: codes most rRNA genes
RNA pol II: all protein-coding genes—> mRNA!!! ←TQ
RNA pol III: tRNA, small RNA’s and 5S rRNA genes
mRNA
what does it correspond to?
Converted into what?
In Eukaryotes it is made by which RNA Pol?
Prokaryotes vs. eukaryotes
Corresponds to a DNA sequence
Converted into amino acid sequence during translation
RNA pol II
Prokaryotes: Polycistronic, occurs in cytoplasm, not always 5’ cap or 3’ tail, transcription is coupled to translation
Eukaryotes: Monocistrionic, occurs in nucleus, 5’ cap and 3’ tail, transcription and translation are separate,
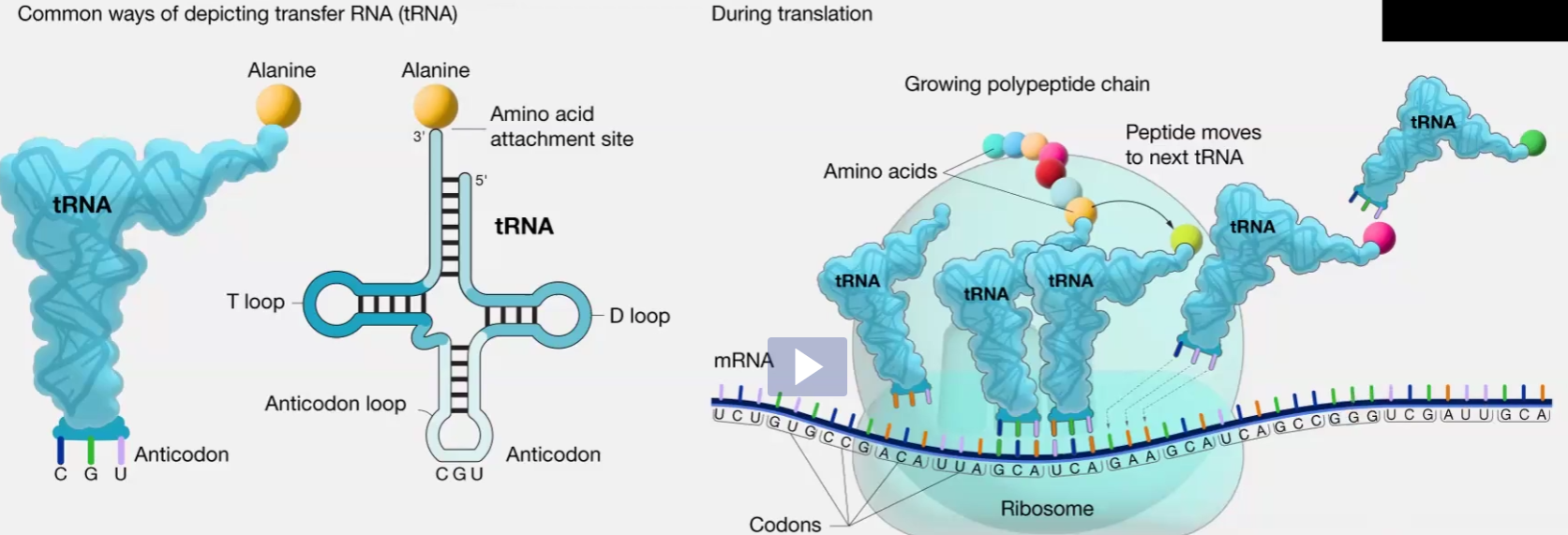
tRNA
Secondary structure
Functions
Synthesized by which RNA pol
Looks like a curved croissant
Function is to carry amino acids into ribosome to make proteins
Anti codon of tRNA reads mRNA codon
DNA Pol III
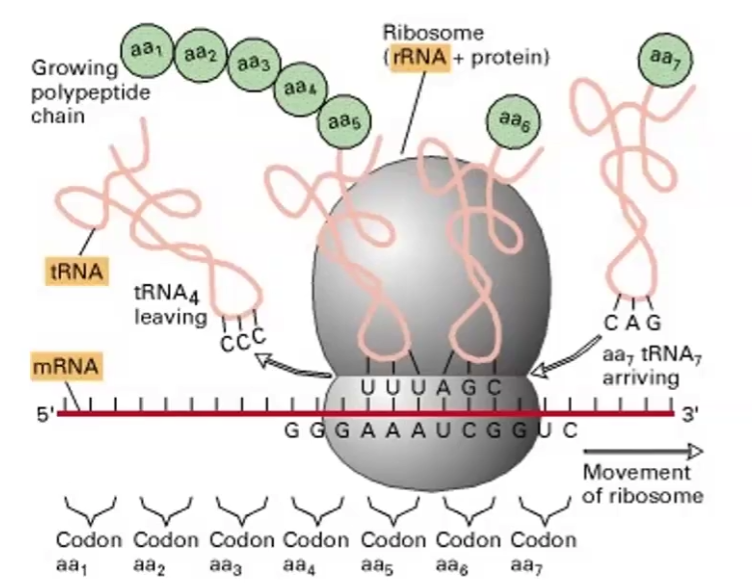
rRNA
What is it?
What does it do?
The unit used to measure the amount of sedimentation of ribosomes is
What DNA pol synthesizes it?
Ribosomal RNA; make up ribosome organelle
It makes proteins during translation
Svedberg unit
DNA Pol I and III
Prokaryotic Promoter
The most upstream component is:
Next follows the ________ region and then the ________ _____ which ,are recognized by the __ _________
The most upstream component is: the upstream promoter element
Next follows the TTGACA region and then the TATA Box, which are recognized by the sigma factor (of the holoenzyme)
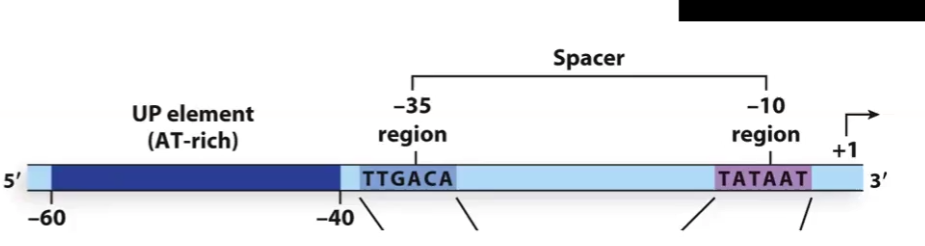
Other Prokaryotic Regulatory Sequences
Operator is located at the end of the promoter
Activator Binding site
Eukaryotic Promoter elements
Short sequence elements that bind transcription factors (sigma factor) and recruit RNA pol
GC box: has increased amount of transcription when there is not TATA box
CAAT box: does 25% of genes transcribed in large quantities
TATA box: core region of the promoter
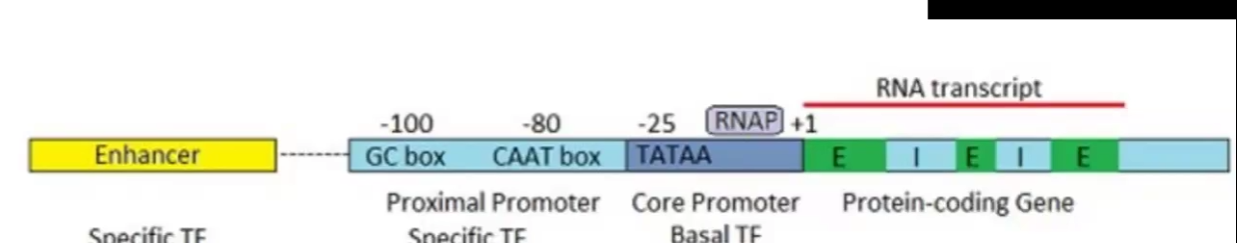
Additional Eukaryotic Regulatory Elements
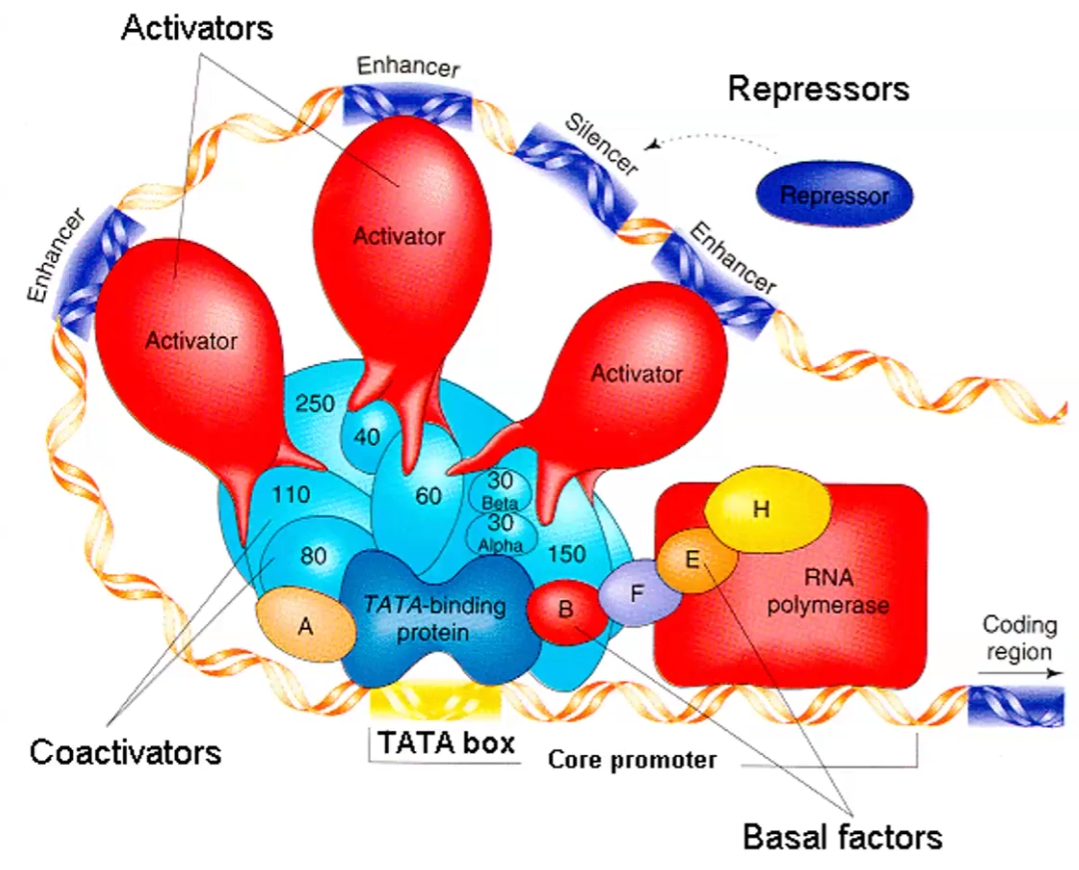
Enhancer vs. Silencer vs Methylation
Methylation vs Acetylation of histones
Can both be found both upstream and downstream
Enhancer sequences: Activator binds it; speeds rate of transcription
Silencer sequence: Repressor binds it; blocks transcription and prevents genes from being expressed
Methylation and demethylation on cytosine+guanine (CpG islands): methylation blocks transcription and demethylation allows transcription
Methylation of histone: blocks transcription so Heterochromatin formed
Acetylation: makes histone negative , which makes DNA get off and loose → Euchromatin
Coding vs. Non-coding strands
Coding/ Sense strand: not used as template for transcription
Non-coding/Antisense strand: used as template
Transcription steps for Eukaryotes
RNA Polymerase II must become phosphorylated to be active
RNA Polymerase II binds to promoter of DNA and forms “closed complex”
RNA Polymerase II melts the DNA duplex near the transcription site and forms transcription bubble “open complex”
RNA Polymerase II catalyzes phosphodiester linkage of the first two rNTPs
Initiation factors are shed and elongation factors are recruited
RNA Polymerase II continues down non-coding DNA 3’-5’ and making a new strand that is growing 5’-3’ → Elongation
At transcription stop site, Polymerase releases DNA and RNA
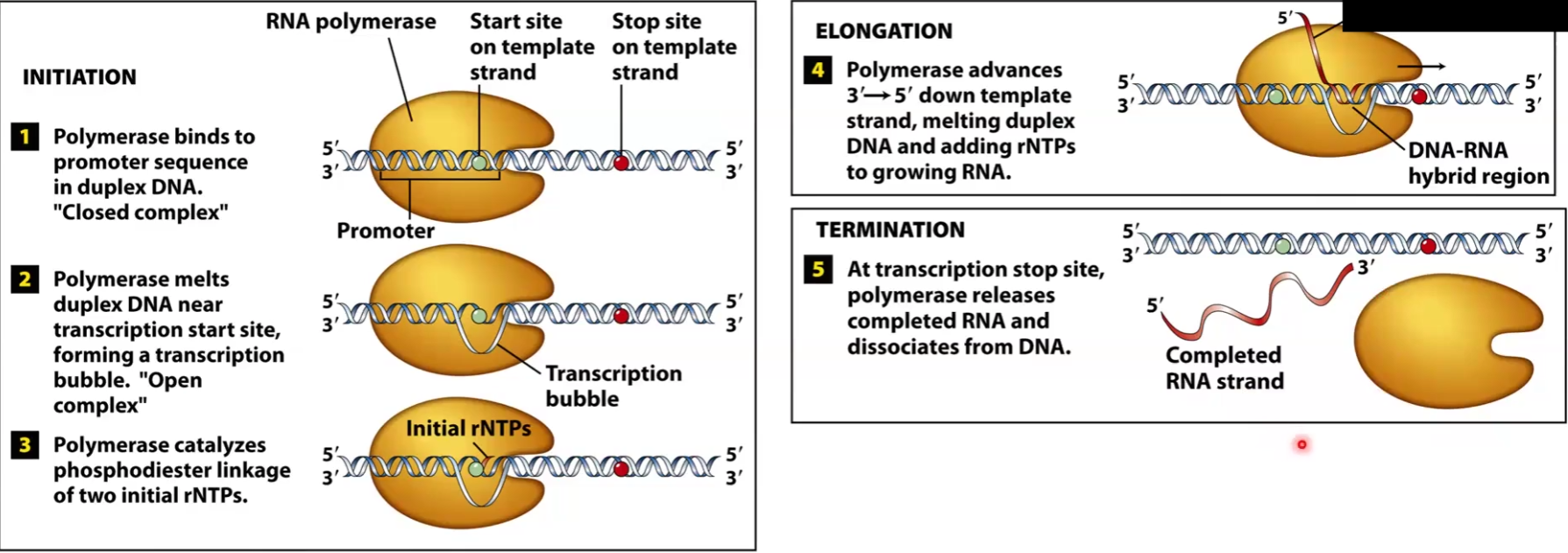
Transcription steps for Prokaryotes
RNA pol holoenzyme binds to promoter of DNA, forming closed complex
Duplex is melted and transcription bubble “open complex” forms
Polymerase starts coding first few nucleotides
Sigma factor is released
Polymerase continues down non-coding DNA 3’-5’ and making a new strand that is growing 5’-3’ —>Elongation
Termination occurs when end gene is reached → Rho dependent or independent
Sigma factor rejoins and finds another promoter
Rho-dependent vs. Rho Independent
Rho independent: when termination sequence is reached, there is a self complementary AAA-UUU segment that forms a hairpin and stops transcription
Rho dependent: Once termination sequence is reached, ir recruits rho helicase, which hydrolyzes RNA away from DNA thus ending transcription
mRNA Post-transcriptional processing methods
3’ Poly A tail and 5’ cap —> their purpose is transport and stability
mRNA Splicing
Components, how does it work
Why do we need splicing
TQ: all of it
Spliceosomes remove introns (noncoding) from sequence and leave the exons (coding)
It’s a way for our cells to make more proteins from less genes; Gives us many alternative combinations
3’ UTRs and 5’ UTRs , microRNAs —> Are there for regulation of gene expression
tRNA Processing
5’ sequence removed
If there are introns, they are spliced out
3’UU is removed and replaced by CCA
Base modification occurs as part of regulatory process ← TQ
What is a Copy number
It’s how many times the same gene can be transcribed and it varies depending on the cell’s needs