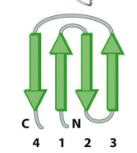
Secondary Structure of Proteins
1/43
Earn XP
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No study sessions yet.
44 Terms
Cheese crystals
in parmesan; tyrosine
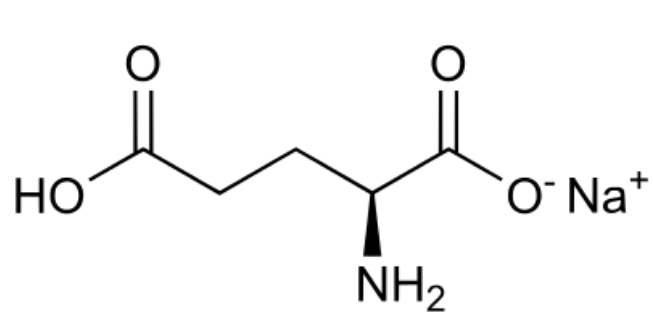
Monosodium glutamate (MSG)
umami taste from specific amino acid receptors; originally isolated from dashi; literally no medical risk
N-terminus to C-terminus
standard for writing primary sequences (don’t be a Knarf)
Levels of protein structure (4)
primary, secondary, tertiary, quaternary
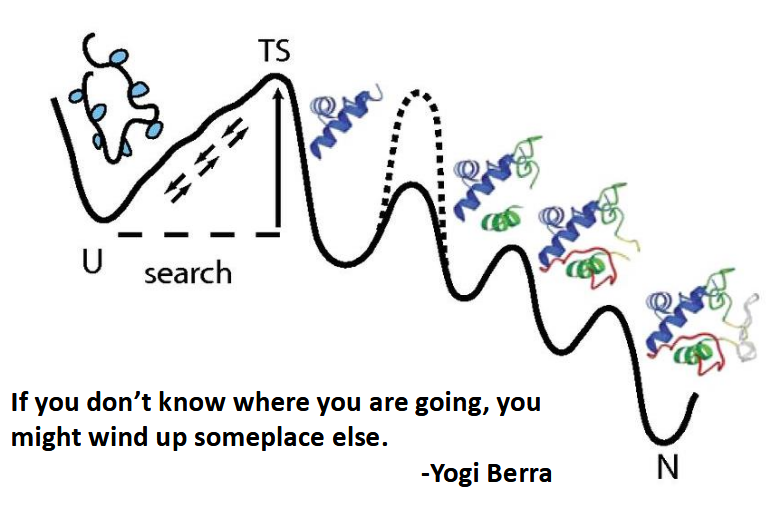
Levinthal's paradox: Do proteins fold by sampling all possible conformations?
protein folding landscape has stable conformations that aren’t its native; lots and lots of testing has to be done then, but proteins fold spontaneously and fast!
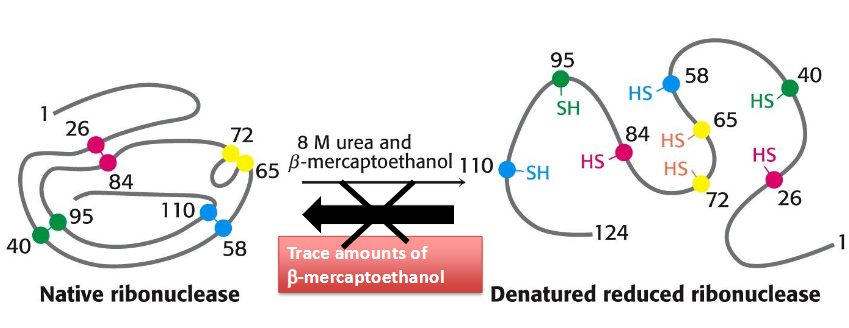
Anfinsen’s Experiment: RNase A Folding
1972 Nobel Prize in Chemistry
postulated that the native structure of a protein is the thermodynamically stable structure; it depends only on the amino acid sequence and on the conditions of solution, and not on the kinetic folding route
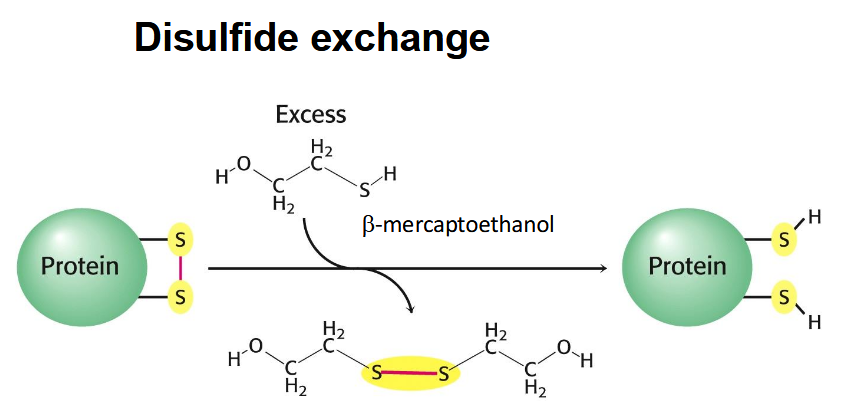
hydrogen bonding disrupted with urea and disulfide bridges disrupted by beta mercaptoethanol → refolds with urea removed (and trace mercaptoethanol added - undos wrong disulfide bonds)
disulfide exchange
exchange salt-bridge for two thiols
Urea hydrogen bond disruption
disrupts the hydrophobic effect, preventing water from properly interacting with the protein during folding
protein folding
alpha helices form
hydrophobic collapse
beta sheets
plus or minus a few kinetic traps
boom native state
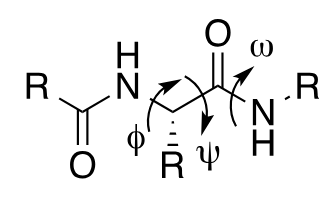
Bond rotations in peptide backbone
specify protein secondary structures
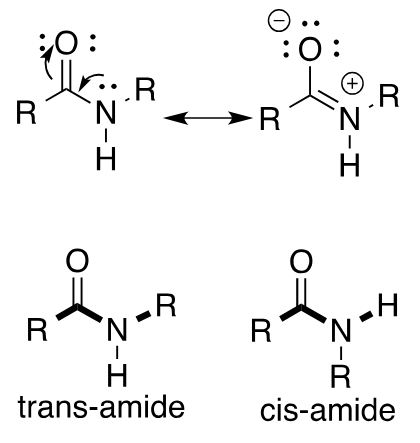
amide bond
always draw trans (more stable by ~ 8 kJ/mol) UNLESS proven to be cis (Proline residues have ~10% cis conformation)
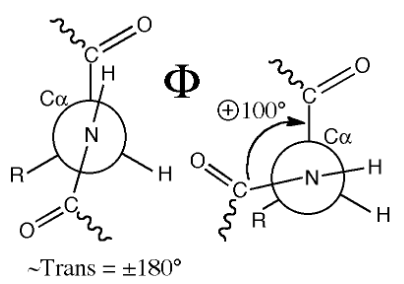
Φ bond rotations
n to alpha carbon; four atoms to track
ψ bond rotation
alpha carbon to carbonyl carbon; four atoms to track
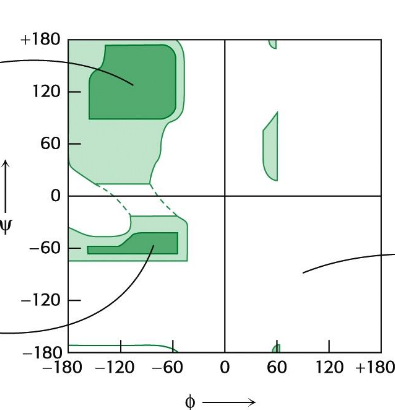
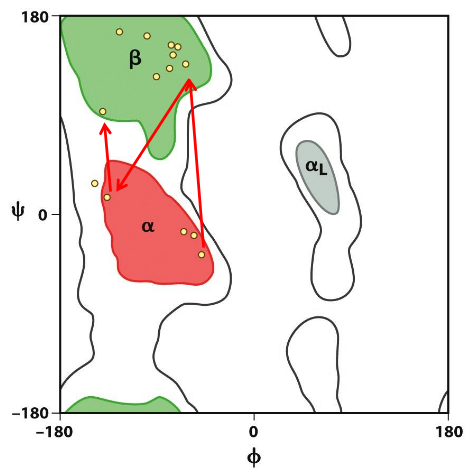
Ramachandaran Plot
shows accessible (low energy) ψ and Φ conformations; Dark areas are “allowed”, white areas denote high energy conformations; Calculate using PDB structures and a calculator (code) or from one of several programs (e.g., pymol, VMD)
Ramachandran plot and secondary structure
match with the dihedrals of specific secondary structures
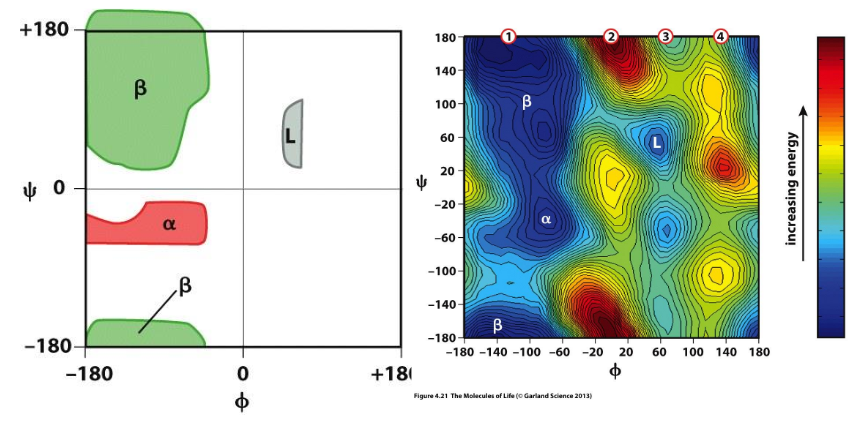
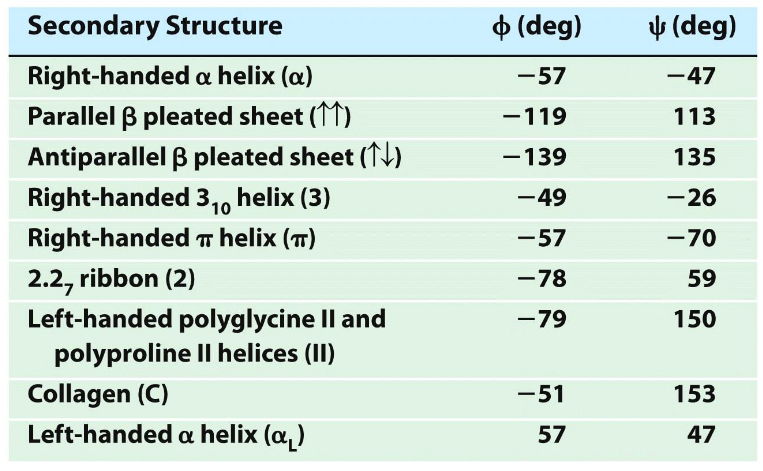
Average dihedral angles for secondary structures
>30% residues have alpha helical values of Φ & ψ; then beta strands; unfolded proteins have lots of PII conformation (collagen)
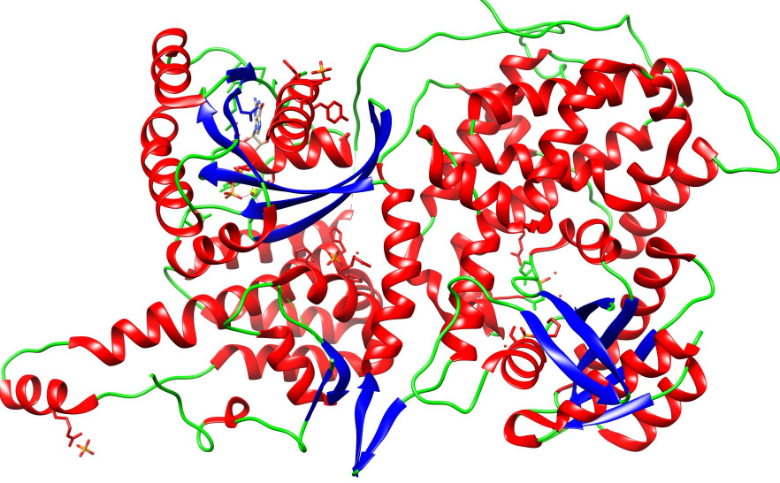
Types of secondary structure
helix
sheets
loops/random coils
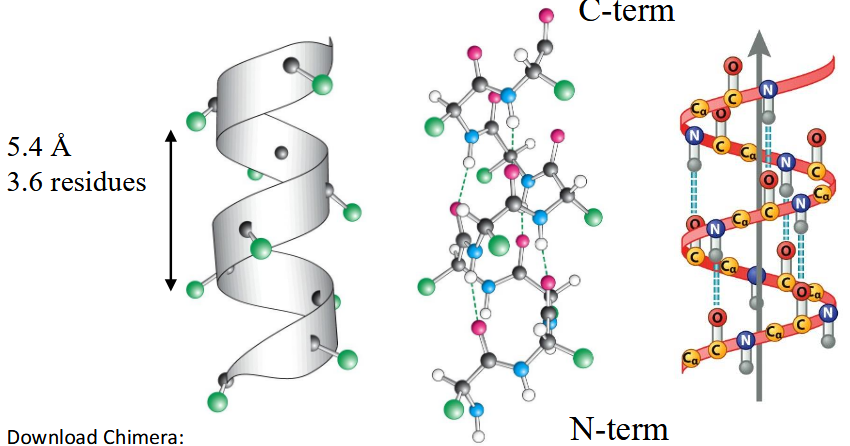
right-handed (alpha) helix
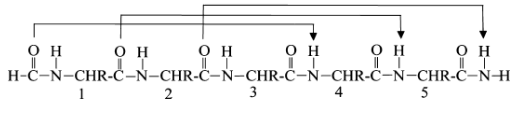
All N-H and C=O pairs make H-bonds except the first 3 N-H (at N-terminus) and last three C=O (at C-terminus); Residue "i" C=O makes H-bond with residue "i+4" N-H; common; i, i+3 and i+4 residues reside on the same face of the alpha-helix so they can interact with side chains at i+3/i+4 positions using salt-bridges, hydrogen bonding, pi-stacking or van der Waals interactions
Secondary structure
regular local folds of the backbone; motifs
guidelines for generating secondary structures in proteins
Avoid steric clashes
Maximise the number of hydrogen bonds
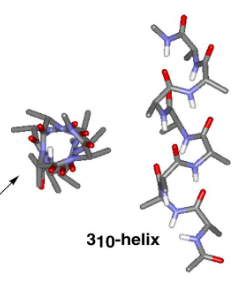
310 helix
10 atom H-bonds, from i to i+3
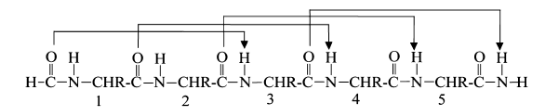
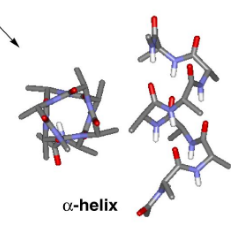
α helix
13 atoms H-bonds; i to i+4; most stable and abundant helix in proteins
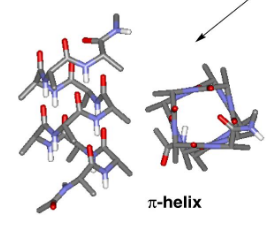
π helix
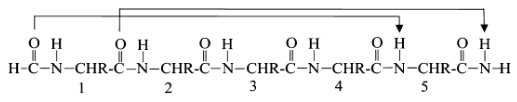
16 atoms H-bonds; i to i+5
helical wheel
represents the alpha helix; Heptad Repeat: left-handed twist makes 3.5 residues/turn
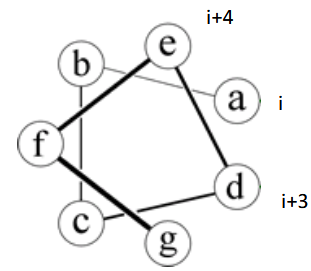
helix preferences
1. Helix propensity is favored by Ala, destabilized by Gly or Pro.
2. Leu stabilizes helix more than I or V. Crowding Cβ is less favorable.
3. Side chains interact at spacing of i,i+3 and i,i+4: acid base bridges (E,D to K,R) stabilize helix, hydrophobic groups too.
P is a tertiary amine - cannot do hydrogen bonding - breaks helices
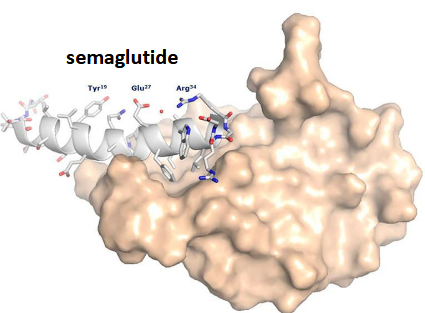
Ozempic (semaglutide)
Modified peptide analog of Glucagon-like peptide-1 (GLP-1) that has stabilised helices (basically is!); Lowers blood glucose; Significant weight loss; development inspired by Gila monsters
β-Strand
often twisted; Distance between adjacent amino acids ~ 3.4 Å

strand/sheet preferences
Enriched in Val, Ile, Thr (branched amino acids); Tyr, Trp, Phe (aromatic); Cys - provides space
Ramachandran plot
Consecutive dihedral angles of (Φ, ψ) of -120° and 120°
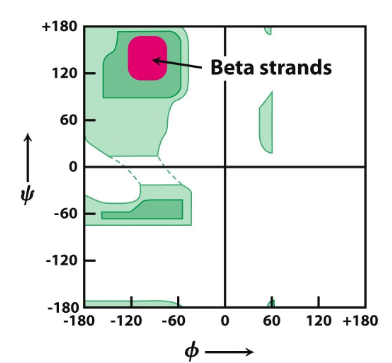
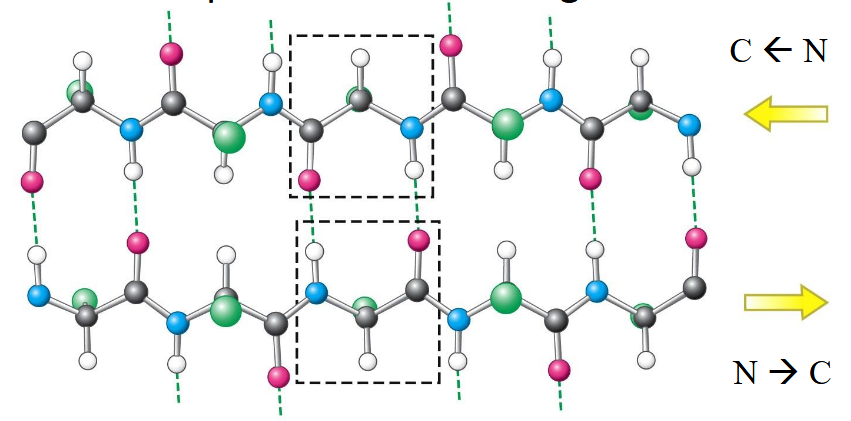
parallel β-Sheet
hydrogen bonds; pleated so atoms can connect
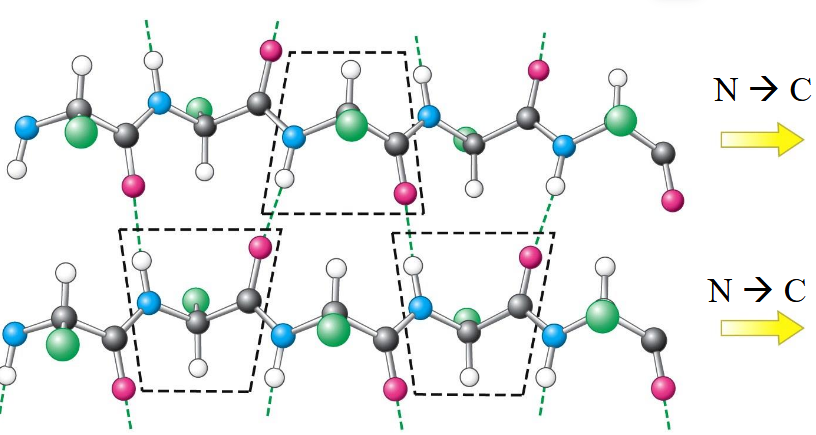
anti-parallel β-Sheet
hydrogen bonds; pleated so atoms can connect; connected by short turns or loops
Silk
fibrous protein consisting of three subunits, one of which consists almost entirely of β-sheets, which collectively give silk its strength; cocoon of the Japanese silkworm
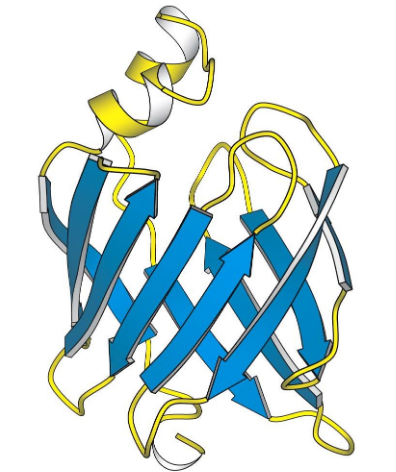
β-Sheet
Connecting β−strands through loops and turns into a channel
Gly and Pro are common in turns. Why?
totally change connectivity and stereochemistry
Random loops/coils Ramachandran plot
various phi/psi angles
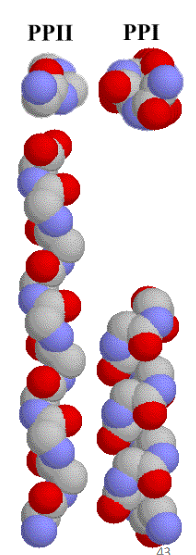
Polyproline Helices
PPII from repeating proline trans amide bonds - 3 residues per turn
PPI from repeating proline cis amide bonds - rare, generally observed only in organic solvents.
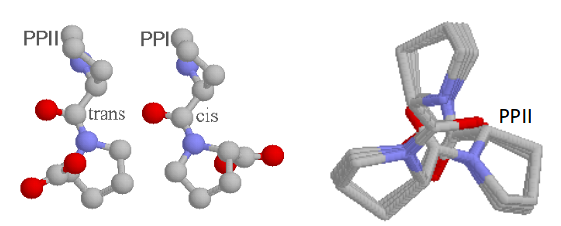
ω deihedral angle
amide bond; can be 0 or 180
“Random” Coil
No well-defined secondary or tertiary structure; Conformations of polypeptide rapidly interconvert between multiple different states; small energy differences among backbone conformations compared with kT (thermal energy)
Coiled-Coils
Helix-helix contacts that wrap around each other in a left-handed spiral
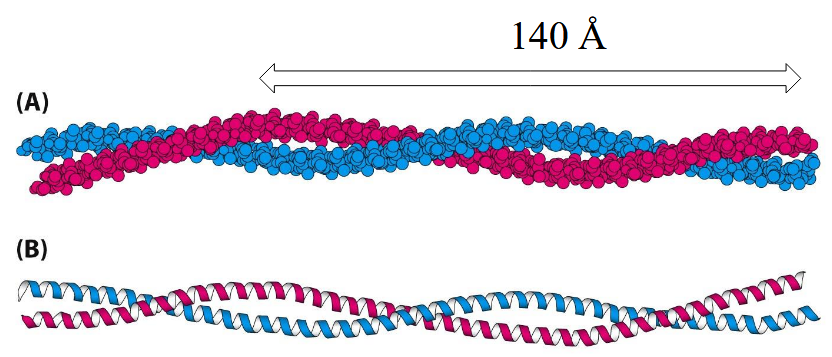
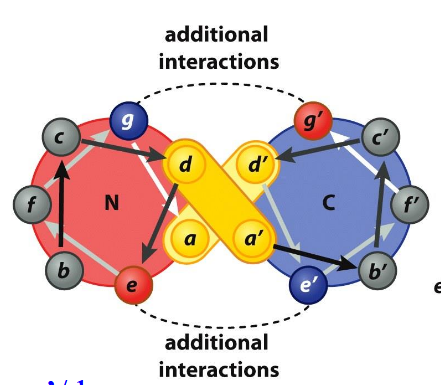
Parallel Coiled-Coil
Heptad Repeat: left-handed twist makes 3.5 residues/turn; a/d always in contact, likely hydrophobic; g/e additional interactions, likely salt-bridge
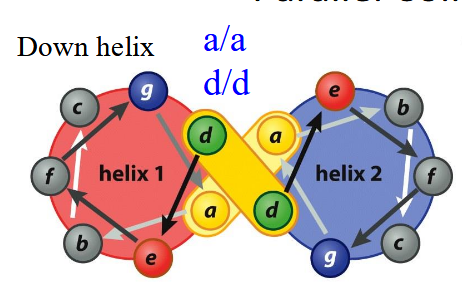
Anti-Parallel Coiled-Coil
a/d’ and d’/a; g/g’, e/e’
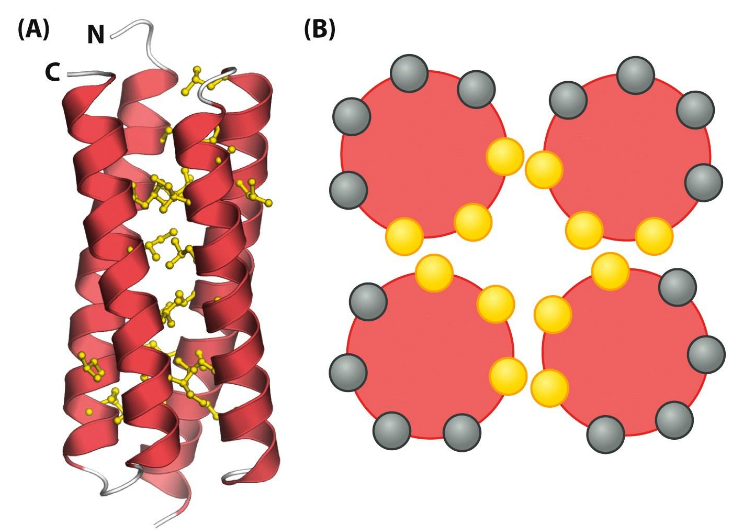
Helix Bundle
hydrophobic core
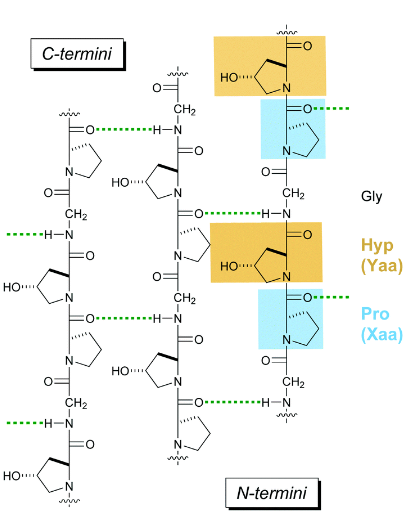
Collagen
most abundant protein in mammals (25 to 35%); main component of connective tissue; triple helix; repeat of Pro-Hyp-Gly
Super-secondary Structures
2+ secondary structures combined
Helix-turn-helix
Helix-loop-helix
β-α-β
Greek key
Beta Barrel