Module 12 - Two-factor ANOVA
1/16
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
17 Terms
Two-factor ANOVA
Used to evaluate the change in a numerical variable across two categorical variables. Builds on the single-factor ANOVA, can look at the effect that each level has on the numerical variable and the interaction between categorical variables
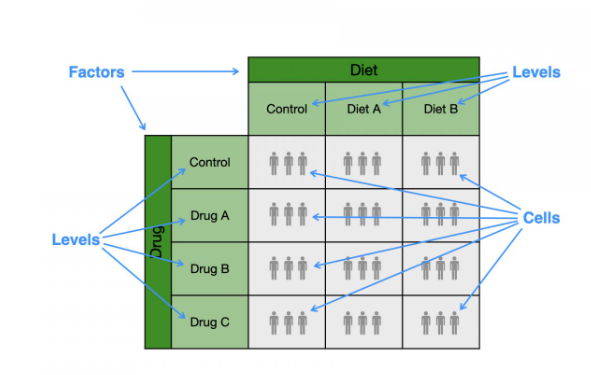
Structure of a two-factor ANOVA
Can be visualized as a two-dimensional grid. One factor can be columns and the other rows
Main effects
Answered in a two-factor ANOVA — can be for factor A: asking about differences among the levels of factor B. These are comparisons among full columns (or full rows) AND for factor B: asking about the differences among the levels of factor B averaging across the levels of factor A. These are comparisons among full rows (or full columns).
Interactions
Questions about the differences among the levels of one factor within each level of the other factor. These are cell-bycell comparisons.
Evaluation of if there is an interaction between the factors
Asks if there is an interaction — any deviation from the assumption that the levels of each factor simply add together (additivity)
Assumption of additivity
The response to the combination of two levels is simply the sum of the two — ie. stacking two 1kg rocks on a scale will result in a measurement of 2kg
Interaction plot
A common way to visualize data - a specialized plot that highlights the interaction pattern between two categorical variables. Use the same information as a box plot but presents the data in a way that showcases the relationship.
Setup for an interaction plot
The y-axis shows the numerical variable in the same way as box plots
The x-axis shows the levels one categorical variable
Lines are used to connect cells across the x-axis according to the levels of the other categorical variable
Representation of categorical variables on an interaction plot
If the categorical variables are additive (no interaction), the lines will be parallel. If the categorical variables are not additive, the lines are not parallel. Antagonistic interactions have lines that cross. Synergistic interactions have non-parallel lines that don’t cross
Null and alternative hypotheses for main effects (ie., A)
Null: The means (μAi) are the same across all levels (i) of factor A: HO: μA1=μA2=...=μAk-1=μAk
Alternative: The means are different somewhere: HA: μA1≠μA2≠...≠μAk-1≠μAk
Null and alternative hypotheses for interaction
Null: the deviation of each cell relative to additivity is zero (notation AiBj refers to the cell that corresponds to the ith level of factor A and the jth level of factor B): HO: δA1B1=δA1B2=...=δAkBk-1=δAkBk=0
Alternative: There is a non-zero deviation from additivity. Does not indicate which cells or how many have non-zero deviations. HA: δA1B1≠δA1B2≠...≠δAkBk-1≠δAkBk≠0
Group variation factor A (MSA)
The variation among the means of the levels of factor A. Calculated as MSA=SSA/dfA, where SSA is the total group variation and dfA is the degrees of freedom. Degrees of freedom are dfA= a-1, where a is the number of levels in factor A.
Group Variation factor B (MSB)
The variation among the means of the levels of factor B. Calculated as MSB=SSB/dfB, where SSB is the total group variation and dfB is the degrees of freedom. Degrees of freedom are dfB= b-1, where b is the number of levels in factor B.
AB interaction (MSAB)
The amount of variation attributable to the deviation from additivity. Calculated as MSAB=SSAB/dfAB, where SSAB is the total variation of the cell deviations from additivity and dfAB is the degrees of freedom. Degrees of freedom are dfAB= (a-1)(b-1), where a is the number of levels in factor A and b is the number of levels in factor B.
Residual Variation (MSE)
The variation among sampling units within a cell. Calculated as MSE=SSE/dfE, where SSE is the total residual variation and dfE is the degrees of freedom. Degrees of freedom are dfE= ab(n-1), where a is the number of levels in factor A, b is the number of levels in factor B and n is the number of sampling units in each cell.
Three F-tests
Main effects A/B: (ie. A) The F-score is F=MSA/MSE, where MSA is the group variation for factor A and MSE is the residual variation. This ratio of variances evaluates mean differences among factor A levels by quantifying the amount of factor A group variation relative to residual variation.
Interactions: The F-score is F=MSAB/MSE, where MSAB is the cell variation attributed to non-additivity and MSE is the residual variation. This ratio of variances evaluates an interaction by quantifying the amount of variation attributable to deviations from additivity relative to residual variation.
Scientific conclusions for a two-factor ANOVA
First: Evaluate the interaction, either:
Reject the null hypothesis and conclude there is evidence that at least one cell deviates from additivity.
Fail to reject the null hypothesis and conclude there is no evidence that the cells deviate from additivity.
If to reject the null hypothesis, main effects should not be evaluates (the presence of interactions says the change in means across levels of one factor differs depending on the level of the other factor)
Second: Evaluate the main effects, if the interaction is not significant, factors are considered additive, can look at main effects for each and conclude:
Reject the null hypothesis and conclude there is evidence that the means of at least two levels are different in the factor.
Fail to reject the null hypothesis and conclude there is no evidence that the means among levels are different in the factor.