Biology - DNA & Genetics: Replication, Gene to Protein, Biotechnology, Relevant Scientists, Mendelian Genetics, Incomplete vs. Co-dominant.
1/20
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
21 Terms
DNA, Nucleotide, Gene, protein?
DNA: carries hereditary (genetic) info of an organism
sequences are codes that allow cells to make proteins
long chain of repeating monomers
Nucleotide: Monomers of DNA/RNA (adenine, thymine (IN DNA), cytosine, guanine, Uracil (IN RNA); A bonds with T/U, C bonds w G)
Gene: segment of the entire DNA polymer
has a unique & particular sequence of nucleotides
the non-coding dna between geneses was considered ‘junk’ but may have importance as switches
sequences of each gene codes for one protein
Protein: polymer made up of monomers called amino acids
chargaffs rule
A’s go w/ Ts, C’s go w/ G’s
amount of adenine nucleotides in any DNA sample is always equal to the amount of thymine, and the amount of cytosine nucleotides is equal to amount of guanine
structure of DNA
covalent bonds hold phosphate and sugars together on the actual strands—difficult to break
hydrogen bonds hold the nucleotides—A and T bond w/ 2 hydrogen bonds, C and G bond w/ 3 hydrogen bonds
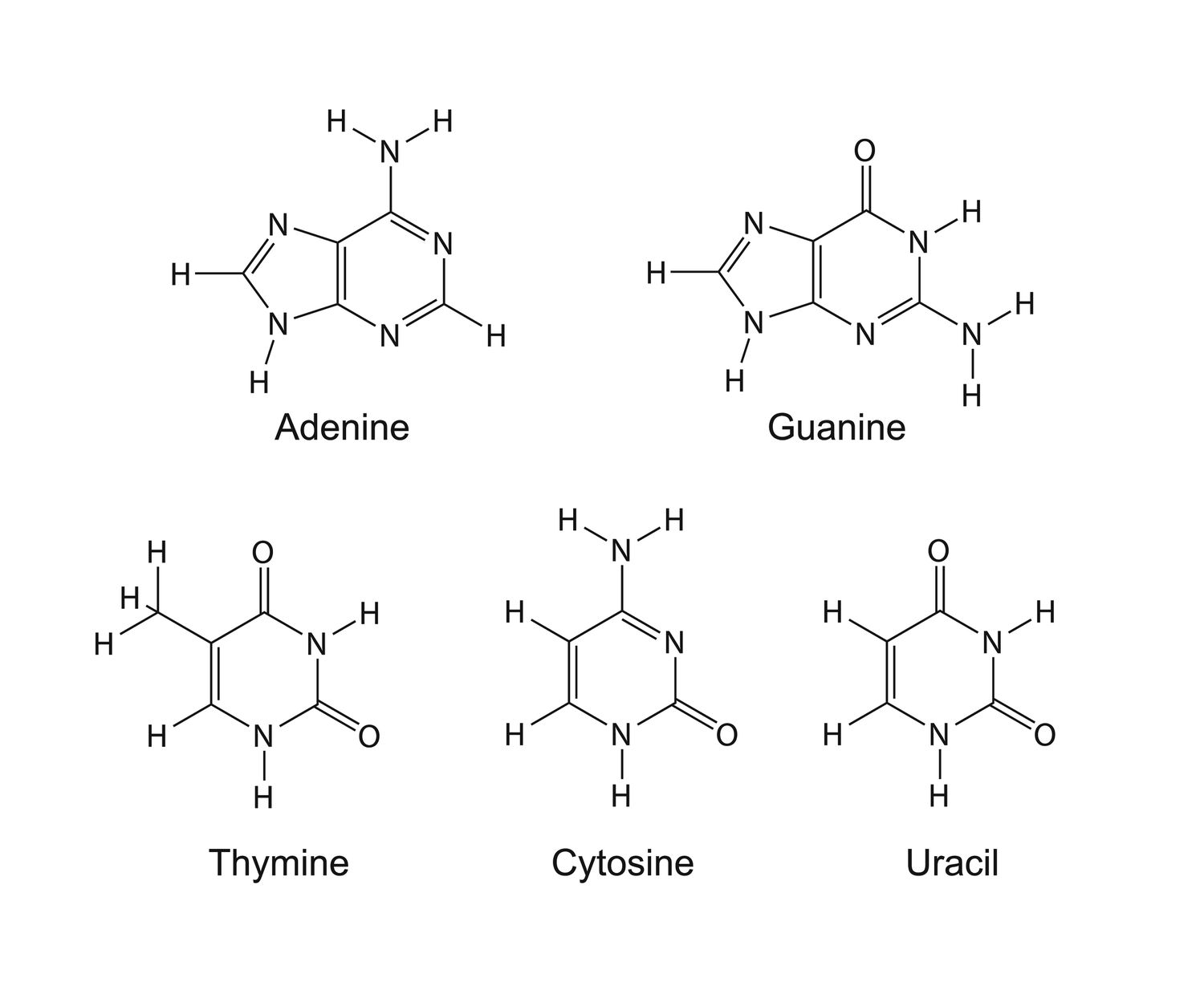
what are the chemical structures of bases
purines: adenine and guanine
double ring structure
Pyrimidines: thymine/uracil and cytosine
single ring structure
what is DNA replication
the process of creating an exact copy of DNA, which preserves the exact order of bases and occurs just before a cell divides.
each DNA strand serves as a template for the creation of a new complimentary strand
describe process of replication
DNA strand serves as a template for the creation of new complimentary strand
complimentary strand is formed by pairing bases A—T and C—G.
Each new molecule of dna contains 1 original (parent strand/conserved strand) and 1 new strand (daughter strand)
Process:
helicase binds to dna and opens the double helix, creating a replication fork
replication begins at specific sequence of dna called the origin
as helicase opens dna strands, replication bubbles are created, and eventually bubbles that begin at different origins will meet up w/ eachother
for the actual replication to occur, after priming, DNA polymerase III inserts itself into a bubble and adds nucleotides one at a time
the two sides of dna are antiparallel, meaning the 2 new strands cannot be synthesized in the same way
Leading strand:
continuous replication
template read in the 3’-5’ direction, polymerase III adds 5’-3’
only requires one primer at the replication origin
for replication/elongation of daughter strand to occur, new nucleotides must have something to attach to
primers are made and attached to template strand by primase enzyme
lagging strand:
fragmented replication
template is opened 5’-3’ direction (last nucleotide has 5’ end available, when the leading has a 3’ end available, thats how you can identify lagging strand)
new dna is synthesized in chunks (okazaki fragments) using the 3’ ends of a series of primers
dna polymerase I then removes RNA primers and fills in the gaps w/ DNA nucleotides
nucleotides are added in the 5’-3’ direction by using 3’ ends of a primer
it kinda looks like its going in the opposite direction
function of each enzyme in replication
helicase: ‘unzips’ dna (breaks hydrogen bonds between bases)
primase: inserts an rna primer (a primer is the molecules to which nucleotides can be added)
DNA polymerase III: adds new nucleotides (elongation)
DNA polymerase I: removes primers and proofreads (recognizes mismatched bases and replaces them)
DNA ligase: seals the new phosphate and sugar backbone together
Describe process of gene expression/protein synthesis
DNA is a code. 3 nucleotides=1 codon, 1 codon=1 amino acid
some amino acids are coded for by 2 or more codons
step one: transcription (DNA to mRNA)
occurs within the nucleus
purpose is to make a copy of a gene when its protein is needed by the cell
genes begin with a sequence of DNA nucleotides called the promoter region
RNA polymerase binds to this region to begin transcription (process similar to replication)
RNA polymerase opens double heliz and moves along the template strand, creating a single sided copy of the gene made of RNA nucleotides (U replaces T)
the template is the non-coding, or non-sense strand, whereas the complimentary copy is the coding stranding or sense strand
the new molecule created is called mRNA (messenger rna) because it carries the copy of the gene from the nucleus to the cytoplasm
EDITING GENES:
before mRNA leaves the nucleus, introns (non-coding regions of the gene sequence) must be removed from the exons (coding regions)
introns all begin and end with similar sequences, allowing them to be recognized and cut out by an enzyme
step two: translation (mRNA to polypeptide chain)
genes (mRNA) begin with the sequence AUG, which is the ‘start’ codon. if AUG is at the start of a sequence, it doesn’t code for anything, it just means start. if it is found within a gene, it codes for the amino acid methionine
genes end with one of the sequences UAA, UAG, UGA, which are the ‘stop’ (terminator) codons, and they do not code for any amino acids no matter where they are
mRNA binds to a ribosome to begin protein synthesis
ribosomes contain a third type of RNA, rRNA, which is responsible for reading the order of amino acids and linking amino acids together
tRNA (transfer RNA) is a strand of RNA folded to have two distinct ends, one has an anticodon and the other spot is to bind an amino acid.
anticodon at one end which has a code that is complimentary to a corresponding codon in a mRNA sequence
other end of tRNA has an amino acid which is formed by the code—matching a codon in an mRNA with the amino acids to make a new protein using data booklet
a ribosome moves along a mRNA strand, exposing one codon at a time
as it moves, a tRNA that has a matching anticodon to codon moves in and drops off its amino acid
peptide bonds form to link amino acids into a polypeptide chain
tRNAS continue to drop off amino acids until a stop codon is reached
ribosome has three slots for tRNAS—tRNAS moves through those sites as they deliver amino acids during translation
what is recombinant dna and genetic transformation?
recombinant dna: fragment of dna composed of sequences originating from at least two different sources
dna that includes genes from 2 (or more) sources
genetic transformation: introduction and expression of foreign dna in a living organism
how can a ‘DNA fingerprint’ be made and what can it be used for?
gel electrophoresis: allows dna samples to be sorted and analyzed
dna fragments are loaded into a gel, and an electric current is appliece
dna is negatively charged, and so pulled through the gel towards the positive charge
shorter pieces move the fastest/farthest
the resulting pattern is called a dna fingerprint
can be used in a variety of ways: paternity, crime scene investigations, etc.
what is DNA sequencing? what technique helps with dna sequencing?
dna sequencing: determines the exact sequence of bases in a particular fragment of DNA (like a gene).
first dna sequencing techniques were simultaneously developed by frederick sanger and his colleagues, and by alan maxim and walter gilbert
polymerase chain reaction
makes many copies of a small quantities of dna
heat is used to denature dna into single stranded pieces, then cooled to allow carefully selected primers to attach
the sample is heated again, and then a heat resistant polymerase, taq polymerase, starts replicating
vector? plasmid? transgenic? multiple-cloning site?
vector: delivery system used to move foreign dna into a cell (vehicle by which foreign dna may be introduced to a cell—useful for gene therapy.
plasmid: small double stranded circular dna molecule found in some bacteria, and is an example of a vector. very useful vector as they can replicate independently and is widely used in gene cloning
introduced to host cells through process called transformation
transgenic: cell organism that is transformed by dna from another species
multiple cloning site: region in a vector (ex. plasmid) that is engineered to contain the recognition site of a number of restriction enzymes
scientists
late 1860s: friedrich Miescher
isolated non-protein substance from nucleus of cells; named it nuclein. found thay nuclein had an acid portion which he called nucleic acid, and an alkaline portion.
1928: frederick griffith
mice experiment
used two different strains of pneumococcus bacteria (virulent and nonvirulent), and observes that when heat-treated virulent pneumococcus was mixed with non-virulent pneumococcus and was injected into healthy mice, death resulted
discovered process of transformation
1943: joachim hammerling
experimented using green alga acetabularia and observed that regneration of new appendages was driven by the nucleus-containing ‘foot’ of the alga
hypothesized that hereditary information is stored in the nucleus
1944: oswald avery, maclyn mccarty, colin macleod
built upon frederick griffiths work and discovered that dna was the transforming prince of pneumococcus bacteria—essentially that dna was indeed the molecular amterial of hereditary
1949: erwin chargaff
discovered that in the dna of numerous organisms, the amount of adenine is equal to the amount of thymine, and the amount of guanine is equal to that of cytosine (developed chargaffs rule that A’s goes with T’s/U’s and that C’s go with G’s)
1952: alfred hershey and martha chase
used radioactively labelled viruses, infected bacterial cells; observed that the infected bacterial cells contained radioactivity originating from dna of the virus, suggesting that dna is hereditary material (meaning genetically passed on from parents)
1953: rosalind franklin
produced X-ray diffraction pattern of dna that suggested it was in the shape of a double helix
1953: james watson and francis crick
deduced the structure of dna using information from the work of chargaff, franklin, and maurice wilkins
What are restriction enzymes
restriction enzymes (endonucleases)
are proteins made by bacteria for defense against infection by foreign DNA (VIRUSES)
these enzymes catalyze the ‘cutting’ of the dna backbone at specific places
molecular scissors that cut double-stranded dna at specific base-pair sequences
each type of restriction enzyme recognizes a particular sequence of nucleotides that is known as its recognition site
most recognition sites are four to eight base pairs long and are usually characterized by a complimentary palindromic sequence
palindromic: read the same backwards and forwards
restriction enzymes are names after the bacteria they are taken from. ex. EcoRI—from E coli
R enzymes cut in the same way every time—most produce ‘sticky ends’
sticky ends: fragment ends of dna molecule w/ short single stranded overhand, resulting from cleavage by a restriction enzyme, however not all enzymes produce sticky ends
restriction enzymes that produce sticky ends are more useful tool bc. they can be joined easier through complimentary base pairing
when fragments are to be sticked together, the segments of dna are joined together by DNA ligase
if 2 fragments generated by the same restriction enzyme, they will be attracted to each other at their complementary sticky ends
hydrogen bonds will form between complimentary base pairs, and dna ligase joins strands together
whats a mutation? what are the 3 possible results of mutation?
mutation: the changing of the structure of a gene, resulting in a variant form that may be transmitted to subsequent generations
beneficial mutation: gives organism a selective advantage and can lead to evolutionary change/can become common trait over time
harmful mutation: reduces individuals fitness and tends to be selected against
Neutral mutation: neither good nor bad, and are not acted on by natural selection
whats a gene mutation and whats a point mutation?
gene mutation: changes the coding for amino acids (can change the number/type of amino acids)
point mutation: affect only one base pair in a DNA sequence
what are the 3 types of point mutations? what changes how the codons are read?
silent mutation: has no effect on cell activities. these mutations often occur in the non-coding region of DNA. if they happen to occu rina areas that code for proteins, the change in base-pair does not affect the amino acid sequence.
missense mutation: when a change in the base sequence of DNA alters a codon, leading to a different amino acid being placed in the polypeptide chain (ex. sickle cell anemia).
nonsense mutation: when a change in the DNA sequence causes a STOP codon to replace a codon that is meant to specify an amino acid.
nonsense mutations are often lethal to the cell
missense and nonsense mutations arise from from the substitution of one base pair for another
a frameshift mutation causes the reading frame of codons to change. this means that it changes how the codons are read.
what does translocation and inversion mean?
translocation (think: transgender, location aka, switch location): the transfer of a fragment of DNA (of groups of base pairs) from one site in the genome to another location.
inversion (think: inverse): the reversal of a segment of DNA within a chromosome. a section of chromosome that has reversed its orientation in the chromosome (has turned itself around)
both involve large segments of DNA and are seen at the chromosomal level.
dihybrid crosses—how to solve?
crosses involving 2 traits (still dominant and recessive genes)
write genotypes and the possible combinations in gametes (these combinations are the ways the chromosomes could split in meiosis during metaphase one)
punnet square possible combinations to see F1 genereation
beyond Mendel genetics
there are some patterns of inheritance that are not simple dominant and recessive
NOTE: when you get beyond Mendel genetics, you no longer use simple ex. B—Dominant, and b—recessive. you begin using ex. Colour: C^R and C^W. this is to show that we are moving beyond mendelian genetics. the C stands for ‘colour” and the R is Red, W is White.
incomplete dominance:
when 2 alleles are equally dominant, they interact to produce a new, intermediate phenotype, that is like a mix between the two. (ex. flower colour: white + red = pink)
if question is incomplete dominance, the heterozygous phenotype is a mix of the ‘recessive’ and ‘dominant’
Co-dominance:
both alleles are fully expressed, but in the different cells (the individual expresses BOTH phenotypes) (ex. blood type AB has IA and IB allele)
if question is co-dominance, the heterozygous phenotype shows both the ‘recessive’ and ‘dominant’ trait
sex-linked traits:
certain genes are carried on the X crms. others on the Y. bc. males only have one X< they show recessive phenotypes more often than females
heterozygous females are called carries
if question says its a sex-linked question, or specifically asks about the sex of offspring (daughters or sons) then its a sex-linked question. if it doesn’t ask about that, or says autosomal, that means its on non-sex chromosomes
Multiple alleles:
many genes have more than 2 alleles (each individual only carries 2 alleles in their genotype but the population as a whole has many different alleles)
ex. rabbit coat colour—there are 4 different genotypes possible, and the order of dominance is shown in a sequence that shows the relevant dominance of each gene
what are linked genes/recombinants?
genes on mom and dad chromatids can sometimes exchange during crossing over (in prophase I) which gives offspring different allele combinations on their chromosomes than their parents had.
offspring with new combinations of mom and dad genes are called recombinants
recombination frequency = # of recombinants (random ones you didn’t expect) / total # of offspring X 100%
the recombination frequency tells us the relative distance that genes are from each other on a chromosome—the farther apart 2 genes are, the more often they will cross over
1% = 1 map unit (mu)