T3M2: Prok Transcriptional Regulation
1/21
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
22 Terms
glucose vs lactose metabolism
E. coli bacteria prefer glucose as an energy source.
When both glucose and lactose are available, E. coli will use up all the glucose first before switching to lactose.
This switch is a highly regulated process — the bacteria only turn on the genes needed for lactose metabolism after glucose runs out.
These genes are controlled at the transcriptional level, meaning that E. coli can turn on or off specific genes depending on nutrient availability.
The bacteria detect two key environmental cues:
Glucose levels (whether it’s high or low)
Presence of lactose
Β-galactosidase and lactose permease proteins are non-detectable when E. Coli is using glucose as its main source of energy
proteins involved in lactose metabolism
When E. coli switches from glucose to lactose:
It starts producing two important proteins:
β-galactosidase – an enzyme that breaks lactose (a disaccharide) into glucose + galactose.
Lactose permease – a membrane transport protein that helps bring lactose into the bacterial cell.
These proteins are not made while glucose is still available — glucose represses their production.
Once glucose is gone, and lactose is present, lactose induces their production.
This ensures E. coli only makes the lactose-digesting enzymes when necessary, saving energy.
operons
⇒ related prokaryotic genes with similar functions can be clustered into operons → allows for the control of transcription of the whole gene cluster in one unit — controlled by a single ON/OFF switch
⤷ in euk… each gene has its own promoter and enhancers
> operon model = discovered by jacob and monod
composed of:
promoter (where RNA polymerase binds to start transcription)
operator (a DNA sequence that acts like an on/off switch)
found near the start of the operon, can be regulated to allow/inhibit transcription
if an operator is NOT bound to inhibitor.. then RNA poly can attach to promoter and transcribe genes in operon
cluster of structural genes (that code for proteins with related functions)
> allows for polycistronic mRNA — where a single mRNA can make multiple proteins bc it contains several start/stop codons
lac operon
operon for specifically managing lactose (for transcriptional regulation)
controls production of:
β-galactosidase (lacZ)
Lactose permease transport proteins (lacY)
> involves regulatory sequences of transcription — w promoter that binds to RNA poly complex and operator (lacO) (binding site for repressor protein that is expressed by lacI coding seq.)
> ⇒ RNA polymerase can bind to and activate the transcription of lacY and lacZ genes… but lacI controls the expression
promoter
where RNA poly binds
operator
where repressor protein can bind to block transcription
» negative regulatory site bound by lac repressors proteins (overlap w the promoter)
» when lac repressor is bound, RNA poly cannot bind to promoter and start transcirption
lacI gene
makes the repressor protein that can bind to the operator
repressor
when the repressor binds to the operator, RNA polymerase cannot transcribe lacZ and lacY — this is negative regulation
coded by lacI gene
negative transcription regulation: the ability of a repressor protein to halt transcription
lacY gene
gene that codes for lactose permease transport proteins
⤷ will embed itself in the cell membrane and allow for the import of lactose into the bacterial cell
lacZ gene
code for β galactosidase
⤷ helps cleave lactose into glucose and galactose
negative regulation of the lac operon
when glucose present:
lacI gene is always expressed at low levels — encoding for repressor protein
repressor protein binds onto the operator… and twists its into a loop (bc of its tetrametric structure) prevening the RNA polymerase to transcript (cannot move downstream — “blocked“)
When the repressor is bound, RNA polymerase is blocked and cannot bind to teh promoter and no transcription occurs
This means no β-galactosidase or lactose permease are produced.
This happens when glucose is available, because the cell doesn’t need lactose-metabolizing enzymes.
when lactose is present, no glucose:
lactose becomes allolactose (isoform of lactose that acts as inducer)
allolactose passes thru the lactose permease channel, and binds onto the operator
binding of allolactose onto operator = causes teh repressor to change shape (conformational change) so the repressor can no longer bind anymore
no repressor on the operator = allows RNA poly to bind ==> allow transcription of beta. galactosidase and permease etc.
positive regulation of lac operon
positive regulation of lac operon occurs when there is no glucose => produces β galactosidase and lactose permease
HIGH glucose
↑ glucose = adenyl cyclase is active = ↑ production of cAMP = cAMP binds onto CRP binding site = ↑ affinity for RNA poly to bind onto promoter = ↑ transcription rates
> when cAMP binds onto CRP binding site = will form CRP-cAMP complex as allosteric activator
LOW glucose
↓ glucose = inhibits adenyl cyclase = ↓ cAMP = ↓ transcription
CRP binding site
positive regulatory site, where CRP binds to promote transcription by helping
CRP/CAP
glucose sensor, transcription factor that binds to DNA in the presence of cAMP to enhance RNA polymerase binding and initate transcription
Glucose present, lactose absent ~
NO transcription of the lac operon (bc lac repressor remains bound to the operator and prevents transcription by RNA polymerase)
cAMP levels are LOW bc glucose levels are HIGH – ∴ CRP is inactive and cannot bind DNA
Glucose present, lactose present ~
Low level transcription of the lac operon
The lac repressor is released from the operator because the inducer (allolactose) is present
cAMP levels = low bc glucose present
∴ CRP remains inactive bc cannot bind to DNA
Glucose absent, lactose absent ~
No transcription of the lac operon
cAMP levels are HIGH bc glucose levels are LOW → ∴ CRP is active and will be bound to DNA
Lac repressor will be bound to the operator (absence of allolactose) → ∴ no RNA poly binding and preventing transcription
Glucose absent, lactose present ~
Strong transcription of the lac operon
The lac repressor is released from the operator because the inducer (allolactose) is present
cAMP levels are high (bc glucose is absent) → ∴ CRP is active and binds to DNA
CRP helps RNA poly to bind to the promoter ⇒ ∴ high levels of transcription
combining negative and positive regulation
Negative regulation: a repressor binds onto DNA operator to block transcription.
Can be turned off by an inducer (like lactose).
Positive regulation: an activator (like CRP–cAMP) binds DNA to stimulate transcription.
Without the activator, transcription won’t happen efficiently.
inducer protein
able to bind to repressor proteins and prevents it to bind onto DNA — reverse its function
transcription activator protein
positive regulation of transcription ⇒ a transcription activator protein binding to an activator binding site on the DNA
the binding site = upstream/downstream or OVERLAP w promoter
Once activator binds the DNA → RNA poly binds to promoter and start transcription
If activator is not present/not able to bind to the activator binding site… then transcription cannot occur
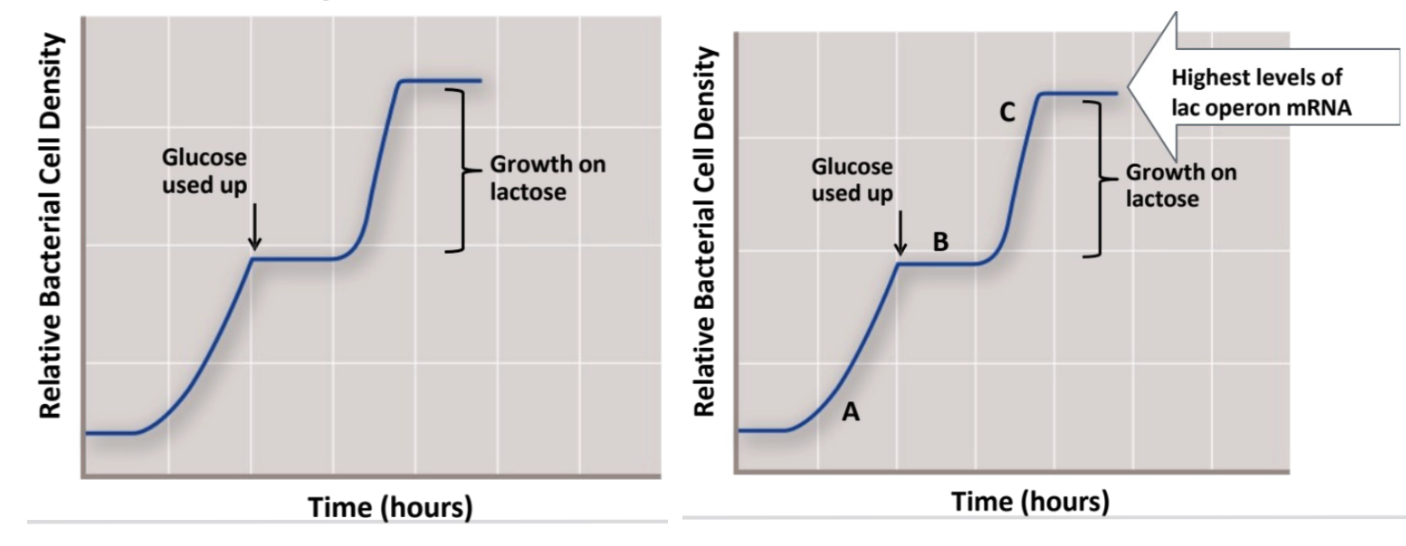
when are levels of lac operon mRNA the highest, lowest, and no lac operon
⇒ the highest level of lac operon mRNA would be @ point C ⇒ bc bacteria are actively utilizing lactose as a nutrient source ⇒ ∴ transcribing the lac operon gene products to be able to produce lactose permease + β galactosidase
⇒ low lac operon @ point B → this is the transitional time point where glucose has been used up and lac operon transcription is initiating
⇒ no lac operon @ point A → bc bacteria is still actively utilizing glucose as a nutrient source (repressed until there is no glucose)